Abstract
By using molecular dynamics simulations, we have examined the binding of a hexaNAG substrate and two potential hydrolysis intermediates (an oxazoline ion and an oxocarbenium ion) to a family 19 barley chitinase. We find the hexaNAG substrate binds with all sugars in a chair conformation, unlike the family 18 chitinase which causes substrate distortion. Glu 67 is in a position to protonate the anomeric oxygen linking sugar residues D and E whereas Asn 199 serves to hydrogen bond with the C2′ N-acetyl group of sugar D, thus preventing the formation of an oxazoline ion intermediate. In addition, Glu 89 is part of a flexible loop region allowing a conformational change to occur within the active site to bring the oxocarbenium ion intermediate and Glu 89 closer by 4–5 Å. A hydrolysis product with inversion of the anomeric configuration occurs because of nucleophilic attack by a water molecule that is coordinated by Glu 89 and Ser 120. Issues important for the design of inhibitors specific to family 19 chitinases over family 18 chitinases also are discussed.
Plants respond to pathogenic attack by producing defense proteins such as chitinases. Chitinases catalyze the hydrolysis of chitin, a β(1,4)-linked N-acetyl-glucosamine (GlcNAc) polysaccharide. Chitin, a fibrous and insoluble polymer, is a major structural component of many organisms including fungi, insects, and crustaceans. Fungal growth is limited by the degradation of fungal cell walls caused by the hydrolytic action of plant chitinases (1, 2). In addition, transgenic tobacco plants expressing a bean endochitinase gene have been reported to resist fungal infection better than nontransformed plants (3) demonstrating the importance of chitinases in the defense mechanism of higher plants.
Chitinases are included in the broad classification of glycosyl hydrolases and have been isolated from many different organisms including plants, insects, and bacteria. Based on amino acid sequence, chitinases are subdivided into two families (families 18 and 19) that differ in structure and mechanism (4–6). Plant chitinases also are divided into classes I-V in which classes III and V belong to family 18 and classes I, II, and IV comprise family 19. Recently, the first prokaryotic family 19 chitinase was isolated from Streptomyces griseus HUT 6037 (7). This is an important addition to the family 19 chitinases, previously found only in higher plants.
Before discussing the hydrolysis mechanism of family 19 chitinases, the focus of this paper, it is useful to review the general understanding of the mechanism for glycosyl hydrolases. Acid-catalyzed glycosidic hydrolysis may proceed to yield a hydrolyzed product with either retention or inversion of the anomeric configuration (at C1′) relative to the starting conformation. All of the family 18 chitinases reported to date (6, 8, 9) yield a β-anomer hydrolysis product (retaining mechanism) whereas family 19 chitinase result in the α-anomer (inverting mechanism) (10).
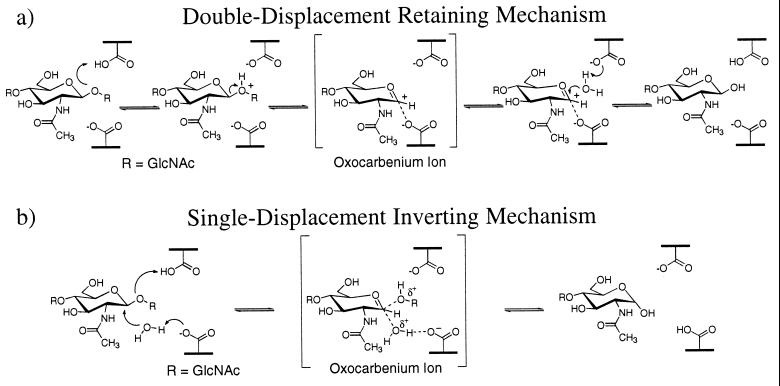
Based on crystallographic structural data (9) and theoretical models (K.B., W. Schrader, B. Imperiali, and W.A.G., unpublished results and ref. 12), the retaining mechanism for family 18 chitinases has been proposed to involve anchimeric assistance by the C2′ N-acetyl group (ref. 9 and K.B., W. Schrader, B. Imperiali, and W.A.G., unpublished results) and substrate distortion of sugar residue D to a boat conformation (the “D-boat” mechanism) (12). Fig. 1a shows how this mechanism requires only one active site carboxylic acid, which acts as both a general acid and general base in addition to stabilizing the oxazoline ion intermediate.
Figure 1.
