Abstract
Chromophore-assisted light inactivation (CALI) offers the only method capable of modulating specific protein activities in localized regions and at particular times. Here, we generalize CALI so that it can be applied to a wider range of tasks. Specifically, we show that CALI can work with a genetically inserted epitope tag; we investigate the effectiveness of alternative dyes, especially fluorescein, comparing them with the standard CALI dye, malachite green; and we study the relative efficiencies of pulsed and continuous-wave illumination. We then use fluorescein-labeled hemagglutinin antibody fragments, together with relatively low-power continuous-wave illumination to examine the effectiveness of CALI targeted to kinesin. We show that CALI can destroy kinesin activity in at least two ways: it can either result in the apparent loss of motor activity, or it can cause irreversible attachment of the kinesin enzyme to its microtubule substrate. Finally, we apply this implementation of CALI to an in vitro system of motor proteins and microtubules that is capable of self-organized aster formation. In this system, CALI can effectively perturb local structure formation by blocking or reducing the degree of aster formation in chosen regions of the sample, without influencing structure formation elsewhere.
The assembly of the cytoskeleton is a prototype for dynamic spatiotemporal phenomena in the cell (1, 2). It can be studied in vivo by perturbing the involved components. Traditionally, such perturbations are achieved by genetic methods; that is, by mutating or deleting genes. This has proven to be a powerful strategy for identifying important components and studying their functions. Conditional mutations additionally enable systematic temporal control of gene activity. However, for a detailed understanding of cellular assembly phenomena, it is crucial to study the roles of protein function locally. For instance, mitotic kinesin-like proteins that participate in spindle assembly change their localization and fulfill different functions at different stages of mitosis (3). One would therefore like to study the function of these proteins by perturbing their activity at particular locations and times. This has been a difficult task so far.
Chromophore-assisted light inactivation (CALI) of proteins is a technique that promises an elegant way to achieve this goal by locally and specifically inactivating one type of protein within a mixture of proteins (4). In CALI, the target protein is labeled with a dye. Upon illumination, the dye generates free radicals that then destroy the labeled protein (5). So far, CALI has been successfully applied to the study of growth processes and guidance of neuronal growth cones (6–9) and of cell fate switching during the development of the visual system (10). Nevertheless, some concerns about the technique remain that must be addressed before it can be used as routinely as genetic techniques are used today.
One such concern is that the molecular mechanism by which the target protein is ultimately inactivated and the characteristics of the inactivated state have not been determined yet. Are there different ways by which CALI can inactivate a protein? For example, it could simply destroy the activity of the protein, analogous to a genetic deletion, or it might arrest the protein in a particular state of its biochemical cycle.
Methods of specific dye attachment form a second impediment to the general use of CALI. Usually, antibodies directed against an epitope of the target protein are used to localize dye molecules within a close range (4). Therefore, each time CALI is applied to a new protein, an antibody that does not block or interfere with the protein’s function must be found. This so far has prevented kinesins from being studied by CALI. More general methods of dye attachment would eliminate this need for new noninhibitory antibodies and are thus desirable.
A third issue in generalizing CALI is the choice of dye. Malachite green has several disadvantages with respect to CALI. The primary disadvantage is that malachite green is hydrophobic and tends to aggregate in solution. In addition, malachite green-mediated inactivation has been thought to require a pulsed laser (4), rather than a more convenient continuous wave light source. A search for improved dyes thus appears worthwhile. In addition, such a search should be of general interest, because it amounts to a systematic study of photodamage, an important effect frequently encountered in fluorescence microscopy.
Here, we address these three points and proceed to implement CALI on a simple in vitro system composed of microtubules and motors, which are of central importance for the spatiotemporal organization of the cytoskeleton (11, 12). Motor proteins are major candidates for CALI experiments, because the use of local (and temporal) perturbation may be crucial for a detailed understanding of many assembly phenomena, including mitosis.
MATERIALS AND METHODS
Construction and Properties of HAkinesin.
We attached a triple hemagglutinin tag (HAtag) to the kinesin construct KinBio401 (13). We amplified the nucleotide sequence for three consecutive copies of the influenza virus hemagglutinin epitope (14) from plasmid pGTEPI (Mark Rose, Princeton University), adding SacII sites at both ends by PCR (Pfu polymerase, Stratagene). The SacII-digested PCR product was inserted into a SacII site of plasmid pEY4 (Jeffrey Gelles, Brandeis University, Waltham, MA), which contains codons 1–401 of Drosophila kinesin heavy chain fused to the last 87 codons of the E. coli biotin carboxyl carrier protein (BCCP). The DNA sequence of the construct was confirmed by sequencing. The expressed HA-kinesin has the amino acid residues VYPYDVPDYAGYPYDVPDYAGSYPYDVPDYAAQSA inserted between codon 3 and 4 of kinesin. We refer to this construct as HAkinesin from now on. The biotinylated HAkinesin was purified as described previously (15) and was dialyzed into 50 mM imidazole/50 mM KCl/4 mM MgCl2/2 mM EGTA/10 mM 2-mercaptoethanol/20% glycerol, pH 6.8, concentrated to 1 mg/ml (Bradford), and stored on ice. The kinesin construct KinBio401 without the tag (13) was purified for control experiments in the same way.
