Abstract
Atomic level structures have been determined for the soluble forms of several colicins and toxins, but the structural changes that occur after membrane binding have not been well characterized. Changes occurring in the transition from the soluble to membrane-bound state of the C-terminal 190-residue channel polypeptide of colicin E1 (P190) bound to anionic membranes are described. In the membrane-bound state, the α-helical content increases from 60–64% to 80–90%, with a concomitant increase in the average length of the helical segments from 12 to 16 or 17 residues, close to the length required to span the membrane bilayer in the open channel state. The average distance between helical segments is increased and interhelix interactions are weakened, as shown by a major loss of tertiary structure interactions, decreased efficiency of fluorescence resonance energy transfer from an energy donor on helix V of P190 to an acceptor on helix IX, and decreased resonance energy transfer at higher temperatures, not observed in soluble P190, implying freedom of motion of helical segments. Weaker interactions are also shown by a calorimetric thermal transition of low cooperativity, and the extended nature of the helical array is shown by a 3- to 4-fold increase in the average area subtended per molecule to 4,200 Å2 on the membrane surface. The latter, with analysis of the heat capacity changes, implies the absence of a developed hydrophobic core in the membrane-bound P190. The membrane interfacial layer thus serves to promote formation of a highly helical extended two-dimensional flexible net. The properties of the membrane-bound state of the colicin channel domain (i.e., hydrophobic anchor, lengthened and loosely coupled α-helices, and close association with the membrane interfacial layer) are plausible structural features for the state that is a prerequisite for voltage gating, formation of transmembrane helices, and channel opening.
Keywords: amphipathic helix, apoptosis, diphtheria toxin, membrane interfacial layer, molten globule
The transition of proteins and peptides from a soluble to a membrane bound state is central to organelle and cellular protein import as well as to the inhibitory and lethal action of toxins and colicins. The characterization of structural changes occurring during this transition not only is key to our understanding of the mode of action for these proteins but also provides insight into the mechanisms by which membrane–protein interactions affect protein structure.
Channel-forming colicins provide an excellent system with which to investigate structural changes in a membrane active protein. The bactericidal colicin E1 belongs to a group of colicins whose cytotoxicity is associated with formation of a voltage-dependent ion channel (1–3) that results in cellular deenergization and depletion of the intracellular potassium pool (4). These colicins are highly soluble in aqueous solution but are able to bind to and insert into membranes without disruption of bilayer integrity. Channel-forming colicins contain three domains associated with functions necessary for channel formation. Binding to the cytoplasmic membrane and channel formation are functions of the C-terminal domain (4). The structure of a 190-residue C-terminal channel polypeptide, P190, has been solved to atomic resolution and found to consist of 10 α-helices (5). This structure is very similar to that determined previously for the channel domain of colicin A (6) and of colicin Ia seen in the structure of the whole molecule (7). It is similar to the pore-forming domain of diphtheria (8) and other toxins such as insecticidal δ-toxin (9). Furthermore, its three-dimensional structure is similar to that of the antiapoptotic protein Bcl-XL (10). This suggests that structural changes observed for the colicins after membrane binding are relevant to a variety of other proteins. Formation of the colicin channel, as shown in studies with colicin Ia, involves the voltage-mediated translocation of several helical segments of the membrane-bound channel domain across the membrane bilayer (11). To understand the mechanism of the voltage-dependent gating and protein insertion into the membrane, it is necessary to extend the structural analysis of the colicin system beyond the soluble proteins to the membrane-bound state.
Previous studies have contributed to a detailed understanding of the energetics of colicin–membrane interaction (12, 13), which are better understood than for the toxins and antiapoptotic proteins. Binding of the channel domain to lipid vesicles involves an initial electrostatic interaction between the positively charged polypeptide and the negatively charged membrane surface followed by a rapid transition to nonelectrostatic binding (12, 13). From the P190 structure, it has been proposed that the charged docking face is formed by helices III–VII and IV–VI and that the transition from electrostatic to nonelectrostatic binding occurs when hydrophobic interactions between these helical segments are replaced by nonpolar interactions between the helices and the membrane surface (5). However, the structural changes that occur on membrane binding have not been defined. Earlier studies that showed a loss of native tertiary structure of soluble C-terminal colicin A (14, 15) or E1 (16) channel polypeptides, along with conservation of their secondary structure at acidic pH (<4.0), suggested the presence of a “molten globule” (10) intermediate. In the present study, using both CD and infrared analysis, it was not possible to detect such an intermediate, but from spectroscopic, biosensor, fluorescence, and calorimetric analysis, the soluble channel domain forms a highly helical, extended, and two-dimensional (2D) flexible net in the membrane interfacial region. Implications of this structural state for the biological function of colicins and other membrane-active proteins are discussed.
MATERIALS AND METHODS
Colicin E1 Channel Polypeptide.
The colicin E1 C-terminal channel polypeptide P190 was expressed from pSKHY(−), a plasmid with the colicin channel domain under control of the colicin E1 promoter (17), and purified as described (18). C-terminal channel polypeptide P178 was prepared by proteolysis with thermolysin of the intact colicin E1 (19). Single Trp (Trp-424, Trp-460, and Trp-495) and “zero Trp” mutants were prepared by the replacement of pertinent Trp residues with Phe by site-directed mutagenesis (20). The single Trp mutant W424/C505A/D509C was constructed by replacing the SmaI–EcoRI fragment in pT7E1C505A with the SmaI–EcoRI fragment from pSKE1W424 (J. B. Heymann, personal communication). Asp-509 was replaced with Cys by PCR amplification of the EcoRI–BamHI region in pT7E1C505A, which was cloned into pT7E1W424 C505A to create pT7E1W424/C505A/D509C.
Unilamellar Lipid Vesicles.
Liposomes equivalent to 10–20 mM phospholipids were prepared from 1,2-dioleoyl-phosphatidylglycerol (DOPG) and 1,2-dioleoyl-phosphatidylcholine (DOPC) (Avanti Polar Lipids) in a 2:3 molar ratio by extrusion through a 0.05-μm (pore size) filter (Nucleopore, Costar), as described (13, 21). Liposomes for IR spectroscopy in H2O, equivalent to 250 mM phospholipid of the same lipid composition, were prepared by sonication using an ultrasonic disintegrator (150 W) equipped with a microprobe, after which the vesicles were centrifuged to remove titanium particles.