(a) The double-displacement hydrolysis mechanism proposed for family 18 chitinases. Protonation of a GlcNAc residue in a boat conformation leads to an oxazoline intermediate, which may be hydrolyzed to form a product with retention of the anomeric configuration. (b) The single displacement hydrolysis mechanism proposed for family 19 chitinases. Two acidic residues are required in the active site and the hydrolysis product has inversion of the anomeric configuration.
The crystal structure of a family 19 plant endochitinase isolated from barley (Hordeum vulare L.) seeds (13) reveals two acidic residues (Glu 67 and Glu 89) in the active site separated by 9.3 Å. A single displacement mechanism for family 19 chitinases is often cited to account for inversion of the anomeric product and the need for two largely separated acidic residues within the active site (see Fig. 1b). The single displacement mechanism requires one acidic residue to act as a general acid (Glu 67) and the other as a general base (Glu 89), activating water for a concerted nucleophilic attack at C1′.
The single displacement mechanism proposed for family 19 chitinases necessarily involves an intermediate with considerable oxocarbenium ion character. A similar intermediate arises for hen egg white lysozyme (HEWL) with the important exception that the second acidic residue is within a few Å of the oxocarbenium ion and stabilizes the charge by forming either a covalent bond at C1′ or an ion pair (14, 15). Theoretical studies of HEWL indicate that this charge stabilization is a critical component of the enzymatic rate enhancement (16–18). The proximity of the acidic residue also has the affect of forcing retention of the anomeric configuration. In solution, without the presence of enzymatic stabilization, ab initio quantum mechanical calculations predict the oxocarbenium ion to be 20 kcal/mol higher in energy than an oxazoline ion (K.B., W. Schrader, B. Imperiali, and W.A.G., unpublished results). Indeed, this provides much of the driving force for the oxazoline ion intermediate proposed for family 18 chitinases. Unlike the HEWL system, the second acidic residue of family 19 chitinases is further considerably from the forming oxocarbenium ion intermediate. Thus, the problem of oxocarbenium ion charge stabilization is not well addressed in the current consensus view of the single displacement mechanism.
To investigate the detailed mechanism of family 19 chitinases, we have carried out molecular dynamics (MD) simulations on the barley seed endochitinase complexed with a hexaNAG substrate and two possible intermediates (triNAG-oxocarbenium ion and triNAG-oxazoline). We conclude: (i) The hexaNAG substrate binds with all sugars in a chair conformation such as to favor protonation by Glu 67 of the anomeric oxygen linking sugar residues D and E; (ii) Asn 199 serves to hydrogen bond with the C2′ N-acetyl group of sugar D, preventing the formation of an oxazoline ion intermediate; (iii) Glu 89 is situated in a flexible loop region allowing a conformational change to occur within the active site to bring the oxocarbenium ion intermediate 4–5 Å closer to Glu 89; (iv) Glu 89 and Ser 120 coordinate with a water molecule that may be activated for nucleophilic attack; this mechanism would yield a product with an inverted anomeric configuration (as observed), and (v) Inhibitors designed to be transition state analogs of the more planar oxocarbenium ion are predicted to be selective against family 19 chitinases over family 18 chitinases.
Simulation methods and results are reported in Methods and Results, respectively. The mechanistic details and suggested biochemical experimental tests of these simulation results are reported in Discussion.
METHODS
Details regarding the simulation methods, charges, solvation calculations, and simulation software were described (12). The starting structure for the hexaNAG/chitinase simulation was based on the crystal structure of NAG-NAM-NAG/HEWL (19). In predicting the structure for barley chitinase, we matched residues 56–68, 112–120, and 148–160 of barley chitinase to residues 24–36, 51–59, and 89–101 of HEWL. This match provided a model for sugar residues B-D (after conversion to all GlcNAc sugar). Sugar residues A, E, and F were built individually, starting from a range of different conformations and optimized through simulated annealing. Each conformation was subjected to 10 annealing cycles during which the temperature was raised from 0 to 350 K and lowered back to 0 K in increments of 50 K every 100 fs for a total of 2.1 ps. This equalibration resulted in only one family of hexaNAG structures. The lowest energy conformation in this family was then energy-minimized before dynamics simulations.‡
Starting structures for the bound oxocarbenium ion and oxazoline ion intermediates were based on the optimized hexaNAG model. GlcNAc residues E and F were removed, and the correct changes in atom hybridization were applied. These conformations were then subjected to annealing dynamics and energy optimization using a protocol analogous to that described above for the hexaNAG substrate.