The new HAkinesin construct moved microtubules in motility assays (see below) with normal speeds, similar to the KinBio401 construct, typically 0.93 μm/sec (pH 7.3) or 0.75 μm/sec (pH 6.8). Addition of anti-HA antibodies (IgGs) inhibited microtubule movement completely, probably because the antibody binds weakly and nonspecifically to microtubules. In the presence of Fabs, however, microtubule gliding was restored, at speeds 30% slower than normal. The antibody fragments bound specifically to the HAtag of HAkinesin, as determined by fluorescence measurements (fluorescein-labeled anti-HA Fabs stained an HAkinesin-coated coverslip, but not a surface covered with the non-HA-tagged KinBio401 construct).
Antibodies, Fab Fragments, and Other Proteins.
A monoclonal anti-HA-antibody was isolated from a cell culture supernatant of the cell line 12CA5 (16) by using a standard procedure based on binding to protein A at high salt (17). The purified anti-HA-antibody was dialyzed into 50 mM KPi, pH 7.3, and part of it was stored at 1.4 mg/ml at −80°C. Most of the remainder was used to prepare Fab fragments by papain digestion (17). The purified anti-HA antibody fragments were dialyzed into 50 mM KPi, pH 7.3/10% glycerol, concentrated to 3.6 mg/ml (UV280), and stored at −80°C.
Tubulin was purified essentially as described previously (18) and stored in liquid nitrogen. Streptavidin was from Molecular Probes, β-galactosidase and BSA were from Sigma, and polyclonal anti-β-galactosidase antibody was from Cappell.
Labeling of Antibodies and Fab Fragments.
For CALI experiments with β-galactosidase, antibodies at 600 μg/ml (IgG) in 500 mM NaHCO3, pH 9.8, were labeled by stepwise addition of fluorescein isothiocyanate or malachite green isothiocyanate (both from Molecular Probes) to 120 μg/ml from a stock solution (2 mg/ml or 20 mg/ml, respectively) in DMSO. After 4 h of incubation on ice, the solution was passed over a desalting column into 150 mM NaCl/50 mM NaPi, pH 7.3 (for malachite green labeling, some precipitate had to be spun down before the buffer change). The final antibody concentration was 220 μg/ml. The molar labeling ratio was 2.3 fluorescein or 2.8 malachite green molecules per antibody as determined by UV absorbance. For control experiments, BSA was labeled the same way, resulting in a similar labeling ratio.
For CALI experiments with HAkinesin, anti-HA Fab fragments were fluorescein-labeled essentially as described above, followed by a buffer change into 80 mM Mops, pH 7.3/5 mM MgCl2 for motility assays or 20 mM Pipes/0.5 mM MgCl2/0.25 mM EGTA/500 mM potassium glutamate, pH 6.8 for aster experiments. The final anti-HA Fab concentration was 1.6 mg/ml; the labeling ratio was 1.5 fluoresceins per protein. For control experiments, fluorescein-labeled BSA was transferred into the same buffers. Streptavidin (Molecular Probes) was fluorescein-labeled to a molar ratio of 4.4.
CALI of β-Galactosidase.
For each CALI experiment, a 20-μl sample containing β-galactosidase (10 μg/ml), dye-labeled antibodies to β-galactosidase (≈200 μg/ml), and BSA (125 μg/ml) was placed in a small well drilled in a Teflon slab. The entire volume of the well was illuminated by a laser beam for various lengths of time. The activity of the samples was measured by an orthonitrophenyl galactoside assay. Pulsed 630-nm light was produced by a frequency-doubled pulsed Nd:YAG laser (Quantel, Santa Clara, CA, model YG592) and an oxygen–helium-filled Raman cell (19), which output 10 ns/100 mJ pulses at 10 Hz. The 620-nm continuous-wave (cw) laser light was produced by a tunable argon-pumped dye laser (Coherent 599 Standing Wave Dye Laser with Rhodamine 6G, Coherent Radiation, Palo Alto, CA). The 488-nm cw laser was an argon–ion laser (Coherent 90–5).
To place an upper limit on the range of the inactivating effect of fluorescein, we illuminated a flat 2-μl sample of β-galactosidase in the presence of 11 μM fluorescein-labeled BSA. No significant inactivation was observed. The average closest distance of β-galactosidase to labeled BSA at these concentrations represents an upper limit for the maximum distance of inactivation with fluorescein. Smaller distances cannot be probed with this simple technique, because at higher concentrations of labeled BSA, the incident light would be absorbed almost completely in the first micrometers of the sample.
Green Fluorescent Protein (GFP)-Mediated CALI.
GFP-β-galactosidase and β-galactosidase-GFP fusion proteins were constructed and purified as follows: DNA containing the full lacZ coding sequence was amplified from pMLB1034 (20) by PCR with primers containing BamHI restriction sites and inserted either C-terminal or N-terminal to GFPmut2 (21), which had been previously inserted, also by PCR, into pQE-8 and pQE-12 His-tagging vectors (Qiagen, Chatsworth, CA). The resulting plasmids expressed either a His6-lacZ-GFPmut2 (pMBE012) or a GFPmut2-lacZ-His6 (pMBE014) fusion protein.