CD.
Spectropolarimeters JASCO-710 (at the Mayo Foundation) equipped with a temperature control system CTC-345 and JASCO-600 (at Purdue University) were used for CD measurements. Quartz cells with 0.01- to 0.2-mm and 10-mm path lengths were used for the far and near UV measurements, respectively. The parameters of the measurements were as follows: time constant, 2 s; scan speed, 20 nm/min; step interval, 0.1 nm; bandwidth, 2 nm and 1 nm, far and near UV, respectively; samples thermostated at 25°C; protein concentration, 0.2–1.0 mg/ml, in 20 mM sodium acetate/0.1 M NaH2PO4, pH 4.0. Lipid/protein ratio, 240 (mol/mol), for membrane-bound P190 (P190m). Secondary structure content was determined as an average of three methods of analysis of CD spectra in the 190- to 240-nm spectral range (22–24), compiled in the program prot cd (25). The difference between the results from the three methods contributes significantly to the error of analysis and the standard deviation of the averaged secondary structure content.
Fourier-Transformed Infrared (FTIR) Transmittance Spectroscopy.
IR spectra in H2O with a resolution of 4 cm−1 were measured with a Digilab FTS-40 FTIR spectrometer (at the Mayo Foundation). One thousand twenty-four spectra were summed for each sample and reference. Matched CaF2 cells, thermostated at 25°C, had path lengths of 3.6 μm (sample) and 3.4 μm (reference) and a sample volume less than 3 μl. A fine path length adjustment (about 0.1 μm) of the reference cell was used to compensate for H2O absorbance in the region of 3,645 cm−1. IR spectra in the amide I–amide II region (1,480–1,800 cm−1) were normalized to zero at 1,800 cm−1, corrected for amino acid side-chain absorbance (26), and analyzed by methods (23, 27) that are discussed elsewhere (refs. 26, 28, and 29; S. Y. Venyaminov, personal communication).
P190 (1.5 mg), lyophilized from water, was dissolved in 5 μl of 2 mM sodium acetate, pH 4.0/10 mM NaCl and mixed with 35 μl of the buffer or liposomes (lipid concentration, 200 mg/ml) prepared in the same buffer. Five microliters of the buffer mixed with 35 μl of liposomes, or the buffer alone, was used as the reference solution. The concentrations of P190 and lipid in the samples were 30–40 and 160–190 mg/ml, respectively (molar ratio, 1:100–160). The ionic strength of the medium was decreased to 12 mM to increase the rate of membrane binding of P190 (13).
Surface Plasmon Resonance Spectroscopy.
A modified surface plasmon resonance apparatus was used, in which two surface resonances were obtained by using light polarized either parallel or perpendicular to the plane of incidence, a technique defined as coupled plasmon-waveguide resonance (30). The reflective surface consisted of an SiO2 film deposited on a silver surface, on which was overlaid a lipid bilayer membrane composed of egg phosphatidylcholine/phosphatidylglycerol, 3:1, in contact with 20 mM dimethylglutaric acid, pH 4.0/10 mM NaCl.
Fluorescence Resonance Energy Transfer (FRET).
C-terminal channel polypeptide (P178) made by thermolysin proteolysis (19) from mutagenized (Trp-460 → Phe; Trp-495 → Phe; Cys-505 → Ala; Asp-509 → Cys) colicin E1 containing a single Trp residue (Trp-424) and Cys residue (Cys-509) was used in FRET analysis with the sulfhydryl-reactive fluorophore 5-(acetylaminoethylamino)naphthalene-1-sulfonic acid (AEDANS), attached to Cys-509 (refs. 31 and 32; but 8 M urea was not used). The side chain of residue 509 protrudes from the protein (5), allowing reaction with IAEDANS (2-iodo-AEDANS) to be conducted under native conditions. Extinction coefficients of 2.2 104 M−1⋅cm−1 at 278 nm, and of 5.7 103 M−1⋅cm−1 at 336 nm were used for determination of the Trp-424 and AEDANS concentrations, respectively. Both mutant P178 and AEDANS-P178 adduct have in vitro channel activity similar to wild-type P178. As a measure of proper folding, the near UV CD signal at 294 nm in the Trp-424 single Trp mutant was not affected in the mutant (Asp-509 → Cys) protein, or by AEDANS conjugation. Fluorescence excitation and emission spectra were measured at 23°C with an SLM-8000C photon counting spectrofluorimeter and corrected for variation of excitation intensity by using a reference channel; half band-width for excitation and emission at 490 nm, 4 nm. Mutant P178-AEDANS adduct (0.2 μM) and liposomes (0.1 mM lipid) were suspended in 20 mM sodium acetate/0.1 M NaCl, pH 4.0. Calculation of donor–acceptor FRET efficiencies and distances was carried out as described (32) by using an R0 of 22 Å. Absorbances at 290 and 337 nm were <0.05. Because Cys-509 is on the outside of the protein, free motion of AEDANS and a corresponding orientation factor of two-thirds were assumed.
Differential Scanning Calorimetry.
Calorimetric measurements were performed with a prototype DASM-4 model microcalorimeter built at the Biocalorimetry Center of The Johns Hopkins University. All calorimetric measurements were conducted at a heating rate of 1°C/min. The protein concentration was 2.0–3.0 mg/ml for soluble P190 (P190s) and P190m. Liposomes consisted of 25 mM DOPG/DOPC, 2:3 (mol/mol). Measurements of P190m were performed against liposome/buffer and buffer solutions. The partial specific heat capacity was determined as described (33), assuming a molecular weight of 21,046 for P190 and a partial specific volume of 0.806 cm/g as calculated from the amino acid composition. The partial heat capacity of the polypeptide chain in the unfolded conformation was calculated as described (34), with known heat capacity values of amino acid residues.
Protein and Lipid Determination.
The protein concentration of P190 samples was determined by using an extinction coefficient of 3.05 × 104 M−1⋅cm−1 at 279.5 nm (18), which was verified by a “micro” method of nitrogen content (35). The absorbance of P190m at 279.5 nm was corrected for solution turbidity according to Winder and Gent (36).
