Abstract
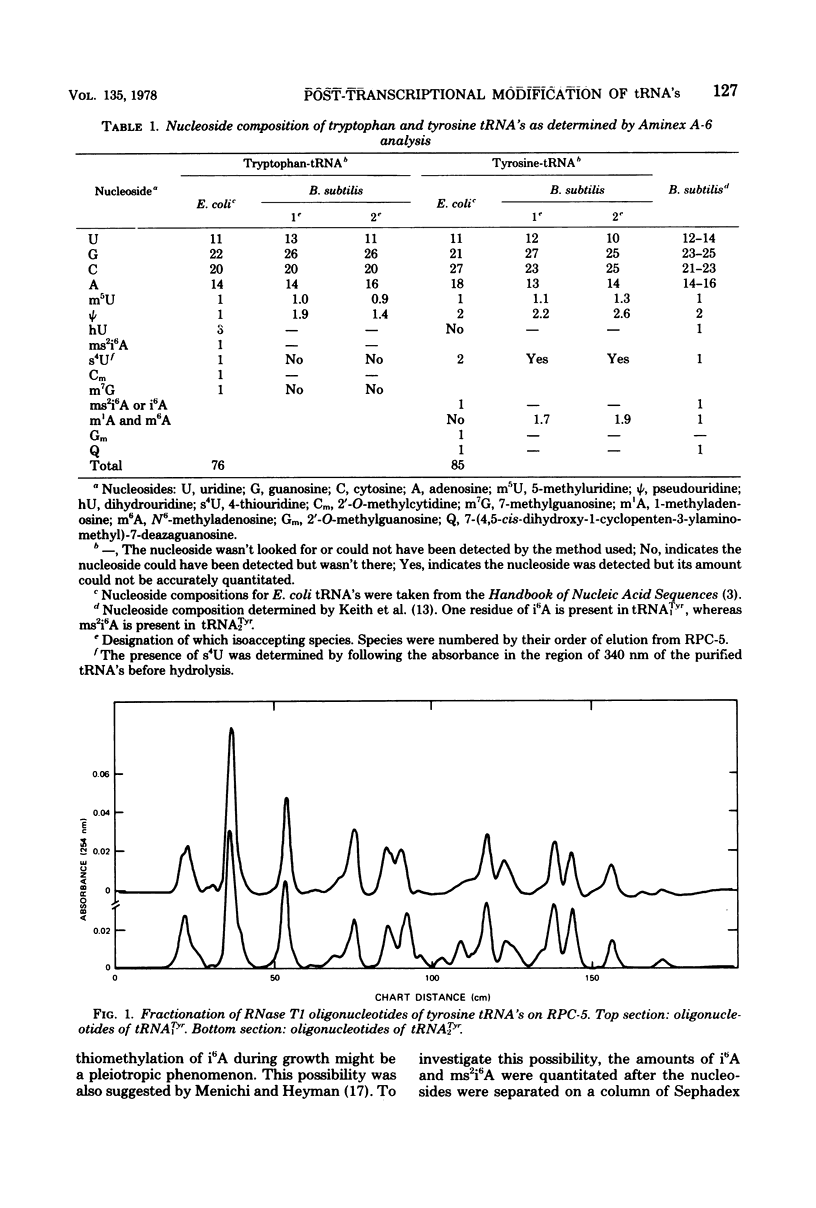
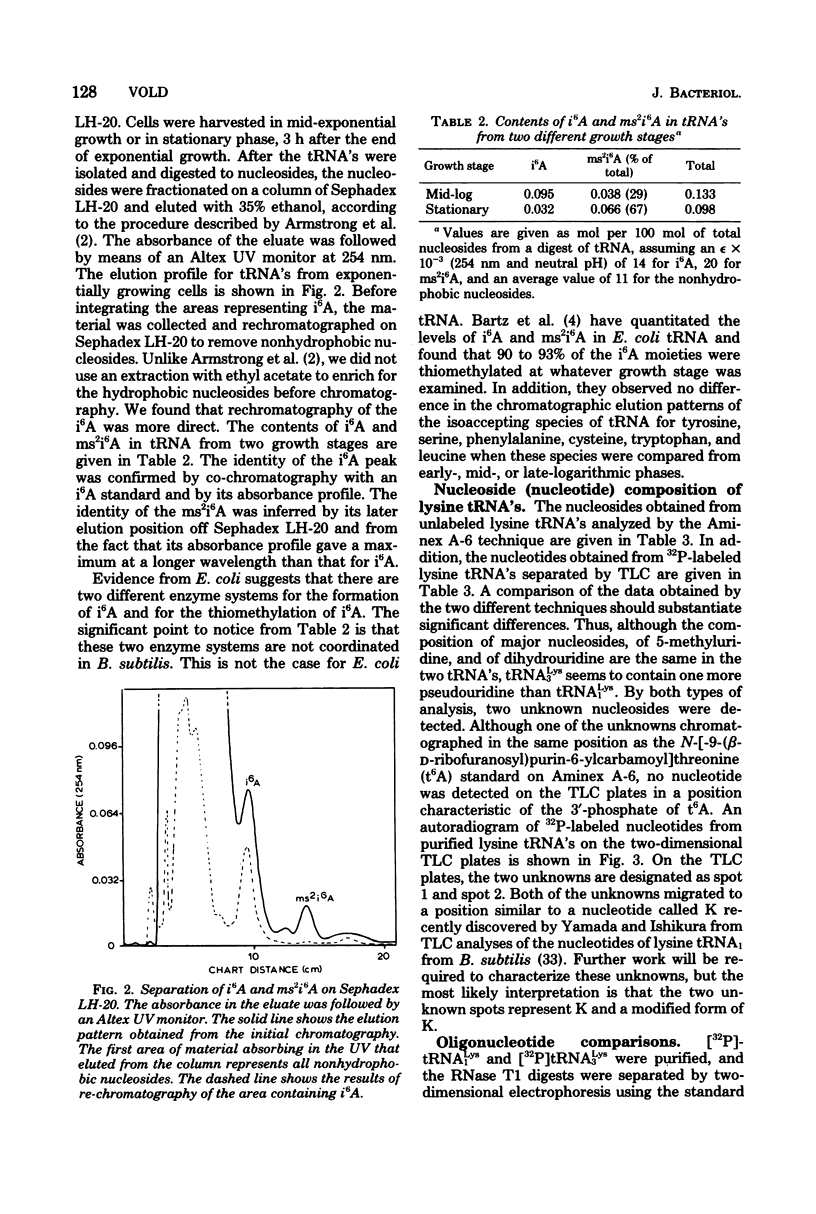
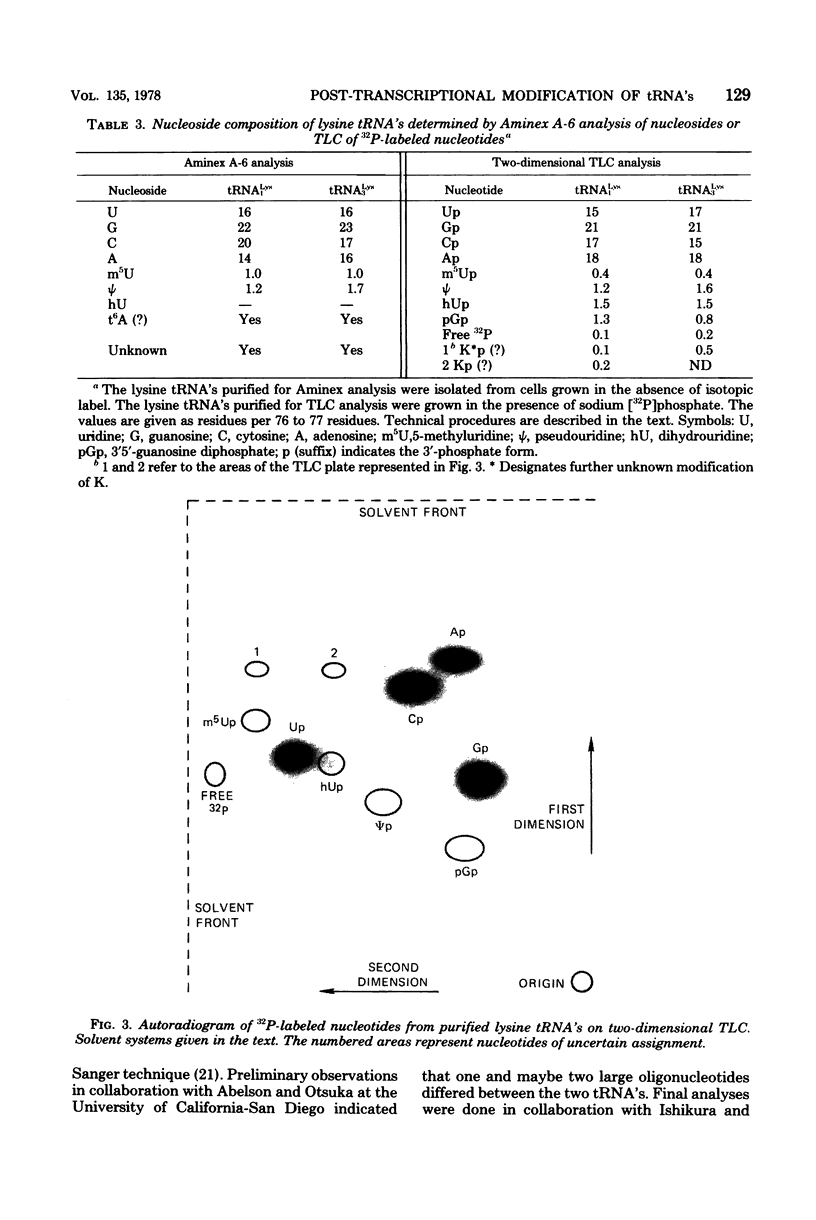
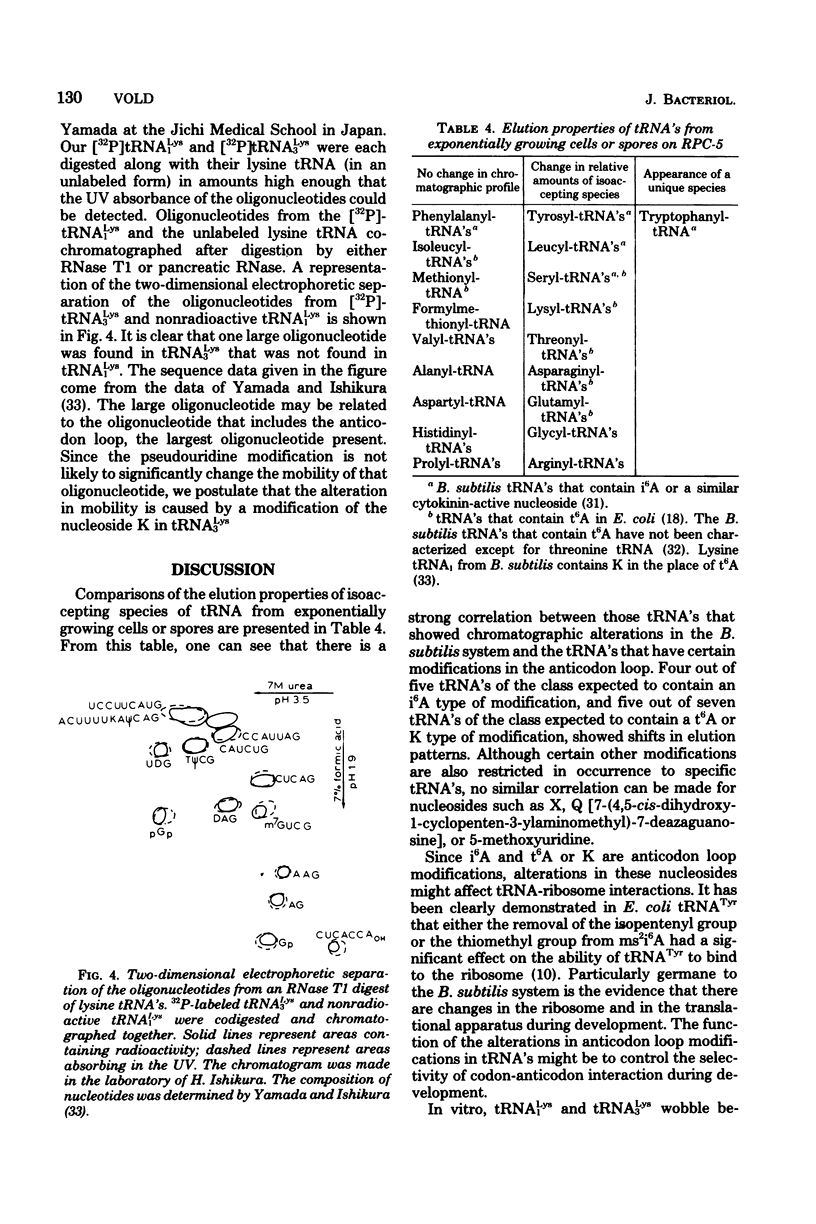
Structural similarities of tRNA's were compared using three sets of isoaccepting species that had previously been shown to undergo significant changes in chromatographic elution properties as a function of developmental stage in Bacillus subtilis. Comparisons of the structures of the tRNA's were based on the composition of their modified nucleosides, comparisons of oligonucleotide elution profiles from RPC-5 columns, and two-dimensional electrophoretic fingerprint analysis of oligonucleotides. The tRNA's studied were tRNALys1 and tRNALys3; tRNATyr1 and tRNATyr2; and tRNATrp1 and tRNATrp2. The results suggest that the difference among these pairs of isoaccepting species is a difference in the degree of post-transcriptional modifications of the anticodon loop region. The nucleosides involved were N6-(Δ2-isopentenyl)adenosine (i6A), 2-methylthio-N6-(Δ2-isopentenyl)adenosine (ms2i6A), and an unknown nucleoside K, which occurred in a position analogous to N-[9-(β-d-ribofuranosyl)purin-6-ylcarbamoyl]threonine. The amounts of i6A and ms2i6A, determined using total tRNA from exponential-or stationary-phase cells, suggest that the thiomethylation of i6A is a pleiotropic phenomenon affecting several tRNA species. As opposed to the situation in Escherichia coli tRNA, where ms2i6A constitutes about 90% of the total hydrophobic nucleosides at all growth stages, B. subtilis tRNA's have i6A as the predominant hydrophobic nucleoside in