Only residues comprising the binding site of barley chitinase were allowed to move during the optimizations and MD simulations. This chitinase binding site included residues: 66–70, 86–96, 110–130, 154–165, 197–203, and 211–214 (based on a 6-Å distance cutoff from the bound hexaNAG substrate). The simulations included crystallographic water molecules plus counterions for solvent exposed residues (leading to a net neutral charge). Water molecules in van der Waals contact with the docked ligand were displaced sufficiently to avoid high energy starting conformations.
RESULTS
Simulations of HexaNAG Substrate Binding.
MD simulations were used to study the binding of a hexaNAG substrate to the family 19 barley chitinase. The conformation of the hexaNAG substrate bound to barley chitinase was based on the reported crystal structure of HEWL complexed with a NAG-NAM-NAG trisaccharide, as described in Methods. Sugar residues A, E, and F (labeled A-F from the nonreducing end, following the convention for lysozyme) were built and optimized to yield the final hexaNAG substrate. We examined a range of conformations for the added sugar residues A, E, and F, but only one low energy global conformation was found to be stable during simulated annealing simulations from 0 to 350 K.
A 100-ps MD simulation was carried out in which the enzyme binding site residues, crystallographic waters, counterions, and hexaNAG substrate were all free to move. During this simulation, the hexaNAG substrate was stable as was the enzyme binding site. The average coordinate rms for all movable residues was 1.31 Å (see Table 1). All of the six GlcNAc residues remained in a chair conformation. Table 2 shows the average coordinate RMS fluctuation of each sugar residue. This result suggests that the D sugar residue was bound most tightly whereas residue A was least strongly bound.
Table 1.
RMS Coordinate difference (Å) for the binding site residues of barley chitinase*
| Substrate | Average RMS† |
|---|---|
| hexaNAG | 1.31 |
| tri-NAG-oxocarbenium | 1.14 |
| tri-NAG-oxazoline | 1.51 |
| None | 1.23 |
Comparing molecular dynamics with the crystal structure obtained without substrate.
Coordinate RMS difference was calculated as the difference between the crystal structure coordinates (13) and the average position for all of the nonhydrogen atoms during the MD interval from 30 to 100 ps.
Table 2.
RMS coordinate fluctuations (Å) from molecular dynamics of sugars bound to barley chitinase*
| Conformation | Binding subsite | |||||
|---|---|---|---|---|---|---|
| A | B | C | D | E | F | |
| hexaNAG | 0.908 | 0.498 | 0.458 | 0.405 | 0.531 | 0.486 |
| tri-NAg-oxocarbenium | 0.501 | 0.447 | 0.384 | 0.411 | — | — |
| tri-NAG-oxazoline | 0.411 | 0.340 | 0.381 | 0.450 | — | — |
The hexaNAG substrate binding and two TriNAG intermediates were considered. RMS coordinate fluctuations were calculated as follows: The average structure for the MD interval from 30 to 100 ps was determined. The RMS coordinate difference between this average structure and snapshots taken every 5 ps from 30 to 100 ps were determined. The average of this RMS is reported separated by sugar residue.
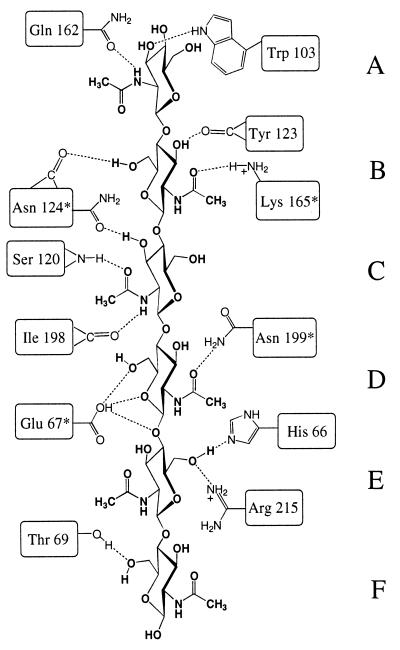
Tight binding of the hexaNAG substrate is achieved through well defined hydrogen bonds, which are constant during the simulations. Fig. 2 shows a schematic of these interactions and highlights residues that are conserved strongly in family 19 chitinases. The N-acetyl amide of sugar A donates a hydrogen bond to the side chain of Gln 162, and O3′ accepts a hydrogen bond from Trp 103. HO3′ and HO6′ of sugar B donate hydrogen bonds to the backbone carbonyls of Tyr 123 and Asn 124, respectively. In addition, the N-acetyl carbonyl forms a hydrogen bond with the charged side chain of Lys 165. The N-acetyl amide of sugar C donates a hydrogen bond to the backbone carbonyl of Ile 198, and the carbonyl of the N-acetyl group accepts a hydrogen bond from the backbone amide of Ser 120. Asn 124 also forms a hydrogen bond with O3′ of sugar C. Two critical hydrogen-bonding interactions are observed for sugar D. The first is between Asn 199 and the N-acetyl carbonyl, which serves to constrain the N-acetyl geometry. The second hydrogen bond is transient forming between the protonated Glu 67 and O5′, O6′, or O1′ (see Fig. 3). Sugar residues E and F make the fewest specific contacts. HO6′ of sugar E donates a hydrogen bond to His 66 whereas O6′ accepts a hydrogen bond from Arg 215. Sugar F forms only one hydrogen bond between O6′ and the side chain of Thr 69.
