Abstract
The matrix metalloproteinase (MMP) family of extracellular proteases is conserved throughout the animal kingdom. Studies of invertebrate MMPs have demonstrated they are involved in tissue remodeling. In Drosophila, MMPs are required for tracheal growth, histolysis, tissue invasion, axon guidance, and dendritic remodeling. Recent work demonstrates that MMPs also participate in Drosophila tumor invasion. In C. elegans an MMP is involved in anchor cell invasion; a Hydra MMP is important for regeneration and maintaining cell identity; and a sea urchin MMP degrades matrix to allow hatching. In worms and in flies, MMPs are regulated by the JNK pathway.
Keywords: extracellular matrix, mutant, TIMP, cancer, review
The matrix metalloproteinase family (MMP) of extracellular proteases has captured attention because of their expression in many human pathologies. However, understanding their normal physiological and developmental functions has been difficult because of the complexity of the mammalian MMP family – with 24 MMP family members that display partially overlapping functions [1]. Fortunately, the MMP family is evolutionarily conserved, with homologs found in genomes of plants [2, 3] and animals including nematodes [4, 5], cnidarians [6, 7], echinoderms [8], arthropods [9], and chordates including vertebrates. Thus it appears that MMPs are conserved among multicellular organisms. These invertebrate models present useful opportunities for understanding MMP gene function. Here I summarize what has been learned about the functions of MMPs in invertebrates; most of this work has been performed in Drosophila melanogaster, and so that is the main focus of this review.
Two MMPs and One TIMP in Drosophila melanogaster
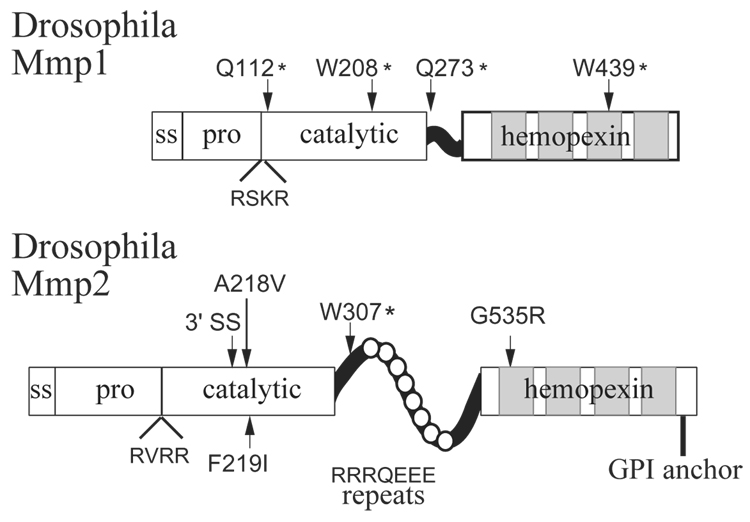
Of the invertebrate models for MMP function, the most developed one to date is the fruitfly Drosophila melanogaster, whose genome includes only two MMPs. Both fly MMPs have the canonical MMP domain structure, with a signal sequence, prodomain, catalytic domain, hinge, and hemopexin domain (Figure 1) [10, 11]. The Drosophila names for the two fly MMP genes are Mmp1 and Mmp2, unfortunate names that resulted from an unintended clash between the Drosophila and MMP nomenclature styles; the MMP community knows these genes also as Dm1-MMP and Dm2-MMP [10, 11]. Despite the similarities of name, it is important to keep in mind that fly Mmp1 is not an ortholog of mammalian MMP1, and fly Mmp2 is not an ortholog of mammalian MMP2; there are not clear orthologous relationships between the Drosophila and vertebrate MMPs [9]. Mmp1 encodes a secreted protein, as it is secreted into the media of cultured S2 cells transfected with the Mmp1 cDNA [9]. Mmp2 has a predicted GPI anchor sequence and its product is localized at the cell membrane of S2 cells transfected with the Mmp2 cDNA; thus Mmp2 is a membrane-associated MMP [10].