exponential growth and ms2i6A as the predominant nucleoside in stationary phase. Thus, the enzyme system which forms i6A and the enzyme system which thiomethylates i6A are not coordinated during growth in B. subtilis as they are in E. coli. It is suggested that these changes in anticodon loop modifications in B. subtilis may be related to changes in the translational apparatus which occur during sporulation.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arceneaux J. L., Sueoka N. Two species of Bacillus subtilis tyrosine transfer ribonucleic acid. Biological properties and alteration in their relative amounts during growth. J Biol Chem. 1969 Nov 10;244(21):5959–5966. [PubMed] [Google Scholar]
- Armstrong D. J., Burrows W. J., Evans P. K., Skoog F. Isolation of cytokinins from tRNA. Biochem Biophys Res Commun. 1969 Oct 22;37(3):451–456. doi: 10.1016/0006-291x(69)90936-x. [DOI] [PubMed] [Google Scholar]
- Bartz J., Söll D., Burrows W. J., Skoog F. Identification of the cytokinin-active ribonucleosides in pure Escherichia coli tRNA species. Proc Natl Acad Sci U S A. 1970 Nov;67(3):1448–1453. doi: 10.1073/pnas.67.3.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang R. Y., Doi R. H. Characterization of lysine transfer ribonucleic acid from vegetative cells and spores of Bacillus subtilis. J Biol Chem. 1972 Jun 10;247(11):3476–3484. [PubMed] [Google Scholar]
- Chuang R., Yamakawa T., Doi R. H. Identification of two lysine tRNA cistrons in Bacillus subtilis by hybridization of lysyl-tRNA with DNA. Biochem Biophys Res Commun. 1971 May 21;43(4):710–716. doi: 10.1016/0006-291x(71)90673-5. [DOI] [PubMed] [Google Scholar]