Lipid concentrations were measured by using the “micro” method of (35), based on the determination of Pi content in ashed samples.
RESULTS
Increase in α-Helical Content in Membrane-Bound State.
Far UV CD spectra. CD spectra of the C-terminal colicin channel domain polypeptide in the soluble state (P190s) at pH 4.0 display a maximum at 191 nm, a minimum at 208.5 nm, and a broad minimum at 220 nm, which are characteristic for α-helical proteins (Fig. 1A, solid trace). The spectra of P190s were not sensitive to ionic strength (10–200 mM) or protein concentration (0.02–40 mg/ml) and were the same at pH 4.0 and 7.0.
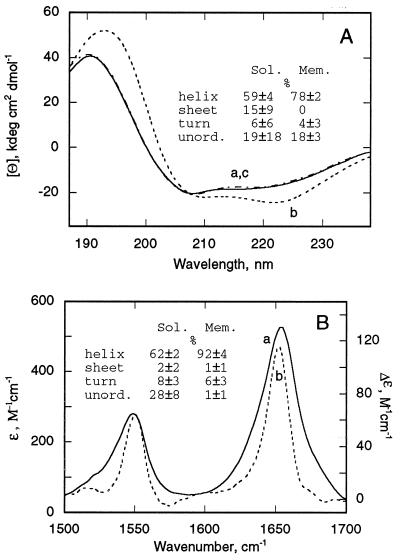
Figure 1.
Increase in α-helical content of P190s on binding to membrane. (A) Far UV CD spectra of P190 in the water-soluble and membrane-bound states: In solution (trace a, solid line), bound to negatively charged liposomes at lipid/P190 molar ratio of 240:1 (trace b, dashed line), and in the presence of neutral liposomes (trace c, dashed–dotted line), DOPG and DOPC at a molar ratio of 2:3 or 100% DOPC were used for preparation of negatively charged or neutral liposomes, respectively. The secondary structure content (P190s vs. P190m) determined by basis spectra (22), ridge regression (23), or variable selection (24) was, respectively, 55 vs. 79%, 63 vs. 79%, or 60 vs. 76% for α-helix; 10 vs. 0%, 26 vs. 0%, or 10 vs. 0% for β-sheet; 0 vs. 7%, 11 vs. 1%, or 8 vs. 5% for β-turns; 35 vs. 14%, 0 vs. 20%, or 22 vs. 19% for unordered structure. (B) IR spectra at pH 4.0 of P190m, lipid/P190 ratio, 145:1 mol/mol, corrected for the absorbance of side chains (trace a, solid line) and difference spectrum, P190m minus P190s (trace b, dotted line). Peaks of amide I band are 1,656 cm−1 (trace a) and 1,654 cm−1 (trace b). Secondary structure components are as follows: α-helix in P190s vs. P190m (62 ± 2% vs. 92 ± 4%), β-turn (8 ± 3% vs. 6 ± 3%), and unordered (28 ± 7% vs. 1 ± 1%). At pH 7, where binding is weak (9), the helix content in the presence of liposomes is 63 ± 4%.
Profound changes of CD spectra take place on binding of P190s to negatively charged (40% DOPG/60% DOPC) liposomes at pH 4.0 [Fig. 1A, dashed trace; lipid/P190, 240:1 (mol/mol)] to form membrane-bound P190m. The absolute value of the molar ellipticity increases, and there is a red shift for all three extrema, to 194 nm, 209.5 nm, and 222 nm, respectively. A pronounced red shift in the 190-nm band associated with trans-membrane helices was previously noted (37). CD spectra obtained with a larger lipid/P190 molar ratio of 500 were essentially identical to that of Fig. 1A, dashed trace (data not shown). To determine whether the observed CD spectral changes were influenced by sample turbidity, CD spectra were obtained (i) with neutral liposomes and (ii) at pH 7.0, conditions for which P190s binds with negligible or small affinity (13). Liposomes prepared from 100% DOPC did not affect the CD spectrum of P190s at pH 4 (Fig. 1A, dash-dotted trace; essentially identical to Fig. 1A, solid trace).
The content of α-helix, β-sheet, β-turn, and unordered structure in P190s, determined by the average of three methods of CD analysis (22–24) was 59 ± 4%, 15 ± 9%, 6 ± 6%, and 20 ± 18%, respectively. The atomic structure of P190s showed the presence of 64% α-helix, no β-sheet structure, 4–8% β-turn, and 28–32% unordered structure (5). Therefore, the prediction of non-zero β-sheet content is an inaccuracy of the method unless it occurred as a result of protein aggregation. CD spectra of P190s at pH 4.0 did not depend on the protein concentration in the range of 0.02–40 mg/ml, ruling out aggregation. It is well known that CD analysis is more accurate in its analysis of α-helix than the other secondary structure components (25). Analysis of the P190m CD spectrum (Fig. 1A, dashed trace) by the same methods showed that the α-helical content increased to 78 ± 2%, β-sheet is absent, and the mean values of β-turn and unordered structure are approximately the same as for P190s.
IR transmittance spectra in H2O.
IR spectra of P190m in the amide I and II regions around 1,650 and 1,550 cm−1 were measured at pH 4.0 and corrected for the absorbance of amino acid side chains (26) and for water perturbed by the presence of membranes by measuring spectra relative to liposomes in suspension (ref. 38 and Fig. 1B, spectrum a]. The difference spectrum of P190m relative to that of P190s shows that the peak of the amide I band shifts from 1,656 cm−1 for P190s to 1,654 cm−1 for P190m (Fig. 1B, spectrum b). Changes in the spectrum of P190s on mixing with neutral liposomes at pH 7.0, where the binding is very weak (13), were much smaller (data not shown).
Analysis of the IR spectra (Fig. 1B, spectrum a; ref. 29) allows determination of the secondary structure content. For P190s these values are as follows: α-helix, 62 ± 2; β-sheet, 1 ± 1; β-turn, 8 ± 3; unordered, 28 ± 7. In contrast to the analysis of the CD spectra, analysis of the IR spectra of P190s does not show any significant β-sheet content, consistent with the crystallographic analysis (5).