Figure 1. The Matrix Metalloproteinases in Drosophila melanogaster.
The domain organization of Mmp1 and Mmp2 is shown. Each has a signal sequence (ss), pro domain (pro), catalytic domain (cat), and four-bladed hemopexin domain. Mmp2 has unusual repeats in its hinge domain, RRRQEEE. Also shown are the positions and lesions of the published alleles of Mmp1 and Mmp2. Allele designation A218V denotes that Ala at position 218 is mutated to Val; an asterix (*) indicates a premature stop codon; one Mmp2 allele is a mutation in a 3’ splice site (3’ SS). All missense alleles indicate residues important for function, as these alleles were identified by mutant phenotype. Adapted from [9, 10]
MMPs are inhibited by endogenous proteins called TIMPs (tissue inhibitor of metalloproteases). Although four TIMP genes are found in vertebrates, only one TIMP gene is found in the Drosophila genome, named Timp, and it is most closely related to mammalian TIMP-3 [12]. In vivo fly Timp can inhibit both fly MMPs, as phenotypes produced by fly MMP overexpression are completely suppressed when Timp is co-expressed [9]. In vitro, the N-terminal domain of recombinant fly Timp inhibits the catalytic activity of both fly Mmp1 and fly Mmp2 on exogenous substrates [12]. Importantly, fly Timp can also inhibit mammalian MMP-1, -2, -3, and -14 [12] and fly Mmp1 is inhibited by mammalian TIMP-2 and TIMP-4 [11]. These cross-inhibition experiments demonstrate the conservation between the mammalian and Drosophila MMPs.
Drosophila MMPs are required for tissue remodeling
Mmp1 and Mmp2 are each required for tissue remodeling. Null mutants for Mmp1 die as larvae with defects in their breathing tubes, or tracheae. The tracheae are a ramified tubular network that grow dramatically in larvae, expanding up to 12-fold in diameter and 14-fold in length. Tracheal growth is accomplished entirely by increasing cell size without division [13]. The inside surface of tracheal tubes is lined with a hard exocuticle to provide structural integrity and a barrier against pathogens, but to accomplish growth the cells must be able to reposition themselves with respect to the cuticle and shed the cuticle at molts. Tubes dilate by cell expansion during the molts, after cell adhesion is released from the old cuticle and before the new cuticle is secreted, and tubes elongate continuously [13]. In Mmp1 mutants, the tracheae appear normal until after the first molt, when the tubes begin to develop breaks and constrictions [9]; closer inspection reveals that patches of tracheal cuticle are not shed at the molt (B.M. Glasheen and A. Page-McCaw, unpublished results). It appears that Mmp1 mutant cells cannot fully release adhesion from the cuticle at the molts and during normal tube elongation, impeding the ability of the tubes to grow. Mmp1 null mutants with defective tracheae exhibit behaviors typical of animals experiencing hypoxia; the defective tracheae cannot support normal growth of internal organs; and eventually larvae die before metamorphosis [9, 14]. Weak alleles of Mmp1 survive to metamorphosis, although they have abnormal body plans: frequently the head cannot evert from the body cavity where it forms, and the epithelial sheets that form the dorsal thorax or notum cannot fuse correctly causing a cleft in the notum (Figure 2) [9]. These phenotypes are discussed in the context of tissue invasion below.
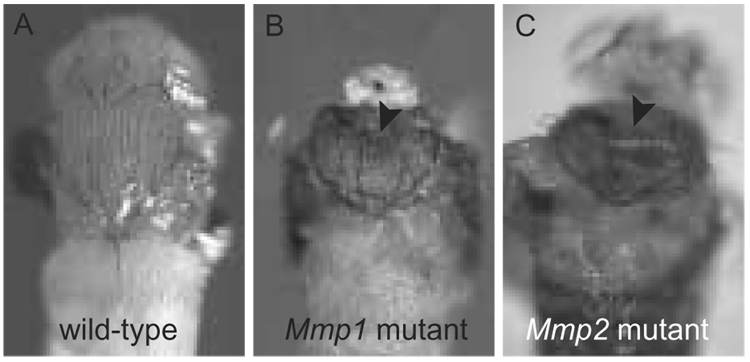
Figure 2. Drosophila MMP pupal phenotypes.