- Cortese R., Landsberg R., Haar R. A., Umbarger H. E., Ames B. N. Pleiotropy of hisT mutants blocked in pseudouridine synthesis in tRNA: leucine and isoleucine-valine operons. Proc Natl Acad Sci U S A. 1974 May;71(5):1857–1861. doi: 10.1073/pnas.71.5.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan B. Z. Separation of oligonucleotides by reversed-phase chromatography. Biochim Biophys Acta. 1973 Mar 19;299(2):245–252. doi: 10.1016/0005-2787(73)90347-x. [DOI] [PubMed] [Google Scholar]
- Gefter M. L., Russell R. L. Role modifications in tyrosine transfer RNA: a modified base affecting ribosome binding. J Mol Biol. 1969 Jan 14;39(1):145–157. doi: 10.1016/0022-2836(69)90339-8. [DOI] [PubMed] [Google Scholar]
- Gefter M. L. The in vitro synthesis of 2'-omethylguanosine and 2-methylthio 6N (gamma,gamma, dimethylallyl) adenosine in transfer RNA of Escherichia coli. Biochem Biophys Res Commun. 1969 Aug 7;36(3):435–441. doi: 10.1016/0006-291x(69)90583-x. [DOI] [PubMed] [Google Scholar]
- Gillam I., Millward S., Blew D., von Tigerstrom M., Wimmer E., Tener G. M. The separation of soluble ribonucleic acids on benzoylated diethylaminoethylcellulose. Biochemistry. 1967 Oct;6(10):3043–3056. doi: 10.1021/bi00862a011. [DOI] [PubMed] [Google Scholar]
- Gross H. J., Raab C. In vivo synthesis of tRNA Tyr 1 and tRNA Tyr 2 : differences in "early" and "late log" E. coli MRE 600. Biochem Biophys Res Commun. 1972 Mar 24;46(6):2006–2011. doi: 10.1016/0006-291x(72)90751-6. [DOI] [PubMed] [Google Scholar]