Figure 2.
A schematic of the hydrogen bonds observed for a hexaNAG substrate bound to barley chitinase. ∗, Residues strongly conserved in family 19 chitinases.
Figure 3.
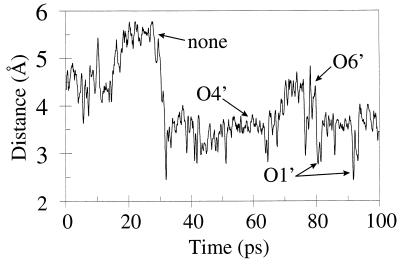
The distance between the proton of Glu 67 and the C1′ anomeric oxygen linking sugar residues D and E. At times, hydrogen bonds are made with O5′ and O6′ as indicated.
Fig. 3 shows the average distance during the MD simulation between the proton of Glu 67 and the β-(1,4)-glycosidic oxygen linking sugar residues D and E. During the early part of the simulation (0–30 ps), only brief hydrogen bonds are formed. However, for the remainder of the trajectory, transient bonds are seen oscillating between O5′, O6′, and O1′ (with O5′ most frequent and O1′ least frequent).
Simulations of Hydrolysis Reaction Intermediates.
Protonation of the anomeric oxygen linking sugar residues D and E followed by subsequent bond cleavage results in a charged intermediate. Because the experimentally observed hydrolysis product has an inverted stereochemistry at C1′, an oxocarbenium ion intermediate is probable. However, we considered both triNAG-oxocarbenium and triNAG-oxazoline intermediates bound to barley chitinase and carried out 100-ps simulations. Starting structures were generated from the hexaNAG/chitinase complex by the deletion of sugar residues E and F followed by conversion of sugar D to the appropriate intermediate.
Binding of the Oxocarbenium Ion.
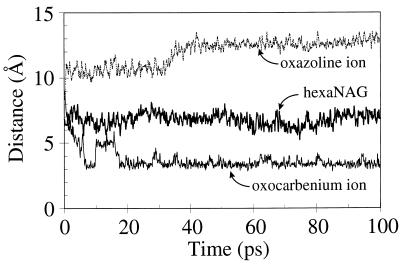
The MD simulation of the triNAG-oxocarbenium ion bound to barley chitinase yielded a stable trajectory with an average enzyme coordinate rms of 1.14 Å. The sugar residues are slightly more stable than observed for the hexaNAG substrate as is evidenced by the smaller coordinate rms fluctuations reported in Table 2. Most surprising was the observed change in conformation for residues 89–92 (the “Glu 89 loop”). These changes do not result in a gross structural change of the enzyme (as indicated by the low total coordinate rms). However, the conformation of the terminus of the acidic side chain of Glu 89 shifts 2 Å toward C1′ of the oxocarbenium ion. Simultaneously, the departure of sugar residues E and F allows the oxocarbenium ion also to move 2–3 Å toward Glu 89. This results in a reduction of the Glu 89-C1′ distance by 4–5 Å (see Fig. 4) and dramatic charge stabilization.
Figure 4.
The distance between the Glu 89 carbonyl oxygen and C1′ of sugar D (or N for the oxazoline case).