Consistent with previous studies (39), it was not possible to detect newly formed β-sheet content in P190m (40). Analysis of the IR spectra of P190 in the membrane-bound state (pH 4.0; ionic strength = 0.012 M, P190/lipid = 1:150, mol/mol) yielded 92% total α-helical content, 6% β-turn, and negligible contents of β-sheet and unordered structure. The fractional increase in amplitude of the amide I and amide II bands (Fig. 1B, spectrum b) is smaller than that of α-helical content because there is also a decrease in the content of the unordered state that absorbs in a similar region of the spectrum. The red shift of the amide I peak from 1,656 cm−1 to 1,654 cm−1 on membrane binding could be explained by an increase in ordered helical structure and concomitant decrease in the unordered component or by an increase in H bond strength of backbone groups on binding.
Increase of Helical Segment Length.
The helical content of P190s and P190m, determined from the average of CD and IR analysis, is 61% and 85%, respectively. The average length of the helical segments in P190s is 12 residues (5). If the number of helical segments in P190m is no larger than in P190s, the average helix length in P190m is increased to 16 or 17 residues, thus potentiating some helices for a transmembrane orientation in the voltage-gated channel.
Loss of Tertiary Structure on Membrane Binding: Near UV CD Spectra.
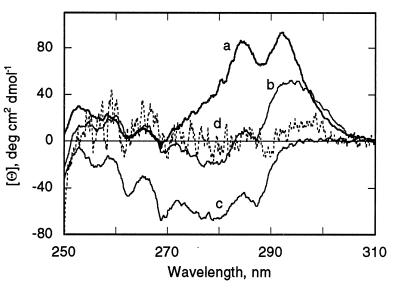
P178s and P190s have a pronounced CD signal in the near UV with two maxima at approximately 285 and 292 nm (Fig. 2, curve a). Characteristic minima associated with the presence of Phe are observed below 270 nm (at about 257, 262, and 268 nm), whereas the signals above 270 nm arise from Trp (Trp-424, Trp-460, and Trp-495) and/or Tyr residues. A major contribution of Trp-424 to the 292-nm positive ellipticity band and association of the short wavelength minima with Phe are inferred from spectra obtained, respectively, with the single Trp (Trp-424; Fig. 2, spectrum b), and zero Trp mutants in P178s (Fig. 2, spectrum c). Despite the noise due to turbidity in the near UV CD spectrum of P178m (Fig. 2, curve d), there is almost total loss of the CD signal of P178s in the near UV on membrane binding. The near UV signals of the channel domain of colicin A are also attenuated on membrane binding (41).
Figure 2.
Decrease in amplitude of UV CD spectrum of P178s on binding to membranes. Near UV CD spectra: a, P178s (bold trace); b, single Trp (Trp-424) mutant of P178s; c, zero Trp mutant of P178s; d, P178m at lipid/P178 molar ratio of 240:1 (dotted trace). Conditions are otherwise as in Fig. 1A.
Increase in Surface Area Subtended by P190 in the Membrane-Bound State.
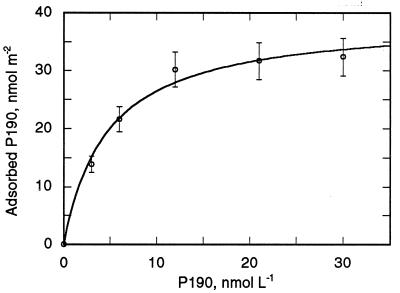
The approximate dimensions of P190s in the atomic structure are 33 Å by 40 Å, so that its cross-section is approximately 1,300 Å2. Binding of P190s to a lipid bilayer surface was determined from coupled plasmon-waveguide resonance spectra obtained under steady-state conditions. A shift of the resonance position to larger incident angles concomitant with an increased amplitude of the resonance showed membrane binding/insertion of P190. By using values of membrane thickness and refractive index obtained by fitting the experimental resonance curves to theoretical curves (30), the mass of P190m can be calculated and a P190 binding isotherm constructed (Fig. 3) from which the following binding parameters can be obtained: Kd = 4.7 nM (in agreement with refs. 12 and 13), and a limiting surface concentration of 39 nmol/m2. This corresponds to a value of 4,200 Å2 for the membrane surface area subtended per molecule of P190m, an increase of more than 3-fold relative to the cross-section of P190s.
Figure 3.
Binding isotherm of P190 to 25 mol % anionic lipid bilayer detected by coupled plasmon-waveguide resonance spectroscopy in the aqueous compartment of the coupled plasmon-waveguide resonance cell obtained with light polarized parallel to the plane of incidence (30).
FRET from Trp-424 to Cys-509-AEDANS.
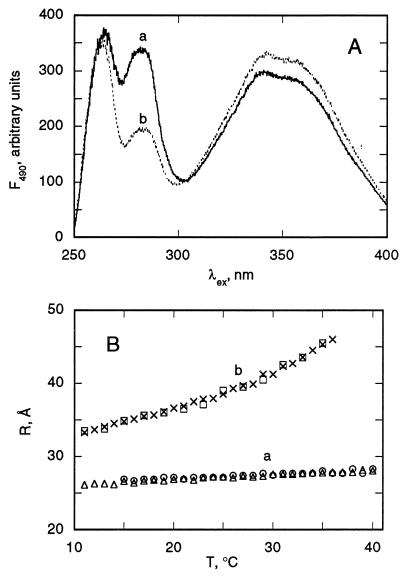
The distance between Trp-424 and Cys-509, located on opposite sides of P190s (4), was determined from the FRET efficiency from Trp-424 in helix V to Cys-509-AEDANS in helix IX. P178s was used in experiments with single Trp mutants. The excitation spectrum of AEDANS in the Trp-424/Cys-509-adduct of P178s has an intense band in the range of 275–295 nm (Fig. 4A, spectrum a). The efficiency of FRET was found to be 29 ± 5% at 2°C, from which a distance, the distance between helices V and IX of P190 (R5–9) = 26 ± 1 Å, between the donor and acceptor groups was calculated based on an R0 value of 22 Å (32). This distance is in good agreement with that between the CE2 atom of Trp-424 and CG atom of Asp-509 (25.3 Å), determined from the atomic structure of P190s (5). The Trp excitation band is significantly lower in the membrane-bound state of Trp-424/Cys-509-adduct (Fig. 4A, spectrum b vs. spectrum a), yielding a FRET efficiency of 10 ± 2%, and a distance, R5–9 = 32 ± 1 Å at 2°C, an increase of 6 Å relative to the soluble state.