- Hasegawa T., Ishikura H. Nucleotide sequence of threonine tRNA from Bacillus subtilis. Nucleic Acids Res. 1978 Feb;5(2):537–548. doi: 10.1093/nar/5.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith G., Rogg H., Dirheimer G., Menichi B., Heyham T. Post-transcriptional modification of tyrosine tRNA as a function of growth in Bacillus subtilis. FEBS Lett. 1976 Jan 15;61(2):120–123. doi: 10.1016/0014-5793(76)81017-4. [DOI] [PubMed] [Google Scholar]
- Kelmers A. D., Heatherly D. E. Columns for rapid chromatographic separation of small amounts of tracer-labeled transfer ribonucleic acids. Anal Biochem. 1971 Dec;44(2):486–495. doi: 10.1016/0003-2697(71)90236-3. [DOI] [PubMed] [Google Scholar]
- McCutchan T. F., Gilham P. T., Söll D. An improved method for the purification of tRNA by chromatography on dihydroxyboryl substituted cellulose. Nucleic Acids Res. 1975 Jun;2(6):853–864. doi: 10.1093/nar/2.6.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillian R. A., Arceneaux J. L. Alteration of tyrosine isoaccepting transfer ribonucleic acid species in wild-type and asporogenous strains of Bacillus subtilis. J Bacteriol. 1975 May;122(2):526–531. doi: 10.1128/jb.122.2.526-531.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menichi B., Heyman T. Study of tyrosine transfer ribonucleic acid modification in relation to sporulation in Bacillus subtilis. J Bacteriol. 1976 Jul;127(1):268–280. doi: 10.1128/jb.127.1.268-280.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura S., Harada F., Narushima U., Seno T. Purification of methionine-, valine-, phenylalanine- and tyrosine-specific tRNA from Escherichia coli. Biochim Biophys Acta. 1967 Jun 20;142(1):133–148. doi: 10.1016/0005-2787(67)90522-9. [DOI] [PubMed] [Google Scholar]