For CALI experiments, the two fusion proteins were purified individually with Ni-NTA agarose (Qiaexpress, Qiagen). They displayed fluorescence spectra similar to GFPmut2 and exhibited levels of β-galactosidase activity comparable to those of β-galactosidase itself. Samples contained GFP-β-galactosidase at ≈44 μg/ml, or β-galactosidase-GFP at ≈120 μg/ml, or β-galactosidase (Sigma) at 68 μg/ml (control). These concentrations were chosen to provide equal β-galactosidase activity in each sample. The control sample also contained His6-GFPmut2, to a fluorescence per ml equal to that in the GFP-β-galactosidase sample. All samples contained 100 μg/ml BSA and 20 mM Tris, pH 7.5. Illumination was performed as described above.
Motility Assays and CALI of Kinesin.
For a motility assay, we constructed a flow chamber with a 5-μl volume by using ethanol-cleaned No. 1 glass coverslips (VWR Scientific) (22) and flowed a series of solutions through it: (i) 5 μl water, (ii) 5 μl 170 μg/ml casein in wash buffer (80 mM Pipes, pH 6.8/2 mM MgCl2/1 mM EGTA), (iii) 30 μl wash buffer, (iv) 8 × 5 μl kinesin solution (70 μg/ml HAkinesin, 18 μg/ml streptavidin, 4 μg/ml casein in wash buffer), each incubated for 1 min, (v) 5 μl motility buffer (80 μM Mops, pH 7.3/10 mM MgCl2/5 mM ATP/40 μM taxol/4 μg/ml casein), (vi) 5 μl Fab solution (45 μg/ml fluorescein-labeled anti-HA Fab, 4.5 μg/ml casein in motility buffer) incubated for 2 min, (vii) 5 μl motility buffer, (viii) 5 μl microtubule solution (1.2 mg/ml tubulin, 3.8 mM GTP in motility buffer, polymerized at 37°C for 20 min, then kept at room temperature) incubated for 4 min, and (ix) 5 μl motility buffer. All solutions were filtered through 0.2-μm filters (the microtubule solution was filtered before tubulin polymerization). When we used anti-HA antibodies instead of Fabs, step vi was modified accordingly. For experiments with fluorescein-labeled streptavidin, the kinesin solution (step iv) was modified accordingly and steps vi and vii were omitted. For simple measurements of the speed of the new HAkinesin construct, we omitted steps v–vii. All motility assays were performed at room temperature.
Microtubules were observed through a ×100 0.9–1.3 variable NA oil objective (Olympus) on a modified Zeiss MPS microscope by dark-field microscopy. A long-pass 570-nm filter was inserted in the illumination path to prevent inadvertent excitation of fluorescein during dark-field illumination. Images were recorded with a cooled, back-illuminated charge-coupled device camera (Princeton Instruments, Trenton, NJ); one 0.5 second exposure was taken every 10 sec. For CALI experiments, a 75-μm diameter disk (defined by the illumination iris) was illuminated with a mercury lamp through a standard fluorescein filter set (Zeiss). In experiments with labeled Fab fragments, we used an OD1 neutral density filter for a total power of 55 μW, whereas in experiments with labeled streptavidin, we omitted the filter and obtained 0.5 mW. Gliding and immobilized microtubules on the surface of illuminated and nonilluminated regions (both 4.4 × 103 mm2) were counted 5 min after adding microtubules to the flow cell.
CALI of Kinesin to Perturb Self-Organization of Microtubules and Motors.
Aster formation of microtubules and motors was performed essentially as described (23). One microliter of a solution containing HAkinesin–streptavidin complexes, tubulin, GTP, ATP, and fluorescein-labeled anti-HA Fab antibody fragments was put between two agarose- and BSA-coated coverslips and heated on the microscope to 35°C to start microtubule polymerization and self-organization of microtubules and motors. The final concentrations were 40 μg/ml HAkinesin, 6 μg/ml streptavidin, 280 μg/ml fluorescein-labeled anti-HA Fab, 4.2 mg/ml tubulin, 3 mM ATP, 1.5 mM GTP, 4 mM MgCl2, 0.5 mM EGTA, 2.5 mM KCl, 2.5 mM imidazole, 30 mM Pipes, 180 mM potassium glutamate, and 1% glycerol, pH 6.8. CALI was performed by illuminating a circular 460-μm diameter area through a fluorescein filter set for 0.2–12 sec with 2.8 mW of light. For control experiments fluorescein-labeled BSA was used instead of the labeled compounds at their respective concentrations. To observe pattern formation, we used a Zeiss Axiovert with a dark-field condenser and 570-nm long-pass filter in the illumination path (see above) and a ×20 objective (Olympus). Pictures were taken with a Nikon 35-mm camera.