Figure 4.
(A) Excitation spectra of AEDANS-Cys-509/Trp-424 single Trp mutant of P178. Spectra: a, P178s; b, P178m. Both were in 20 mM dimethylglutaric acid, pH 4.0/0.1 M NaCl at 2°C. Emission was measured at 490 nm. Liposomes contained a DOPG/DOPC molar ratio of 2:3. P178 and lipid concentrations were 0.2 μM and 0.1 mM, respectively. (B) Temperature (T) dependence of FRET efficiency for P178s (trace a) and P178m (trace b). Two measurements with increasing T are shown: P178s, (▵) T increased to 50°C, decreased to 15°C, followed by a second scan (○); P178m (×), 10°C → 50°C, followed by a second scan (□). Heating rate, 1.5–2.0°C/min.
The flexible nature of arrangement of helical segments in the membrane-bound state is illustrated by the increase in the R5–9 distance as a function of temperature. Thus, the energy transfer efficiencies are 7.9 and 1.3% at 10°C and 35°C, respectively, corresponding to values of R5–9 = 33 Å and 45 Å. This temperature effect is reversible, as heating to 50°C, cooling and reheating yields essentially the same values of R5–9. R5–9 has a much smaller temperature dependence in P178s. Thus, at 35°C, R5–9 increases from 27 Å in P178s to 45 Å in P178m, an increase of 18 Å.
Thermal Denaturation of P190s and P190m: Absence of a Hydrophobic Core in P190m.
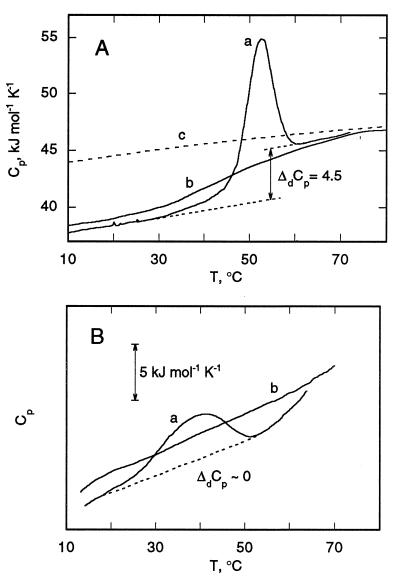
Calorimetric melting profiles for P190s and P190m are shown (Figs. 5). The partial specific heat capacity (Cp) of P190s (37 kJ/mol⋅K at 20°C; Fig. 5A, curve a) is close to values of other compact globular proteins with native tertiary structure (42). Because this Cp value is characteristic of a compact protein structure, it is inferred that P190s at pH 4.0 has a globular structure with a developed hydrophobic core. On heating, P190s denatures with extensive heat absorption and an increased heat capacity (Fig. 5, curve a), reaching values expected for completely unfolded P190 calculated from its amino acid composition (Fig. 5A, curve c, and ref. 34). The pronounced phase of heat absorption (Fig. 5A, curve a) takes place in the same temperature range (52–60°C), where there are major decreases in the amplitude of the near UV CD ellipticity (S. Y. Venyaminov and S.D.Z., unpublished results) that are indicative of the melting of tertiary structure. The change of Cp associated with denaturation between the starting temperature and the high temperature denatured state is ΔdCp = 4.5 kJ·mol−1⋅K−1 (Fig. 5A, curve a). After heat-denatured P190 was cooled to room temperature, its heat capacity was close to that of the native state. A second heating is characterized by a very small heat absorption and the same increase in heat capacity between low and high temperature states (Fig. 5A, curve b). This implies that, after heating to 70°C and recooling, P190 is in a compact denatured state, with a significant fraction of the nonpolar residues shielded from solvent in the protein interior. This compact state with residual secondary, but no tertiary structure, has features of a molten globule (43, 44).
Figure 5.
Cp temperature dependence of P190s (A) in 100 mM NaCl/20 mM sodium acetate, pH 4.0, and P190m (B) in the same buffer. (A) Curves: a and b, first and second calorimetric scans; c (dashed line) theoretical temperature dependence of Cp of P190s calculated from amino acid content. ΔdCp for transition from low to high temperature states of P190s, determined by extrapolation from these states, is shown. (B) Curves a and b are the first and second scans.
P190m has dramatically altered thermodynamic characteristics (Fig. 5B). The data were obtained by subtracting the Cp curve for liposomes from that of P190m with liposomes. The initial temperature-induced unfolding of P190m is characterized by a broad transition centered at 40°C (Fig. 5B, curve a) whose heat absorption is smaller than that of P190s (Fig. 5A, curve a) and may arise from residual helix–helix interactions. After cooling, a second calorimetric scan (Fig. 5B, curve b) indicates no significant thermal transition and shows a featureless monotonic temperature dependence of the Cp of P190m. Applying this temperature dependence to the initial calorimetric scan of P190m, it can be seen that the ΔCp between high and low temperature domains is small. This indicates that the ΔCp between predenatured and post-denatured states is not significant, implying that a developed proteinaceous hydrophobic core is absent in P190m.
DISCUSSION
The Membrane-Bound State of P190 as the Precursor State for Channel Formation.
Consideration of the availability of conformations for general polymer chains has shown that three-dimensional chains tend to develop internal structural organization (e.g., helices and sheets) when they approach a surface (45). The α-helical state is a thermodynamically favorable secondary structure for an isolated or noninteracting polypeptide backbone in the hydrophobic core or in the interfacial layer of the membrane (46, 47). A transition from unordered to α-helical structure on binding to membranes has been previously detected for small peptides (e.g., ref. 48) and polypeptides, e.g., α-lactalbumin (49). However, in these previous studies the magnitude of the increase in helical structure was either not calculated or determined only qualitatively. Binding of the P190 polypeptide of colicin E1 to membranes provides a unique case of the use of existing helical segments as nuclei for extension of α-helices in the 2D environment of the membrane surface. The extension of the average helix length to 16–17 residues in P190m implies that some segments have a length close to the 20 residues that readily span the bilayer (46).