- Nishimura S. Minor components in transfer RNA: their characterization, location, and function. Prog Nucleic Acid Res Mol Biol. 1972;12:49–85. [PubMed] [Google Scholar]
- Palatnik C. M., Katz E. R. Isolation and characterization of transfer RNAs from Dictyostelium discoideum during growth and development. J Biol Chem. 1977 Jan 25;252(2):694–703. [PubMed] [Google Scholar]
- Sanger F., Brownlee G. G., Barrell B. G. A two-dimensional fractionation procedure for radioactive nucleotides. J Mol Biol. 1965 Sep;13(2):373–398. doi: 10.1016/s0022-2836(65)80104-8. [DOI] [PubMed] [Google Scholar]
- Singhal R. P., Griffin G. D., Novelli G. D. Separation of transfer ribonucleic acids on polystyrene anion exchangers. Biochemistry. 1976 Nov 16;15(23):5083–5087. doi: 10.1021/bi00668a021. [DOI] [PubMed] [Google Scholar]
- Singhal R. P., Vold B. Changes in transfer ribonucleic acids of Bacillus subtilis during different growth phases. Nucleic Acids Res. 1976 May;3(5):1249–1262. doi: 10.1093/nar/3.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skjold A. C., Juarez H., Hedgcoth C. Relationships among deoxyribonucleic acid, ribonucleic acid, and specific transfer ribonucleic acids in Escherichia coli 15T - at various growth rates. J Bacteriol. 1973 Jul;115(1):177–187. doi: 10.1128/jb.115.1.177-187.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. W. Reticulocyte transfer RNA and hemoglobin synthesis. Science. 1975 Nov 7;190(4214):529–535. doi: 10.1126/science.1103288. [DOI] [PubMed] [Google Scholar]