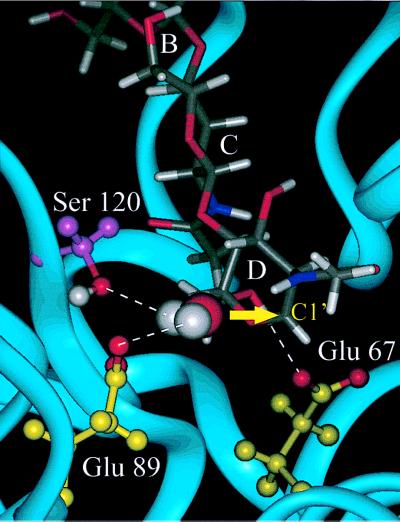
In addition to stabilizing the oxocarbenium ion, part of Glu 89 is well solvated and a specific water molecule is observed to coordinate with Ser 120, Glu 89, and the positive charge of the oxocarbenium ion. Fig. 5 shows a snapshot from the simulation, which highlights this bridged water molecule. This result shows that the conformational change of Glu 89 and of the oxocarbenium ion intermediate place the Glu 89-H2O-C1′ atoms appropriately to complete the hydrolysis reaction. This will result in inversion of the D sugar.
Figure 5.
A snapshot from the dynamics simulations of a tri-NAG oxocarbenium ion intermediate bound to barley chitinase. An arrow marks the α face of the oxocarbenium ion, which will result in inversion of the anomeric configuration upon nucleophilic attack by water. Ser 120 and Glu 89 are positioned to coordinate with this water molecule as shown with dashed lines. Glu 67 forms a hydrogen bond with HO6′.
No changes in the hydrogen bonds for sugar residues A-C took place as a result of the shift in conformation at residue D. However, Asn 199 no longer forms a hydrogen bond with the N-acetyl carbonyl of sugar D, which now becomes solvent accessible. A new hydrogen bond is formed between the deprotonated Glu 67 and HO6′. The proximity of Glu 89 and the bridging water molecule to C1′ prevents the formation of an oxazoline ion because of steric constraints between the bound water molecule and the N-acetyl group. Such an intermediate possibly could occur if the formation of the oxazoline ion was sufficiently favorable enough to displace the bound water molecule.
Binding of the Oxazoline Ion.
The MD simulations of triNAG-oxazoline bound to barley chitinase indicate that an oxazoline reaction intermediate is extremely unlikely. Although the net charge is the same for the oxazoline ion and oxocarbenium ion intermediates, these intermediates complexed with barley chitinase behave very differently. The oxazoline ion is not stabilized by Glu 89, in contrast to the oxocarbenium ion. Indeed, throughout the simulation, the average distance between the positive oxazoline nitrogen and Glu 89 increases, eventually converging to a distance of 12–13 Å (see Fig. 4). This distance is >8 Å greater than the oxocarbenium ion, resulting in a significant loss of charge stabilization. The oxazoline ion is not charge-stabilized by other binding site residues and is exposed predominately to solvent.
The calculation of relative binding energies for small ligands to proteins is fraught with peril caused by complications resulting from solvation, polarization, and entropic affects. However, such calculations may reveal trends useful in obtaining mechanistic insights. Consequently, we calculated the relative binding energies for the triNAG-oxazoline compared with the triNAG-oxocarbenium ion. Because of the charge stabilization by Glu 89, the molecular mechanics ligand binding energy (internal energy) favors the oxocarbenium ion by 81 kcal/mol. The differential solvation energy for each bound and unbound ligand was calculated using the Poisson–Boltzmann continuum solvation approximation (20). Because of the greater solvent accessibility, the oxazoline ion is expected to lose less solvation energy upon binding. Indeed, the oxazoline ion was found to have a more favorable solvation energy of >51 kcal/mol. Combining these contributions, we estimate the oxocarbenium ion to be more stable by 30 kcal/mol. The validity of these methods for determining binding energies has not been established, and the 30 kcal/mol difference should be considered as a qualitative estimate. These results do indicate a preference for the oxocarbenium ion intermediate.
DISCUSSION
These MD simulations on a family 19 barley chitinase complexed with a hexaNAG substrate and two possible hydrolysis intermediates lead to a single displacement mechanism with an inverted anomeric configuration for the hydrolysis product. These simulation results are in excellent agreement with experimental results, offering considerable insight to the specific interactions that stabilize the oxocarbenium ion intermediate. We propose a modified single displacement reaction mechanism in which Glu 89 serves to charge stabilize the oxocarbenium ion in addition to recruiting and activating a nucleophilic water. Next, we will discuss the roles of other important enzyme residues within the active site and indicate some biochemical tests of these predictions.
Protonation of the HexaNAG Substrate.