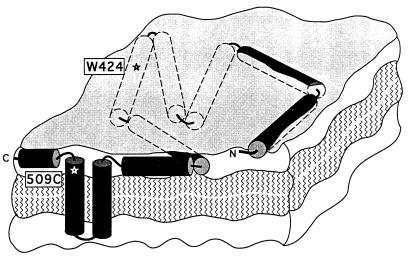
The membrane-bound state of the colicin E1 channel domain is a flexible highly helical (80–90%) extended (4,200 Å2 per molecule) 2D array of weakly or noninteracting helices anchored by a hydrophobic hairpin (50, 51) near the C terminus (Fig. 6). Much of the extended 2D array appears to reside buried within the membrane interfacial layer (53, 54), as determined by solid-state 15N NMR analysis of the helix orientation (50), resistance to external protease probes (18), and rapid and efficient quenching (k ∼ 20–30 s−1) of Trp fluorescence by lipid brominated in the acyl chains (S.D.Z. and W.A.C., unpublished results). Through helix extension and immersion in the interfacial region of low dielectric constant where significant electric fields can be maintained, the helical segments upstream of the hydrophobic anchor (Fig. 6) could respond to imposition of a gating voltage that would result in formation of transmembrane helices and channel opening (11). The flexibility and decoupling from other helices are properties of the closed channel state that would conceivably facilitate helix insertion.
Figure 6.
Schematic of membrane-bound state of C-terminal colicin E1 channel domain (P190m) as an extended highly helical 2D array. Ten helices are shown, mostly buried in membrane interfacial region. Anchoring hydrophobic helical hairpin is near the C terminus. Except for region near N terminus (18), surface-bound helices are shown buried in interfacial layer, in contrast to those depicted in refs. 18 and 52.
Application of the Concept of Amphipathicity.
Peptides that bind avidly to charged membrane surfaces commonly have an amphipathic nature, a variation of charged and hydrophobic residue that follows the periodicity of the helix turn. The segments of P190 with an amphipathic nature are residues 341–359 (overlapping helix I of P190s) (7), 385–403 (helix III), and 422–440 (helix V) (55). These three segments contain 30% of P190 and 8 of the 24 Lys residues and 2 His residues in the sequence. The question then arises as to how the charged side chains of the other 16 Lys and 1 Arg residues in the 2D array of P190m are thermodynamically compensated. Ten of the remaining Lys residues are in a position in the extended α-helices of P190m to form salt bridges with carboxylates displaced by n ± 3–4 residues. Charge compensation of carboxylates is less of an energetic problem (56). The energetic cost of burying the remaining 6 Lys residues in the interfacial layer depends on the actual values of the dielectric constant of intermediate polarity in the interfacial layer and on the detailed positions of the basic side chains. It is relevant that the hydrocarbon side chains of Lys and Arg residues are long enough to extend through a significant thickness of the interfacial layer to a more polar environment.
Nonobservable Molten Globule Intermediate.
An increase of ellipticity in the far UV (14) and a loss of ellipticity in the near UV CD spectrum on membrane binding (14, 15) were observed previously for the channel domain of colicin A. These data were interpreted as a conformational change associated with reorientation of helix pairs that does not involve “drastic unfolding” (14) and also as formation of a molten globule in which there is no significant change of α-helical content (15). The latter has been suggested as an intermediate in translocation of membrane proteins (57). The significant increase in helical content and the loss of the hydrophobic core do not fit the criteria for a molten globule state associated with the formation of P190m. Thus, there does not appear to be any observable membrane-bound intermediate that is spectroscopically consistent with a molten globule. However, we do not preclude the possibility of a short-lived transient intermediate molten globule state in the pathway from P190s to P190m.
Consequences of P190m-Like Structures for Membrane Infrastructure: Mechanism of Some Antiapoptotic Proteins.
The surface structure of P190m (Fig. 6) bears some qualitative resemblance to the “carpet-like” structures that result from adsorption to membranes of high concentrations of charged or amphipathic peptides like melittin (26 residues) or pardaxin (33 residues). It has been proposed that a large surface density of these small peptides, bound to the membrane surface or in the interfacial layer, might exert a lytic effect (58). On the other hand, a comparable density of tightly bound anchored multihelical molecules such as P190m that occupy a significant fraction of the surface area in the interfacial layer could stabilize the membrane, as has been shown by differential scanning calorimetry for the binding of surface-bound proteins to liposomes (59).
It should be emphasized that a high surface density of P190m is not required for toxic action of colicin E1, which is exerted by a single molecule per cell and per liposome (60). It is suggested, however, that a high density of extended 2D arrays may be relevant to the function of certain antiapoptotic proteins. The atomic structure of the antiapoptotic protein Bcl-XL, in the Bcl-2 family, resembles the membrane insertion domain of diphtheria toxin and the colicin A and E1 channel domains (61). Consistent with this structural resemblance, the intriguing finding was recently made that Bcl-XL and Bcl-2 proteins have ion channel activity (61, 62). However, it is presently not clear how this activity can explain the ability to exert an antiapoptotic function of preventing the permeability transition in the mitochondrial outer membrane. We would hypothesize that (i) the structural similarity between the soluble forms of Bcl and P190 implies that the structure of the surface-bound form of some Bcl proteins is similar to P190m (Fig. 6). Such a structure implies a tendency toward channel formation that may not be an accurate indicator of the real function of the Bcl proteins. Rather, Bcl proteins may prevent a membrane permeability transition through the establishment of 2D arrays as shown in Fig. 6.
Acknowledgments
We thank S. Y. Venyaminov for state-of-the-art measurements and analyses of CD and IR spectra, T. Wu for skilled technical assistance, J. Hollister for expert help in the completion of the manuscript, and S. L. Schendel for helpful discussions. This study was supported by grants from the National Institutes of Health, GM-18457 (W.A.C.), GM-34847 (F.G.P.), and RR04328 (to E. Friere and P. Privalov), and from the National Science Foundation, MCB940702 (G.T. and Z.S.).
