Abstract
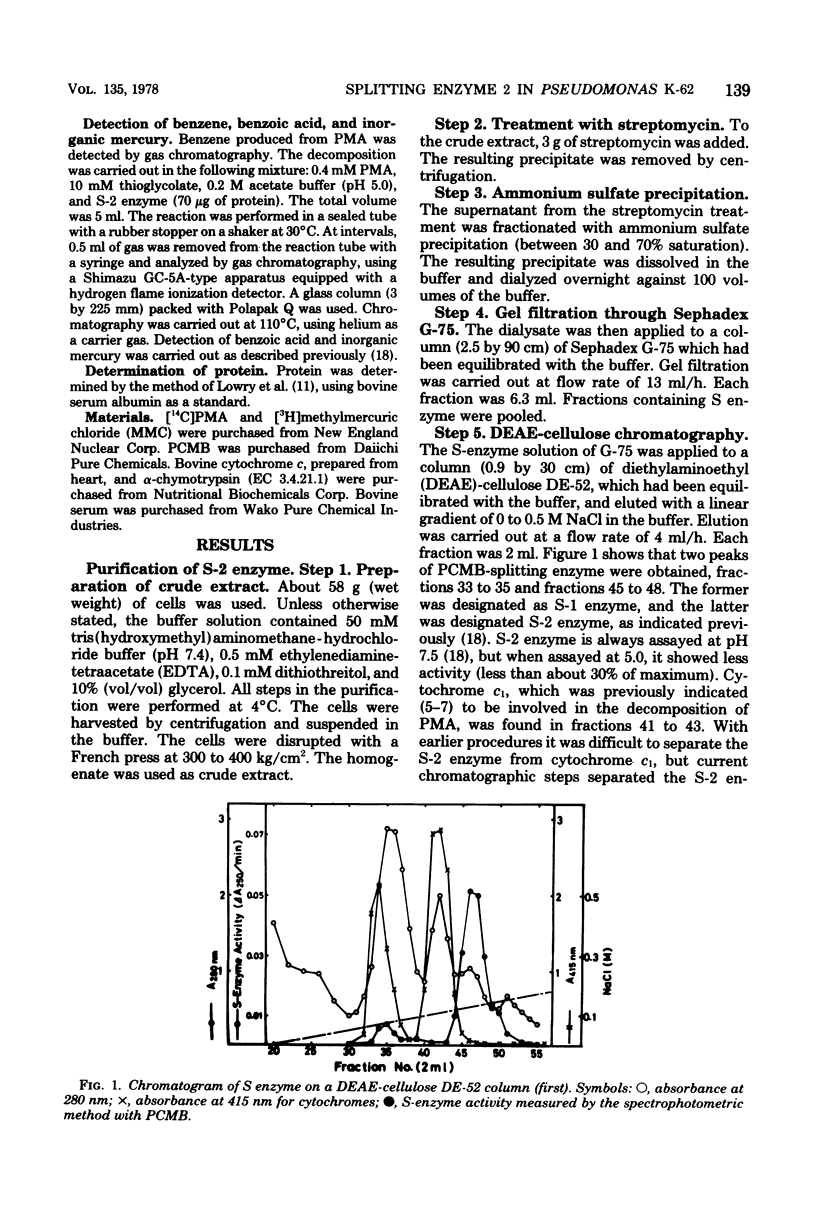
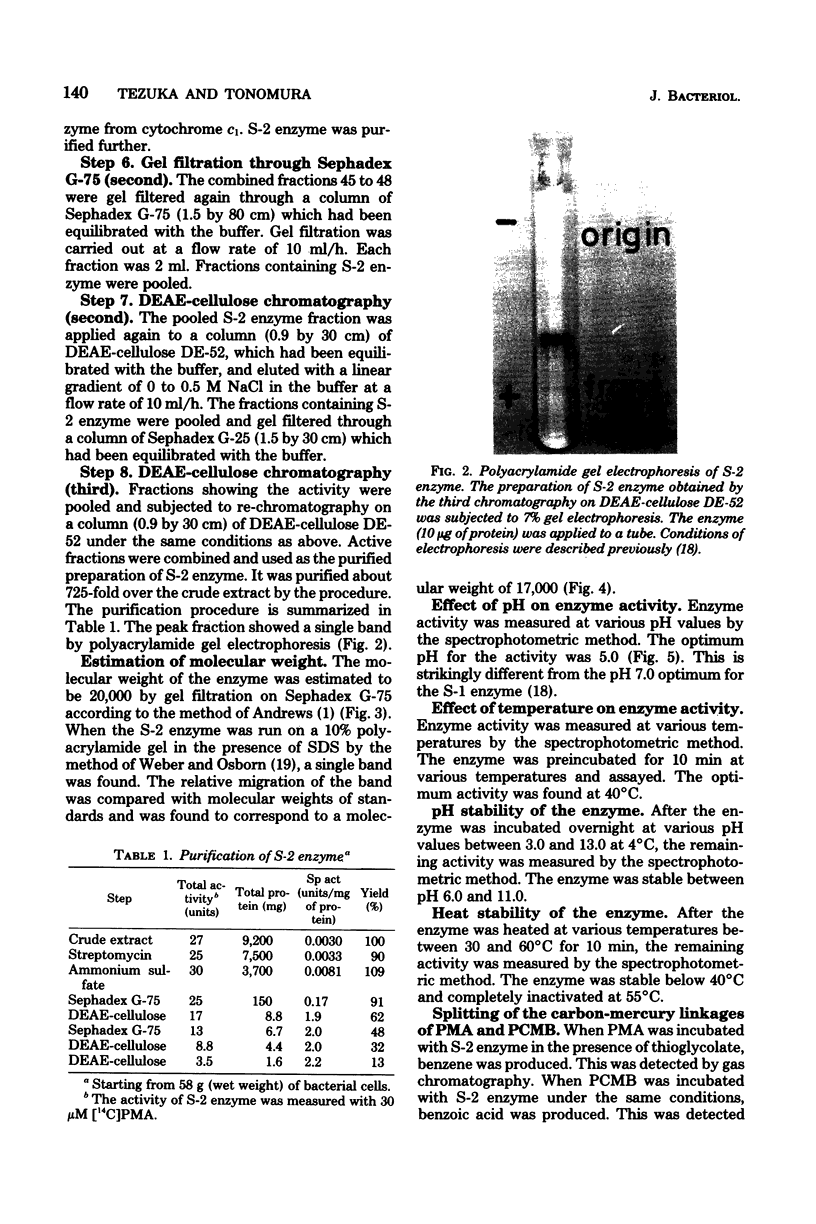
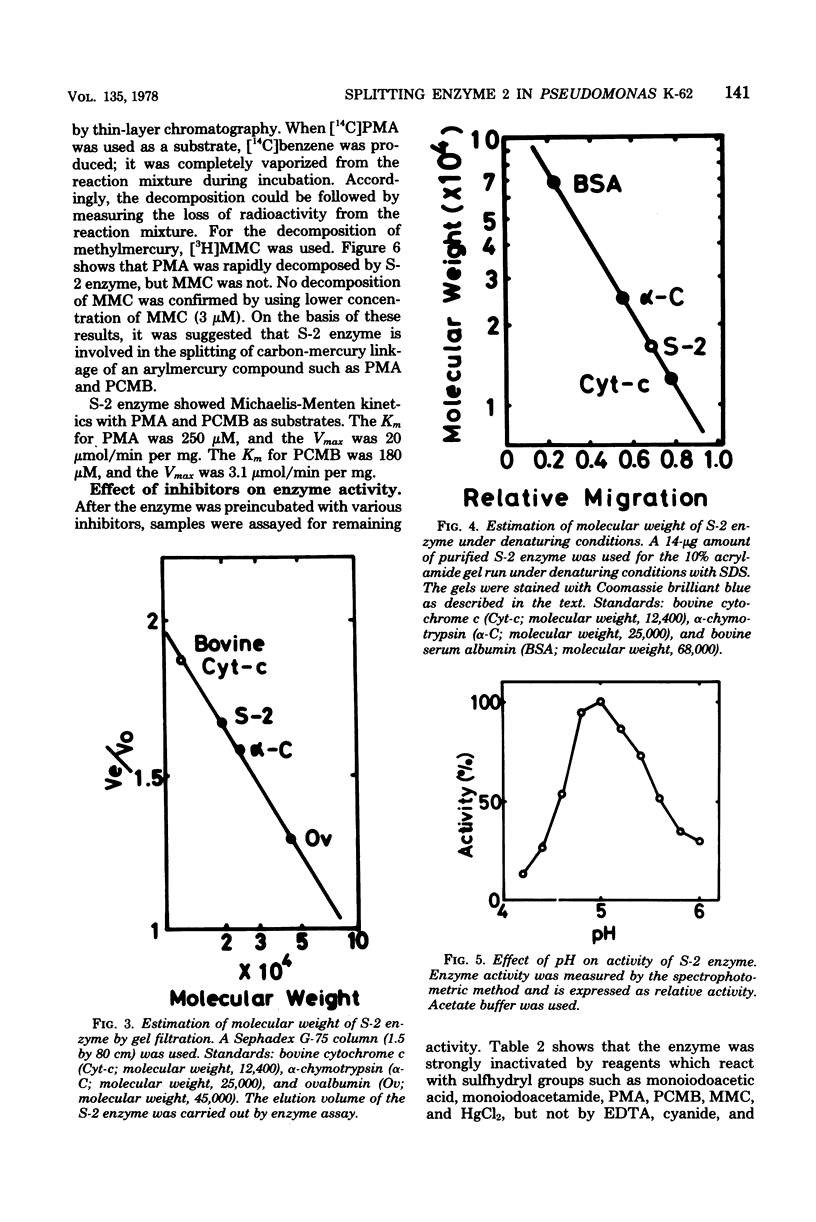
An enzyme (splitting enzyme 2) which catalyzes the splitting of carbon-mercury linkage of arylmercury compounds was found in extracts of mercury-resistant Pseudomonas K-62. This enzyme was purified about 725-fold by treatment with streptomycin, precipitation with ammonium sulfate, and successive chromatography on Sephadex G-75 and diethylaminoethyl-cellulose. A purified preparation of the enzyme showed a single band in electrophoresis either on polyacrylamide or sodium dodecyl sulfate-containing polyacrylamide gels. The molecular weight of the enzyme was estimated to be 20,000 (determined by Sephadex G-75 gel filtration) 17,000 (determined by sodium dodecyl sulfate-polyacrylamide disc gel electrophoresis). The enzyme showed a Km of 180 micron and a Vmax of 3.1 mumol/min per mg for p-chloromercuribenzoic acid and a Km of 250 micron and a Vmax of 20 mumol/min per mg for phenylmercuric acetate. The optimum temperature and pH for the reaction were 40 degrees C and 5.0, respectively.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews P. Estimation of the molecular weights of proteins by Sephadex gel-filtration. Biochem J. 1964 May;91(2):222–233. doi: 10.1042/bj0910222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A. M., Friello D. A. Dissociation and interaction of individual components of a degradative plasmid aggregate in Pseudomonas. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3410–3414. doi: 10.1073/pnas.71.9.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D. L., Weiss A. A., Silver S. Mercury and organomercurial resistances determined by plasmids in Pseudomonas. J Bacteriol. 1977 Oct;132(1):186–196. doi: 10.1128/jb.132.1.186-196.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIS B. J. DISC ELECTROPHORESIS. II. METHOD AND APPLICATION TO HUMAN SERUM PROTEINS. Ann N Y Acad Sci. 1964 Dec 28;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- Furukawa K., Tonomura K. Cytochrome c involved in the reductive decomposition of organic mercurials. Purification of cytochrome c-I from mercury-resistant Pseudomonas and reactivity of cytochromes c from various kinds of bacteria. Biochim Biophys Acta. 1973 Dec 14;325(3):413–423. doi: 10.1016/0005-2728(73)90202-8. [DOI] [PubMed] [Google Scholar]
- Izaki K., Tashiro Y., Funaba T. Mechanism of mercuric chloride resistance in microorganisms. 3. Purification and properties of a mercuric ion reducing enzyme from Escherichia coli bearing R factor. J Biochem. 1974 Mar;75(3):591–599. doi: 10.1093/oxfordjournals.jbchem.a130427. [DOI] [PubMed] [Google Scholar]
- Kondo I., Ishikawa T., Nakahara H. Mercury and cadmium resistances mediated by the penicillinase plasmid in Staphylococcus aureus. J Bacteriol. 1974 Jan;117(1):1–7. doi: 10.1128/jb.117.1.1-7.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Loutit J. S. Investigation of the mating system of Pseudomonas aeruginosa strain I. VI. Mercury resistance associated with the sex factor (FP). Genet Res. 1970 Oct 2;16(2):179–184. doi: 10.1017/s0016672300002408. [DOI] [PubMed] [Google Scholar]
- Nakahara H., Ishikawa T., Sarai Y., Kondo I., Kozukue H. Mercury resistance and R plasmids in Escherichia coli isolated from clinical lesions in Japan. Antimicrob Agents Chemother. 1977 Jun;11(6):999–1003. doi: 10.1128/aac.11.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Roth C. Plasmid-linked resistance to inorganic salts in Staphylococcus aureus. J Bacteriol. 1968 Apr;95(4):1335–1342. doi: 10.1128/jb.95.4.1335-1342.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ORNSTEIN L. DISC ELECTROPHORESIS. I. BACKGROUND AND THEORY. Ann N Y Acad Sci. 1964 Dec 28;121:321–349. doi: 10.1111/j.1749-6632.1964.tb14207.x. [DOI] [PubMed] [Google Scholar]
- Schottel J., Mandal A., Clark D., Silver S., Hedges R. W. Volatilisation of mercury and organomercurials determined by inducible R-factor systems in enteric bacteria. Nature. 1974 Sep 27;251(5473):335–337. doi: 10.1038/251335a0. [DOI] [PubMed] [Google Scholar]