RESULTS
In what follows we first compare malachite green, the standard dye for CALI, with two potential alternatives: fluorescein and GFP. We show that fluorescein can mediate specific and complete photodamage of proteins at considerably lower energies of illumination than malachite green or GFP. Then we demonstrate that CALI is effective when a chromophore is attached to an artificial epitope tag inserted in the target protein. This renders proteins such as kinesin readily accessible for perturbation by CALI. Because motor activities are easily observable with in vitro motility assays, we are able to demonstrate that inactivation can result in different effects depending on the conditions of the experiment. CALI is then shown to be useful for locally perturbing the self-organization of microtubules and motors.
The Effectiveness of CALI Depends on the Choice of Dye and Light Source.
We tested whether fluorescein could be used in place of malachite green, the dye most commonly used for CALI (4). This choice was motivated by the widespread use of fluorescein in microscopy and by a previous report of its use for specific protein inactivation (24). We also tested GFP as a CALI dye because it can be easily expressed endogenously as a fusion with a target protein (25, 26). We used β-galactosidase as a test enzyme for comparison with previous studies (4).
First, specific inactivation with malachite green-labeled antibodies upon illumination with a high-intensity pulsed light source at 630 nm was reproduced (4) (see Table 1). However, in contrast to earlier reports (4), inactivation with cw light was found to be highly effective. We illuminated with 225 mW (7.2 W/cm2) at 488- and 620-nm wavelengths near the two major peaks in malachite green’s absorption spectrum. As shown in Table 1, the rate of inactivation with these cw light sources was between 9 and 14 times higher than with the pulsed laser. Even in terms of inactivation per unit energy, continuous-wave illumination was at least as efficient as pulsed. Nonspecific inactivation of β-galactosidase in cw experiments was at least as low as in experiments with pulsed light (data not shown). In addition, we found that the degree of inactivation with all three light sources depended solely on the total energy and was independent of the power used (over a 10-fold range, data not shown).
Table 1.
Malachite green, fluorescein, and GFP as dyes for CALI of β-galactosidase
| Malachite green | Fluorescein | GFP | |||
|---|---|---|---|---|---|
| Wavelength in nm | 630 | 620 | 488 | 488 | 488 |
| Laser type | Pulsed | cw | cw | cw | cw |
| Power in mW (flux in W/cm2) | ≈80* (2.6*) | 225 (7.2) | 225 (7.2) | 2 (0.064) | 225 (7.2) |
| Inactivation time, t1/2 in sec | ≈120 | 8.5 | 13 | 18 | ≈900† |
Average power of flux. ≈80 mJ/pulse; 10-nsec pulse length.
Only 40% inactivation.
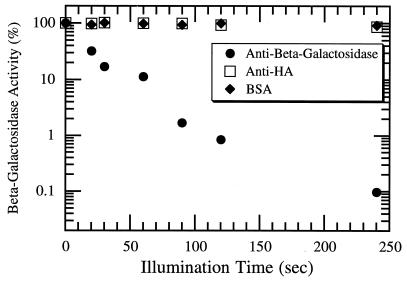
We compared these results obtained for malachite green labeling with similar experiments made with fluorescein-labeled antibodies. As shown in Fig. 1, fluorescein was also capable of inactivating β-galactosidase: enzyme activity decreased exponentially with illumination time when anti-β-galactosidase antibodies labeled with fluorescein were included. Moreover, the energy required for fluorescein-mediated inactivation is far less than that required by malachite green. The power used in fluorescein experiments was 100-fold lower than that used in malachite green experiments, but inactivation still proceeded at about half the rate. This implies a 50-fold-higher inactivation efficiency for fluorescein (Table 1). This energy scale for protein inactivation via fluorescein is comparable to the energy scale required for photobleaching under the same conditions.
Figure 1.
Inactivation of β-galactosidase activity by fluorescein-labeled antibodies against β-galactosidase after being exposed to 2 mW (64 mW/cm2) of light at 488 nm. Controls with the same level of fluorescein fluorescence were also performed: in one case fluorescein was bound to anti-HA antibodies; in the other case, it was attached to BSA. Each point is the average of at least three independent activity determinations using an orthonitrophenyl galactoside colorimetric assay.
Futhermore, the fluorescein CALI effect was local. Illumination in the presence of labeled antibodies to other proteins did not lead to significant inactivation of β-galactosidase (Fig. 1). In one control experiment, we mixed fluorescein-labeled BSA at a high concentration with β-galactosidase (see Materials and Methods) and found that the enzyme activity was reduced by only 10% after 4 min of illumination at 64 mW/cm2 (data not shown). By calculating the average distance of a β-galactosidase molecule to its closest neighboring BSA molecule, we conclude that the range of the inactivating effect of fluorescein under our CALI conditions is less than 30 nm.