- Tockman J., Vold B. S. In vivo aminoacylation of transfer ribonucleic acid in Bacillus subtilis and evidence for differential utilization of lysine-isoaccepting transfer ribonucleic acid species. J Bacteriol. 1977 Jun;130(3):1091–1097. doi: 10.1128/jb.130.3.1091-1097.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vold B. S. Analysis of isoaccepting transfer ribonucleic acid species of Bacillus subtilis: changes in chromatography of transfer ribonucleic acids associated with stage of development. J Bacteriol. 1973 Apr;114(1):178–182. doi: 10.1128/jb.114.1.178-182.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vold B. S. Analysis of isoaccepting transfer ribonucleic acid species of Bacillus subtilis: chromatographic differences between transfer ribonucleic acids from spores and cells in exponential growth. J Bacteriol. 1973 Feb;113(2):825–833. doi: 10.1128/jb.113.2.825-833.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vold B. S., Clinton G. M., Spizizen J. An effect of temperature on the Bacillus subtillis transfer RNA's which respond to codons beginning with U and A correlation with cytokinin activity. Biochim Biophys Acta. 1970;209(2):396–404. doi: 10.1016/0005-2787(70)90737-9. [DOI] [PubMed] [Google Scholar]
- Vold B. Modified nucleosides of Bacillus subtilis transfer ribonucleic acids. J Bacteriol. 1976 Jul;127(1):258–267. doi: 10.1128/jb.127.1.258-267.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Ishikura H. Nucleotide sequence of a lysine tRNA from Bacillus subtilis. Nucleic Acids Res. 1977 Dec;4(12):4291–4303. doi: 10.1093/nar/4.12.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]