ABBREVIATIONS
- DOPC and DOPG
1,2-dioleoyl-phosphatidylcholine and -phosphatidylglycerol, respectively
- IAEDANS
5-(2-iodoacetyl)aminoethylaminonaphthalene-1-sulfonic acid
- P190s and P190m
soluble and membrane-bound P190, respectively
- FRET
fluorescence resonance energy transfer
- 2D
two-dimensional
References
- 1.Schein S J, Kagan B L, Finkelstein A. Nature (London) 1978;276:159–163. doi: 10.1038/276159a0. [DOI] [PubMed] [Google Scholar]
- 2.Bullock J O, Cohen F S, Dankert J R, Cramer W A. J Biol Chem. 1983;258:9908–9912. [PubMed] [Google Scholar]
- 3.Cleveland M V, Slatin S, Finkelstein A, Levinthal C. Proc Natl Acad Sci USA. 1983;80:3706–3710. doi: 10.1073/pnas.80.12.3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cramer W A, Heymann J B, Schendel S L, Deriy B N, Cohen F S, Elkins P A, Stauffacher C V. Annu Rev Biophys Biomol Struct. 1995;24:611–641. doi: 10.1146/annurev.bb.24.060195.003143. [DOI] [PubMed] [Google Scholar]
- 5.Elkins P A, Bunker A, Cramer W A, Stauffacher C V. Structure. 1997;5:443–458. doi: 10.1016/s0969-2126(97)00200-1. [DOI] [PubMed] [Google Scholar]
- 6.Parker M W, Postma J P M, Pattus F, Tucker A D, Tsernoglou D. J Mol Biol. 1992;224:639–657. doi: 10.1016/0022-2836(92)90550-4. [DOI] [PubMed] [Google Scholar]
- 7.Wiener M, Freymann D, Ghosh P, Stroud R M. Nature (London) 1997;385:461–464. doi: 10.1038/385461a0. [DOI] [PubMed] [Google Scholar]
- 8.Bennett M J, Choe S, Eisenberg D. Protein Sci. 1994;3:1444–1463. doi: 10.1002/pro.5560030911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Carrol J, Ellar D J. Nature (London) 1991;353:815–821. doi: 10.1038/353815a0. [DOI] [PubMed] [Google Scholar]
- 10.Muchmore S W, Sattler M, Liang H, Meadows R P, Harlan J E, Yoon H S, Nettesheim D, Chang B S, Thompson G S, Wong S-L, et al. Nature (London) 1996;381:335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- 11.Qiu X-Q, Jakes K S, Kienker P K, Finkelstein A, Slatin S L. J Gen Physiol. 1996;107:313–328. doi: 10.1085/jgp.107.3.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heymann J B, Zakharov S D, Zhang Y-L, Cramer W A. Biochemistry. 1996;35:2717–2725. doi: 10.1021/bi951535l. [DOI] [PubMed] [Google Scholar]
- 13.Zakharov S D, Heymann J B, Zhang Y-L, Cramer W A. Biophys J. 1996;70:2774–2783. doi: 10.1016/S0006-3495(96)79847-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van der Goot F G, Gonzalez-Mañas J M, Lakey J H, Pattus F. Nature (London) 1991;354:408–410. doi: 10.1038/354408a0. [DOI] [PubMed] [Google Scholar]
- 15.Muga A, Gonzalez-Mañas J M, Lakey J H, Pattus F, Surewicz W K. J Biol Chem. 1993;268:1553–1557. [PubMed] [Google Scholar]
- 16.Schendel S L, Cramer W A. Protein Sci. 1994;3:2272–2279. doi: 10.1002/pro.5560031212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elkins P A, Song H Y, Cramer W A, Stauffacher C V. Proteins. 1994;19:150–157. doi: 10.1002/prot.340190208. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y-L, Cramer W A. Protein Sci. 1992;1:1666–1676. doi: 10.1002/pro.5560011215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bishop L J, Bjes E S, Davidson V L, Cramer W A. J Bacteriol. 1985;164:237–244. doi: 10.1128/jb.164.1.237-244.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merrill A R, Cramer W A. Biochemistry. 1990;29:8529–8534. doi: 10.1021/bi00489a004. [DOI] [PubMed] [Google Scholar]
- 21.Hope M J, Bally M B, Webb G, Cullis P R. Biochim Biophys Acta. 1985;812:55–65. doi: 10.1016/0005-2736(85)90521-8. [DOI] [PubMed] [Google Scholar]
- 22.Chang C T, Wu C-S, Yang J T. Anal Biochem. 1978;91:12–31. doi: 10.1016/0003-2697(78)90812-6. [DOI] [PubMed] [Google Scholar]
- 23.Provencher S W, Glöckner J. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 24.Manavalan P, Johnson W C., Jr Anal Biochem. 1987;167:76–85. doi: 10.1016/0003-2697(87)90135-7. [DOI] [PubMed] [Google Scholar]
- 25.Venyaminov S Y, Yang J T. In: Circular Dichroism and the Conformational Analysis of Biomolecules. Fasman G D, editor. New York: Plenum; 1996. pp. 69–107. [Google Scholar]
- 26.Venyaminov S Y, Kalnin N N. Biopolymers. 1990;30:1243–1257. doi: 10.1002/bip.360301309. [DOI] [PubMed] [Google Scholar]
- 27.Hennessey J P, Johnson W C., Jr Biochemistry. 1981;20:1085–1094. doi: 10.1021/bi00508a007. [DOI] [PubMed] [Google Scholar]
- 28.Venyaminov S Y, Kalnin N N. Biopolymers. 1990;30:1258–1271. [Google Scholar]
- 29.Kalnin N N, Baikalov I A, Venyaminov S Y. Biopolymers. 1990;30:1273–1280. doi: 10.1002/bip.360301311. [DOI] [PubMed] [Google Scholar]