To investigate the effectiveness of GFP for protein inactivation, we studied fusion proteins composed of β-galactosidase and GFP domains. We found that GFP was capable of reducing the β-galactosidase activity of the samples by up to 40% under conditions where a control sample, containing β-galactosidase and GFP mixed together, showed no measurable activity reduction at all. Specifically, at 225 mW (7.2 W/cm2) of power, GFP required about 15 min to reach saturating inactivation effects. The energy required by GFP for inactivation is comparable to that required by malachite green (Table 1), and similar to the energy necessary for photobleaching. However, we were unable to produce complete inactivation with GFP. Based on these results, we decided to use fluorescein as a dye for CALI experiments on kinesin.
CALI Can Inhibit Kinesin in Motility Assays in Different Ways.
We fused a triple HA tag (14) to the N terminus of the motor domain of the KinBio401 kinesin construct (13). The new HAkinesin moved microtubules with normal velocities (see Materials and Methods). Fluorescein-labeled Fab fragments bound to the HA tag still allowed microtubule movement, thus making CALI experiments on this tagged kinesin construct possible.
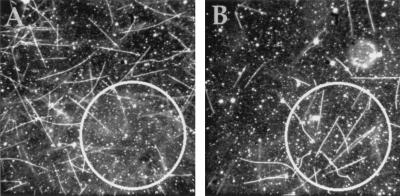
Therefore, we performed CALI experiments with fluorescein-labeled anti-HA Fab fragments in motility assays (22). We observed different types of HAkinesin inactivation depending on whether microtubules were present during the inactivation period or were introduced afterward. A circular area of immobilized HAkinesins bound to labeled Fab fragments was first illuminated in the absence of microtubules (Fig. 2A). The motors were inactivated and did not bind to microtubules subsequently injected into the chamber. Outside the illuminated area microtubules moved normally (see Table 2). When microtubules moved into the illuminated area, they detached (moved out of focus), and very few became immobilized. (In control experiments, HAkinesin was not inactivated by illumination in the presence of BSA labeled with a similar amount of fluorescein.)
Figure 2.
CALI of HAkinesin using fluorescein-labeled anti-HA Fabs in motility assays. (A) An area with a diameter of 75 μm (indicated by a circle) was illuminated through the fluorescein filter set in the absence of microtubules (only immobilized HAkinesin and HA-kinesin-bound, fluorescein-labeled Fab fragments were present). Ten minutes after adding the microtubules and washing the flow cell (see Materials and Methods), microtubules were observed. (B) As in A, but the area was illuminated after adding the microtubules. (Because some gliding microtubules detach from the surface after washing the flow cell, but hardly any immobilized ones do so, the density of microtubules is higher in the illuminated area of B. In experiment B a lower concentration of microtubules was used than in A. Typical microtubule densities are in Table 2.)
Table 2.
CALI of HAkinesin in motility assays (see Fig. 2)
| CALI of kinesin in the absence of microtubules
|
CALI of kinesin in the presence of microtubules
|
|||
|---|---|---|---|---|
| Illuminated | Not illuminated | Illuminated | Not illuminated | |
| Total no. of microtubules* | 2 | 17.5 | 12 | 6 |
| No. of gliding microtubules* | 0 | 17 | 0.5 | 6 |
The variability of these numbers is typically 15%.
In a different experiment, we first put microtubules inside the chamber and observed them gliding. Then we illuminated a circular area as in the previous experiment. In this case, microtubules became immobilized in the illuminated area (Fig. 2B, Table 2). Thus, illumination in the presence of microtubules leads to a different type of inactivation. Whereas illumination performed on unbound motors caused kinesin to lose its ability to propel and even bind to microtubules, illumination of microtubule-bound motors either crosslinked the motors and microtubules together or locked the motors in their rigor state (22), in which they bind microtubules but fail to release them.
Finally, taking advantage of the biotinylation of the kinesin construct (see Materials and Methods) we indirectly attached a dye to kinesin in another way: by binding fluorescein-labeled streptavidin to biotin. In CALI experiments with fluorescein-labeled streptavidin (and without fluorescein-labeled Fabs) kinesin was inactivated specifically (data not shown). However, 50 times more energy was needed than in comparable experiments with labeled Fabs (data not shown). This increased energy requirement for inactivation can be partially explained by the different stoichiometries. Streptavidin can bind up to four kinesin dimers, whereas each kinesin dimer can bind two Fab fragments. Geometric factors involving the arrangement of dyes on their respective molecules might be another cause for different inactivation efficiencies. We were able to use this method of attachment again in later experiments (see below).
CALI Perturbs in Vitro Self-Organization of Asters.