Our model for binding of the hexaNAG substrate to barley chitinase is in general agreement with the model proposed by Hart et. al. (13). Minor differences were observed in the positioning of sugar residues A and F (which make few hydrogen bonds and are not bound tightly). It is likely that Glu 67 donates a proton to the β-(1, 4)glycosidic oxygen linking sugar residues D and E. Fig. 3 shows the distance between the Glu 67 proton and this anomeric oxygen. After an equilibration period of 30 ps, the proton remains within 4 Å of O1′. At times, a hydrogen bond is made directly with the anomeric oxygen (O1′) although much of the time the proton is involved in a hydrogen bond with either O5′ or O6′.
Ab initio quantum mechanical calculations (21) predict that protonation at O1′ of 2-oxanol in the chair conformation leads to an oxonium ion. There is a significant energy barrier (≈0.82 kcal/mol) preventing formation of an oxocarbenium ion directly from the oxonium ion (this is the barrier to glycosidic bond cleavage). The magnitude of this barrier will depend on the local enzyme structure. Our simulations assume a stepwise mechanism in which protonation is followed by bond cleavage and formation of a charged intermediate (oxocarbenium or oxazoline ion). Certainly, there will be oxocarbenium character in the intermediate, but we cannot be certain that the lifetime for this intermediate will be sufficiently long to be observed experimentally.
Charge Stabilization by Glu 89.
Simulations of the oxocarbenium ion intermediate bound to barley chitinase yielded the surprising result that Glu 89 serves to charge stabilize the oxocarbenium ion through a conformational change in both substrate and enzyme. Commonly, the role of the second acidic residue in a single displacement reaction mechanism (the first having donated a proton as discussed above) is referred to in general terms as accommodating a water molecule that can participate in nucleophilic attack at C1′ (22, 23). Although this result also is observed in our simulations, charge stabilization is likely to be of comparable importance for this mechanistic pathway.
The distance of Glu 89 from C1′ (or the protonated oxazoline nitrogen) of sugar residue D depends on the exact nature of the substrate. Fig. 4 shows a plot of this distance during our MD simulations. There are three distinct responses observed: (i) HexaNAG substrate binding has little affect on the Glu 89 loop region, and the average distance of 8 Å remains fairly constant throughout the simulation; (ii) On departure of sugar residues E and F, the oxocarbenium ion is allowed to move toward Glu 89. A simultaneous change in the conformation of Glu 89 results in a 4-Å reduction in the separation distance and considerable charge stabilization, and (iii) In contrast, for the oxazoline ion (which has the same net charge) the Glu 89 moves away from the charge center eventually reaching a distance of 12 Å. This outcome indicates that the oxazoline gains no stabilization from Glu 89. This lack of stabilization by Glu 89 provides additional evidence against a role for the oxazoline ion intermediate. In these calculations, the free energy change associated with this change in enzyme structure has not been determined explicitly and the assumed stabilization is a qualitative estimate.
Crystallographic Data Supports a Flexible Glu 89 Loop.
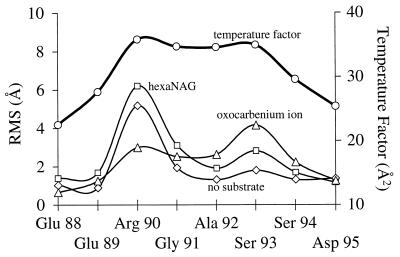
Our model of a flexible Glu 89 loop is supported by crystallographic data. Residues 89–95 have the highest temperature factors (25–30 Å; ref. 2) within the entire enzyme (excluding end effects) (13). This data is in agreement with the average rms coordinate differences between the simulation and crystal structure as shown in Fig. 6. In addition, this indicates considerable thermal instability of the Glu 89 loop and the lack of a well defined packed structure.
Figure 6.
Comparison of the rms (RMS) coordinate difference of various residues from MD (30–100 ps) with the experimental temperature factors. This data supports the flexibility of the Glu 89 loop region. The average temperature factors for all of the other residues (except the amino and C-termini) are <22 Å (2).
Soaking of monoclinic barley chitinase crystals with tetraNAG caused the crystals to crack and dissolve, only to reform 24 hr later. Based on these observations, it was suggested by Hart et al. (13) that, “TetraNAG substrate binding causes the molecule to undergo a conformational change incompatible with maintenance of crystal contacts.” The crystals could reform in the native conformation upon cleavage of tetraNAG to the disaccharide, which remains unbound. Our simulations indicate that binding of hexaNAG does not cause a change in conformation. However, during the hydrolysis mechanism, a conformational change occurs to stabilize the oxocarbenium ion. Such a change could cause the crystals to dissolve.