- Summers A. O., Lewis E. Volatilization of mercuric chloride by mercury-resistant plasmid-bearing strains of Escherichia coli, Staphylococcus aureus, and Pseudomonas aeruginosa. J Bacteriol. 1973 Feb;113(2):1070–1072. doi: 10.1128/jb.113.2.1070-1072.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers A. O., Sugarman L. I. Cell-free mercury(II)-reducing activity in a plasmid-bearing strain of Escherichia coli. J Bacteriol. 1974 Jul;119(1):242–249. doi: 10.1128/jb.119.1.242-249.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezuka T., Tonomura K. Purification and properties of an enzyme catalyzing the splitting of carbon-mercury linkages from mercury-resistant Pseudomonas K-62 strain. I. Splitting enzyme 1. J Biochem. 1976 Jul;80(1):79–87. doi: 10.1093/oxfordjournals.jbchem.a131261. [DOI] [PubMed] [Google Scholar]
- Weber K., Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969 Aug 25;244(16):4406–4412. [PubMed] [Google Scholar]
- Weiss A. A., Murphy S. D., Silver S. Mercury and organomercurial resistances determined by plasmids in Staphylococcus aureus. J Bacteriol. 1977 Oct;132(1):197–208. doi: 10.1128/jb.132.1.197-208.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]