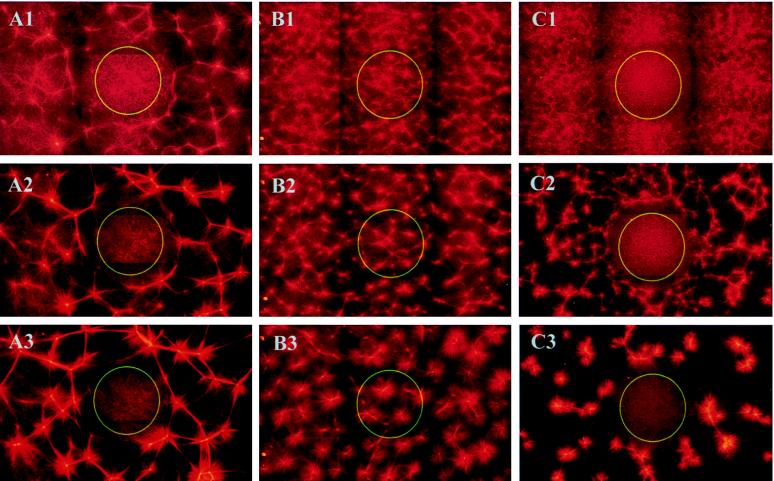
Multiheaded kinesin–streptavidin constructs can organize microtubules into a variety of different large-scale structures in vitro (23). The motor constructs form moving plus end-directed crosslinks between microtubules and can organize the microtubules into asters of various sizes. Fig. 3 shows the evolution of self-assembling structures of microtubules and multiheaded motor constructs after preilluminating a central region with blue light and immediately shifting the temperature to 35°C to start microtubule polymerization. In the presence of fluorescein-labeled Fabs (Fig. 3A), no organized structures formed in the illuminated area, although microtubules still polymerized. In contrast, outside the illuminated area a net of asters slowly formed. In the control experiment with fluorescein-labeled BSA (Fig. 3B), aster formation appeared the same inside and outside of the illuminated area, demonstrating again the specificity of inactivation. (Note that the final pattern outside of the illuminated spot in Fig. 3A differed from that observed in the control experiment in Fig. 3B, because the dynamic parameters of the motors and the microtubule nucleation were changed by the presence of the antibody fragments.) In the experiment shown in Fig. 3C, the streptavidin component of the motor construct was labeled with fluorescein and no Fabs were introduced. In this case, CALI also resulted in local inhibition of aster formation, but 60-fold-higher energies were necessary.
Figure 3.
Perturbation of aster formation by CALI of HAkinesin using fluorescein-labeled anti-HA Fabs (A), fluorescein-labeled BSA (B), or fluorescein-labeled streptavidin (C). A central area with a diameter of 460 μm (indicated by a circle) was illuminated through the fluorescein filter set. The temperature was immediately shifted to 35°C (t = 0) to start microtubule polymerization and organization of microtubules by motors. (Under these conditions, motors should immediately bind to polymerizing microtubules and therfore no longer diffuse freely.) In the presence of fluorescein-labeled anti-HA Fab fragments, perturbed aster formation is shown 18 min (A1), 22 min (A2), and 27 min (A3) after an illumination period of 0.2 sec. Unperturbed aster formation in the control experiment (labeled BSA instead of labeled Fabs) is shown 4 min (B1), 6 min (B2), and 10 min (B3) after 0.4 sec of illumination. Because the presence of Fabs slows down the speed of kinesin on microtubules, aster formation in the presence of Fabs is comparatively slow. When the motor constructs contained fluorescein-labeled streptavidin (no addition of labeled Fabs), aster formation was locally inhibited by 12 sec of illumination, as shown by the evolving structures after 4 min (C1), 6 min (C2), and 10 min (C3). Each picture was assembled from nine individual, overlapping photographs.
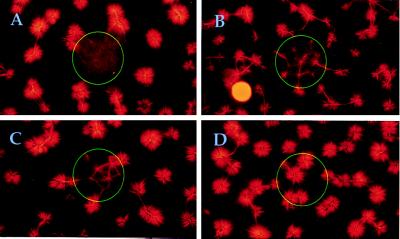
Fig. 4 shows that CALI affects pattern formation differently depending on the extent and timing of inactivation. Final states of CALI experiments with fluorescein-labeled streptavidin (as in Fig. 1C) are shown. We first illuminated the central spot before microtubule polymerization. Although inactivation of kinesins after 12 sec of illumination was sufficient to prevent any pattern formation (Fig. 4A), a shorter illumination period of only 6 sec allowed aster formation to occur (Fig. 4B). However, in this case asters took more time to form and were smaller, which is typical of experiments with lower kinesin concentrations (23), indicating that one can locally “tune” the degree of enzyme inactivation by reducing the illumination time.
Figure 4.
Different perturbations of aster formation generated by varying the timing and extent of CALI illumination. In A, illumination proceeded for 12 sec. In B, 6 sec of illumination was used. In C, 12 sec of illumination was used, as in A, but only after microtubule polymerization had proceeded for 4 min. D shows a control experiment with fluorescein-labeled BSA (12 sec of illumination, as in A). In each case, end states 10 min after the start of microtubule polymerization are shown. Each picture is composed of nine overlapping photographs.
The final structures in these experiments also depend on the time point chosen for the start of inactivation. For instance, when we illuminated the spot for the same length of time, 12 sec, beginning 4 min after the start of microtubule polymerization (instead of just before it), we observed the formation of small asters even in the illuminated area (Fig. 4C). This indicates that illumination reduces the kinesin activity sufficiently to prevent aster formation ab initio, but not sufficiently to halt the further development of preorganized areas into small asters.