- 30.Salamon Z, MacLeod H A, Tollin G. Biophys J. 1997;73:2791–2797. doi: 10.1016/S0006-3495(97)78308-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merrill A R, Cohen F S, Cramer W A. Biochemistry. 1990;29:5829–5836. doi: 10.1021/bi00476a026. [DOI] [PubMed] [Google Scholar]
- 32.Steer B A, Merrill A R. Biochemistry. 1994;33:1108–1115. doi: 10.1021/bi00171a009. [DOI] [PubMed] [Google Scholar]
- 33.Privalov P L, Potekhin S A. Methods Enzymol. 1986;131:4–51. doi: 10.1016/0076-6879(86)31033-4. [DOI] [PubMed] [Google Scholar]
- 34.Makhatadze G I, Privalov P. J Mol Biol. 1990;213:375–389. doi: 10.1016/S0022-2836(05)80197-4. [DOI] [PubMed] [Google Scholar]
- 35.Jaenicke L. Anal Biochem. 1974;61:623–627. doi: 10.1016/0003-2697(74)90429-1. [DOI] [PubMed] [Google Scholar]
- 36.Winder A F, Gent W L G. Biopolymers. 1971;10:1243–1251. doi: 10.1002/bip.360100713. [DOI] [PubMed] [Google Scholar]
- 37.Park K, Perczel A, Fasman G D. Protein Sci. 1992;1:1032–1049. doi: 10.1002/pro.5560010809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Venyaminov S Y, Prendergast F G. Anal Biochem. 1997;248:234–245. doi: 10.1006/abio.1997.2136. [DOI] [PubMed] [Google Scholar]
- 39.Rath P, Bousche O, Merrill A R, Cramer W A, Rothschild K J. Biophys J. 1991;59:516–522. doi: 10.1016/S0006-3495(91)82268-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suga H, Shirabe K, Yamamoto T, Tusumi M, Umeda M, Nishimura C, Nakazawa A, Nakanishi M, Arata Y. J Biol Chem. 1991;266:13537–13543. [PubMed] [Google Scholar]
- 41.Lakey J H, Massotte D, Heitz F, Dasseux J-L, Faucon J-F, Parker M W, Pattus F. Eur J Biochem. 1991;196:599–607. doi: 10.1111/j.1432-1033.1991.tb15855.x. [DOI] [PubMed] [Google Scholar]
- 42.Privalov P L. Adv Protein Chem. 1979;33:167–241. doi: 10.1016/s0065-3233(08)60460-x. [DOI] [PubMed] [Google Scholar]
- 43.Griko Y V, Freire E, Privalov P P. Biochemistry. 1994;33:1889–1901. doi: 10.1021/bi00173a036. [DOI] [PubMed] [Google Scholar]
- 44.Griko Y V, Privalov P L. J Mol Biol. 1994;235:1318–1325. doi: 10.1006/jmbi.1994.1085. [DOI] [PubMed] [Google Scholar]
- 45.Wattenbarger M R, Chan H S, Evans D F, Bloomfield V A, Dill K A. J Chem Phys. 1991;93:8343–8351. [Google Scholar]
- 46.Engelman D M, Steitz T A. Cell. 1981;23:411–422. doi: 10.1016/0092-8674(81)90136-7. [DOI] [PubMed] [Google Scholar]
- 47.Jacobs R E, White S H. Biochemistry. 1989;28:3421–3437. doi: 10.1021/bi00434a042. [DOI] [PubMed] [Google Scholar]
- 48.Briggs M S, Cornell D G, Dluhy R A, Gierasch L M. Science. 1986;233:206–208. doi: 10.1126/science.2941862. [DOI] [PubMed] [Google Scholar]
- 49.Bañuelos S, Muga A. J Biol Chem. 1995;270:29910–29915. doi: 10.1074/jbc.270.50.29910. [DOI] [PubMed] [Google Scholar]
- 50.Kim Y, Valentine K, Opella S J, Schendel S L, Cramer W A. Protein Sci. 1998;7:342–348. doi: 10.1002/pro.5560070214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kienker P K, Qiu X-Q, Slatin S L, Finkelstein A, Jakes K S. J Membr Biol. 1997;157:27–37. doi: 10.1007/s002329900213. [DOI] [PubMed] [Google Scholar]
- 52.Palmer L R, Merrill A R. J Biol Chem. 1994;269:4187–4193. [PubMed] [Google Scholar]
- 53.Cherepanov D A, Krishtalik L I. Bioelectrochem Bioenerg. 1990;24:113–127. [Google Scholar]
- 54.White S H, Wimley W C. Curr Opin Struct Biol. 1994;4:79–86. doi: 10.1016/0959-440x(95)80038-7. [DOI] [PubMed] [Google Scholar]
- 55.Shiver J W, Peterson A A, Widger W R, Cramer W A. Methods Enzymol. 1989;172:439–461. doi: 10.1016/s0076-6879(89)72028-0. [DOI] [PubMed] [Google Scholar]
- 56.Krishtalik L I, Cramer W A. FEBS Lett. 1995;369:140–143. doi: 10.1016/0014-5793(95)00756-y. [DOI] [PubMed] [Google Scholar]
- 57.Bychkova V E, Pain R H, Pitsyn O B. FEBS Lett. 1988;238:231–234. doi: 10.1016/0014-5793(88)80485-x. [DOI] [PubMed] [Google Scholar]
- 58.Oren Z, Shai Y. Biochemistry. 1997;36:1826–1835. doi: 10.1021/bi962507l. [DOI] [PubMed] [Google Scholar]
- 59.Papahadjopoulos D, Moscarello M, Eylar E H, Isac T. Biochim Biophys Acta. 1975;401:317–335. doi: 10.1016/0005-2736(75)90233-3. [DOI] [PubMed] [Google Scholar]
- 60.Peterson A A, Cramer W A. J Membr Biol. 1987;99:197–204. doi: 10.1007/BF01995700. [DOI] [PubMed] [Google Scholar]
- 61.Minn A J, Velez P, Schendel S L, Liang H, Muchmore S W, Fesik S W, Fill M, Thompson C B. Nature (London) 1997;385:353–357. doi: 10.1038/385353a0. [DOI] [PubMed] [Google Scholar]
- 62.Schendel S L, Xie Z, Montal M O, Matsuyama S, Montal M, Reed J C. Proc Natl Acad Sci USA. 1997;94:5113–5118. doi: 10.1073/pnas.94.10.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]