Predicted Mechanistic Details.
The MD simulations also offer considerable insight about the role of specific active site residues during substrate binding and hydrolysis. Some of these residues are conserved strictly in all of the family 19 chitinases. Fig. 2 shows the contacts made during hexaNAG binding and highlights conserved residues. Particularly critical are the interactions with Lys 165, Asn 124, Asn 199, and Glu 67, all of which are strongly conserved in family 19 chitinases, including the recently isolated bacteria chitinase. We found that Asn 199 serves a unique role of hydrogen bonding to the N-acetyl group of sugar D. This role forces an extended geometry for the N-acetyl group and prevents the formation of an oxazoline ion intermediate. As a test of these results, mutations of any of these residues would be predicted to reduce hydrolysis activity. Indeed, some of these mutagenesis studies recently have been reported after the development of a heterologous expression system for a barley endochitinase (24). The mutations of Glu 67 and Glu 89 to Gln both resulted in complete loss of catalytic activity as would be expected for a single displacement reaction mechanism. Furthermore, the mutation of Asn 124 to Ala reduced the enzyme activity to 0.82% of the wild-type. These findings are consistent with our model for substrate binding in which Asn 124 forms two hydrogen bonds to the hexaNAG substrate.
In addition to stabilizing the positively charged oxocarbenium ion, Glu 89 coordinates with several water molecules. One of these water molecules becomes bridged between Ser 120, Glu 89, and the oxocarbenium ion (Fig. 5). The positioning of this water ensures nucleophilic attack from the α-face of the oxocarbenium ion to yield only an inverted stereochemistry at C1′. A mutation that would position Glu 89 close enough to the oxocarbenium ion to form a covalent bond to C1′ would force an alternative mechanism to occur (the double displacement mechanism of HEWL) leading to retention of the anomeric configuration. An analysis of the HEWL structure matched to barley chitinase suggests a Gly 113 to Asp mutation (and the complementary Glu 89 to Gly mutation) would result in an active site architecture similar to HEWL and the associated change in mechanism. However, our simulation of this mutation shows there is still sufficient space to accommodate a water molecule between the oxocarbenium ion and Asp 113. A mutation to Glu 113 would be predicted to occupy a large enough space to prevent coordination of a water molecule and force a double displacement retaining mechanism.
Design of Family-Specific Chitinase Inhibitors.
Family 18 and 19 chitinases use different hydrolysis mechanisms: family 18 involving an oxazoline intermediate whereas family 19 involves an oxocarbenium ion intermediate. As a result, specific transition state analogs could be designed for each family of chitinase. Indeed, the allosamidins mimic an oxazoline ion intermediate (ref. 9 and K.B., W. Shrader, B. Imperiali, and W.A.G., unpublished results), making them potent family 18 chitinase inhibitors (25–27). Family 19 inhibitors could be designed to mimic the more planar oxocarbenium ion geometry with the delocalized positive charge. Some such inhibitors have been synthesized including amidines, amidrazones (28), and nojiritetrazoles (11). We expect that selective inhibition of family 19 chitinases can be achieved by condensation of one of these transition state analogs with the NAG-NAG-NAG glycone specificity of chitinase.
CONCLUSION
We have applied the methods of MD simulations to examine the hydrolysis mechanism of a family 19 barley chitinase. These simulations are consistent with experimental results, which indicate a single displacement reaction mechanism. In addition, the oxocarbenium ion intermediate may be stabilized through electrostatic interactions with Glu 89 following a conformational change both in the binding geometry of the tri-NAG oxocarbenium ion intermediate and the enzyme active site. A similar conformational change for the oxazoline ion intermediate was not observed and the oxazoline ion is not likely to be part of the hydrolysis mechanism.
Acknowledgments
This research was funded by the Department of Energy-Biological and Chemical Technologies Research Program (DE-FG36–93CH105 81). The facilities of the Materials and Process Simulation Center also are supported by grants from National Science Foundation (CHE 95–22179 and ASC 92–100368), Chevron Petroleum Technology Co., Saudi Aramco, Asahi Chemical, Owens-Corning, Exxon, Chevron Chemical Company, Chevron Research and Technology Co., Avery-Dennison, Hercules, BP Chemical, and Beckman Institute.
ABBREVIATIONS
- HEWL
hen egg white lysozyme
- MD
molecular dynamics
Footnotes
Atomic coordinate files (Brookhaven Protein Data Bank format) for the optimized hexaNAG/chitinase structures and triNAG-intermediate/chitinase structures may be downloaded from the publications section of the http://www.wag.caltech.edu/website.