DISCUSSION
For a detailed understanding of dynamic spatiotemporal phenomena in the cell, such as self-organization of the cytoskeleton, it is crucial to study local protein functions. CALI is an attractive approach that permits one to remove a protein activity in a temporally and locally defined way (4). We have addressed here several aspects of this technique that are of general importance for its application to the study of dynamic processes and their regulation in cell biology.
We first showed that CALI can be simplified in two ways. Specific inactivation can be achieved with cw light at least as easily as with pulsed light. This observation calls into question the recent hypothesis that a two-photon process is required for radical generation (5). Additionally, using the more soluble fluorescein instead of malachite green reduces the energy required for inactivation by orders of magnitude, making microscope lamps instead of high-energy pulsed lasers suitable tools for CALI experiments. This demonstrates at the same time the high degree of photodamage to be expected in experiments in which fluorescein is used simply as a reporter.
The locality of CALI is largely determined by the lifetime of the photogenerated radicals. For fluorescein this range is below 30 nm (see Materials and Methods). Further studies are required to provide a more precise estimate of this range and to determine whether it is comparable to the 6-nm range previously obtained for malachite green (27). However, the established limit is already sufficiently short for a class of in vitro experiments such as the self-organization studies of microtubules and motors described here. One disadvantage of fluorescein, however, should be mentioned. The use of blue instead of red light for CALI can pose problems when proteins with natural blue-absorbing chromophores are present. It is therefore still worthwhile to look for other CALI dyes that exhibit good solubility, require low photon energies for radical generation, and produce short-range radicals for best local specificity.
The standard technique of dye attachment is the use of dye-labeled, noninhibitory antibodies (28). Finding such an antibody for each protein one wishes to inactivate is a time-consuming search that, in many cases, comes up empty handed. For example, so far all antibodies against motor domains of kinesins have been reported to be inhibitory (e.g., refs. 3, 29, and 30). Attaching an epitope tag close to the protein’s active site can circumvent this problem, as shown for HAkinesin. This provides a more general way of attaching chromophores to proteins without having to find a new antibody for each new protein to be studied by CALI.
The difficulty of introducing dyes into living cells could potentially be circumvented by the use of GFP as a dye. If effective in this capacity, it would have tremendous advantages over all other dyes: attached to a target protein by gene fusion, it forms a covalently bound label. We found that GFP can inactivate proteins specifically, but it does so only at energy doses much higher than those required by fluorescein. Furthermore, its inactivating effects are incomplete under the present conditions. GFP’s relatively poor efficiency as a CALI dye is consistent with the general observation that it is relatively nonphototoxic in fluorescence microscopy. GFP variants with higher radical photogeneration efficiencies and longer wavelength absorption features are desirable for GFP-CALI.
Alternatively, smaller molecules might be of advantage to deliver dyes to target proteins inside cells, because they could be more easily introduced. Therefore, we synthesized fluorescein-nitrilotriacetic acid, which bound to His tags on proteins as expected (31). But unfortunately, it formed more stable nickel complexes with itself (unpublished data). Therefore, a large excess of the dye had to be used for labeling His tags that created problems of nonspecific binding to other proteins and thus reduced the specificity of CALI. Further development of such His-tag-binding dyes could be another promising possibility to make CALI more general and more easily applicable to living cells. Alternatively, one might use modified proteins in which fluorescent tags have been directly incorporated (32).
For a correct interpretation of CALI experiments it is crucial to know whether different types of enzyme inactivation are possible. Inactivation of kinesin, for example, can lead either to detachment or immobilization of microtubules in motility assays. The results of inactivation depend on the timing of illumination and on the molecular interactions. Inactivation sometimes results in switching off of enzyme function but can also cause more complicated effects when the target protein is part of a protein complex (e.g., kinesin bound to a microtubule). These last effects might be due to chemical crosslinking or permanent arrest of the target protein in one state of its biochemical cycle.
CALI is useful as a tool to locally perturb the self-organization of microtubules and motors in vitro. It can be used to gradually attenuate the activity of kinesins, resulting in formation of different structures. This opens the possibility of quantitatively specifying local enzyme activities, thereby expanding the versatility of the aster system and enabling it to mimic more types of cytoskeletal self-organization phenomena. Because the timing of inactivation can influence the final structure chosen by the system, CALI also provides information not just about the local role of a protein but also about its changing temporal importance in the regulation of complex dynamic biological systems.
Acknowledgments
We thank Jeff Gelles for plasmid pEY4, Mark Rose for plasmid pGTEPI, Brendan Cormack for GFP mutants, and Jill Johnson for taxol. We also thank Joseph Forkey for running the pulsed laser apparatus and Jerome Zawadsky and Bob Austin for help in the cw laser experiments. We acknowledge Martha Fonseca for preparing C12A5 cell culture supernatants. The partial support of the National Institutes of Health, the National Science Foundation, and the Human Frontier Science Program Organization is gratefully acknowledged. T.S.’s research was also supported by the Deutsche Forschungsgemeinschaft.
ABBREVIATIONS
- CALI
chromophore-assisted light inactivation
- Fab
antibody fragment
- HA
hemagglutinin
- GFP
green fluorescent protein
- cw
continuous wave
References
- 1.Kirschner M, Mitchison T. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- 2.Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Nature (London) 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- 3.Vernos I, Raats J, Hirano T, Heasman J, Karsenti E, Wylie C. Cell. 1995;81:117–127. doi: 10.1016/0092-8674(95)90376-3. [DOI] [PubMed] [Google Scholar]
- 4.Jay D G. Proc Natl Acad Sci USA. 1988;85:5454–5458. doi: 10.1073/pnas.85.15.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liao J C, Roider J, Jay D G. Proc Natl Acad Sci USA. 1994;91:2659–2663. doi: 10.1073/pnas.91.7.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang H Y, Takel K, Sydor A M, Born T, Rusnak F, Jay D G. Nature (London) 1995;376:686–690. doi: 10.1038/376686a0. [DOI] [PubMed] [Google Scholar]
- 7.Wang F-S, Wolenski J S, Cheney R E, Mooseker M S, Jay D G. Science. 1996;273:660–663. doi: 10.1126/science.273.5275.660. [DOI] [PubMed] [Google Scholar]
- 8.Sydor A M, Su A L, Wang F-S, Xu A, Jay D G. J Cell Biol. 1996;134:1197–1207. doi: 10.1083/jcb.134.5.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Müller B K, Jay D G, Bonhoeffer F. Curr Biol. 1996;6:1497–1502. doi: 10.1016/s0960-9822(96)00754-3. [DOI] [PubMed] [Google Scholar]
- 10.Schmucker D, Su A L, Beermann A, Jäckle H, Jay D G. Proc Natl Acad Sci USA. 1994;91:2664–2668. doi: 10.1073/pnas.91.7.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyman A A, Karsenti E. Cell. 1996;84:401–410. doi: 10.1016/s0092-8674(00)81285-4. [DOI] [PubMed] [Google Scholar]
- 12.Vernos I, Karsenti E. Trends Cell Biol. 1995;5:297–301. doi: 10.1016/s0962-8924(00)89045-5. [DOI] [PubMed] [Google Scholar]
- 13.Berliner E, Young E C, Anderson K, Mahtani H K, Gelles J. Nature (London) 1995;373:718–721. doi: 10.1038/373718a0. [DOI] [PubMed] [Google Scholar]
- 14.Wilson I A, Niman H L, Houghten R A, Cherenson A R, Connolly M L, Lerner R A. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 15.Young E C, Berliner E, Mahtani H K, Perez-Ramirez B, Gelles J. J Biol Chem. 1995;270:3926–3931. doi: 10.1074/jbc.270.8.3926. [DOI] [PubMed] [Google Scholar]
- 16.Field J, Nikawa J-I, Broek D, MacDonald B, Rodgers L, Wilson I A, Lerner R A, Wigler M. Mol Cell Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1988. p. 311. , 628–629. [Google Scholar]
- 18.Mitchison T, Kirschner M. Nature (London) 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 19.Zhang B, Lempert W R, Miles R B, Diskin G. Optics Lett. 1993;18:1132–1134. doi: 10.1364/ol.18.001132. [DOI] [PubMed] [Google Scholar]
- 20.Silhavy T J, Berman M L, Enquist L W. Experiments with Gene Fusions. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1984. p. 250. [Google Scholar]
- 21.Cormack B P, Valdivia R H, Falkow S. Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 22.Howard J. Annu Rev Physiol. 1996;58:703–729. doi: 10.1146/annurev.ph.58.030196.003415. [DOI] [PubMed] [Google Scholar]
- 23.Nedelec F J, Surrey T, Maggs A C, Leibler S. Nature (London) 1997;389:305–307. doi: 10.1038/38532. [DOI] [PubMed] [Google Scholar]
- 24.Jean B, Schmolz M W, Schöllhorn V G. Med Biol Eng Comput. 1992;30:CE17–CE20. doi: 10.1007/BF02446173. [DOI] [PubMed] [Google Scholar]
- 25.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 26.Kain S R, Adams M, Kondepudi T-T, Yang W, Ward W W, Kitts P. BioTechniques. 1995;19:650–655. [PubMed] [Google Scholar]
- 27.Linden K G, Liao J C, Jay D G. Biophys J. 1992;61:956–962. doi: 10.1016/S0006-3495(92)81902-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müller B K, Bonhoeffer F. Curr Biol. 1995;5:1255–1256. doi: 10.1016/s0960-9822(95)00251-x. [DOI] [PubMed] [Google Scholar]
- 29.Boleti H, Karsenti E, Vernos I. Cell. 1996;84:49–59. doi: 10.1016/s0092-8674(00)80992-7. [DOI] [PubMed] [Google Scholar]
- 30.Walczak C E, Verma S, Mitchison T J. J Cell Biol. 1997;136:859–870. doi: 10.1083/jcb.136.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McMahan S A, Burgess R A. Anal Biochemistry. 1996;236:101–106. doi: 10.1006/abio.1996.0137. [DOI] [PubMed] [Google Scholar]
- 32.Thorson J S, Chapman E, Schultz P G. J Am Chem Soc. 1995;117:9361–9362. [Google Scholar]