References
- 1.Boller T, Gehri A, Mauch F, Vogeli U. Planta. 1983;157:22–31. doi: 10.1007/BF00394536. [DOI] [PubMed] [Google Scholar]
- 2.Schlumbaum A, Mauch F, Vögeli U, Boller T. Nature (London) 1986;324:365–367. [Google Scholar]
- 3.Broglie K, Chet I, Holliday M, Cressman R, Biddle P, Knowlton S, Mauvais C J, Broglie R. Science. 1991;254:1194–1197. doi: 10.1126/science.254.5035.1194. [DOI] [PubMed] [Google Scholar]
- 4.Henrissat B. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henrissat B, Bairoch A. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iseli B, Armand S, Boller T, Neuhaus J-M, Henrissat B. FEBS Lett. 1996;382:186–188. doi: 10.1016/0014-5793(96)00174-3. [DOI] [PubMed] [Google Scholar]
- 7.Ohno T, Armand S, Hata T, Nikaidou N, Henrissat B, Mitsutomi M, Watanabe T. J Bacteriol. 1996;178:5065–5070. doi: 10.1128/jb.178.17.5065-5070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armand S, Tomita H, Heyraud A, Gey C, Watanabe T, Henrissat B. FEBS Lett. 1994;343:177–180. doi: 10.1016/0014-5793(94)80314-5. [DOI] [PubMed] [Google Scholar]
- 9.Terwisscha van Scheltinga A C, Armand S, Kalk K H, Isogai A, Henrissat B, Dijkstra B W. Biochemistry. 1995;34:15619–15623. doi: 10.1021/bi00048a003. [DOI] [PubMed] [Google Scholar]
- 10.Fukamizo T, Koga D, Goto S. Biosci Biotech Biochem. 1995;59:311–313. [Google Scholar]
- 11.Ermert P, Vasella A, Weber M, Rupitz K, Withers S G. Carbohydr Res. 1993;250:113–129. [Google Scholar]
- 12.Brameld, K. & Goddard, W. A., III (1998) J. Am. Chem. Soc., in press.
- 13.Hart P J, Pfluger H D, Monzingo A F, Hollis T, Robertus J D. J Mol Biol. 1995;248:402–413. [PubMed] [Google Scholar]
- 14.Phillips D C. Proc Natl Acad Sci USA. 1967;57:484–495. [Google Scholar]
- 15.Sinnott M L. Chem Rev. 1990;90:1171–1202. [Google Scholar]
- 16.Warshel A, Levitt M. J Mol Biol. 1976;103:227–249. doi: 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]
- 17.Warshel A. Computer Modeling of Chemical Reactions in Enzymes and Solution. New York: Wiley; 1991. [Google Scholar]
- 18.Dao-Pin S, Liao D, Remington S J. Proc Natl Acad Sci USA. 1989;86:5361–5365. doi: 10.1073/pnas.86.14.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly J A, Sielecki A R, Sykes B D, James M N G, Phillips D C. Nature (London) 1979;282:875–878. doi: 10.1038/282875a0. [DOI] [PubMed] [Google Scholar]
- 20.Honig B, Nicholls A. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 21.Smith B J. J Am Chem Soc. 1997;119:2699–2706. [Google Scholar]
- 22.McCarter J D, Withers S G. Curr Opin Struct Biol. 1994;4:885–892. doi: 10.1016/0959-440x(94)90271-2. [DOI] [PubMed] [Google Scholar]
- 23.Davies G, Herissat B. Structure (London) 1995;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 24.Andersen M D, Jensen A, Robertus J D, Leah R, Skriver K. Biochem J. 1997;322:815–822. doi: 10.1042/bj3220815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakuda S, Isogai A, Matsumoto S, Suzuki A. Tetrahedron Lett. 1986;27:2475–2478. [Google Scholar]
- 26.Koga D, Isogai A, Sakuda S, Matsumato S, Suzuki A, Kimura S, Ide A. Agric Biol Chem. 1987;51:471–476. [Google Scholar]
- 27.Kinoshita M, Sakuda S, Yamada Y. Biosci Biotechnol Biochem. 1993;57:1699–1703. doi: 10.1271/bbb.57.2067. [DOI] [PubMed] [Google Scholar]
- 28.Papandreou G, Tong M K, Ganem B. J Am Chem Soc. 1993;115:11682–11690. [Google Scholar]