Weak alleles of Mmp1 and Mmp2 display defects in the adult body plan. A. The head and thorax of a wild-type fly just before eclosion. B. Mmp1 homozygote displays a clefted notum (arrow) and appears headless because head eversion failed. C. Mmp2 homozygote displays a clefted notum (arrow).
Mmp2 mutants are defective in histolysis, the large-scale destruction of tissues required for metamorphosis as the fly transitions from a larval to adult body plan. Histolysis of larval tissues occurs via autophagic cell death [15, 16]. In Mmp2 null mutants, however, larval tissues such as the midgut persist aberrantly – histolysis appears to be initiated, as the organ shrinks in size, but the tissue is not eliminated [9]. In some sense, this phenotype was anticipated: the original identification of the first MMP was based on its upregulation in histolyzing tadpole tales during frog metamorphosis [17]. Interestingly, a genome-wide expression study demonstrates that Mmp1 is highly upregulated – almost 200-fold – in autophagic tissues during metamorphosis in Drosophila, and Timp is downregulated 25-fold in these tissues. Mmp2 does not show nearly such a striking change [18]. Although Mmp1 null mutants die as larvae before a pupal histolysis phenotype could be observed, the weak alleles of Mmp1 should allow its role in histolysis to be determined in future studies. Like weak Mmp1 mutants, weak Mmp2 mutants also display clefted notums (Figure 2), discussed below [9, 19]
Drosophila Timp mutants have defects in bonding their wing surfaces together. The wing is made of cuticle, secreted by two epithelial sheets that form the two flat surfaces of the wing. After the adult wing has adopted its final form, these cells undergo an epithelial-to-mesenchymal transition and migrate out of the wing; the two cuticle surfaces then become bonded together. In Timp mutants, however, although the cells are able to exit the wing, the cuticle surfaces are unable to form a stable bond, and instead become blistered [20, 21]. This phenotype is similar to an integrin mutant phenotype [22], and it suggests that there are defects in the extracellular matrix (ECM) that normally cements the surfaces together. It is unclear if this ECM defect in Timp mutants is caused by unregulated proteolysis or if Timp has some other function in the wing blade. Interestingly, the genomic structure of TIMP loci is highly conserved between vertebrates and invertebrates, as Drosophila Timp and most vertebrate TIMP genes are each nested within conserved exons of a synapsin gene [23–25].
The phenotypes observed in MMP and Timp mutants in Drosophila explain why these genes have not been isolated in mutant screens. Classic F2 homozygous mutant screens identified embryonic lethal mutants, but fly MMP mutants die later, as larvae. Late onset processes can often be probed via clonal screens, where mutant clones are introduced into heterozygous tissue; yet because the MMPs are extracellular proteases, they do not act cell autonomously, and clones of MMP or Timp mutant cells do not display phenotypes in homozygous tissue [20, 26]. Interestingly, the only mutant screen to date that has identified an MMP is a gain-of-function screen, where random genes were overexpressed in the nervous system [27]. Although MMP alleles have not been identified in loss-of-function screens, MMP function in Drosophila has been explored and will continue to be explored through a combination of reverse genetic and candidate gene approaches.
Drosophila MMPs in axon guidance
The only function known yet for fly MMPs in the embryo is in nervous system development. Although embryos double mutant for both MMPs are viable and hatch at rates similar to controls (even when any maternal contribution is also removed), the expression patterns of Mmp1 and Mmp2 suggested the existence of unknown embryonic functions [9]. Recent work demonstrates a requirement for Mmp2, and to a lesser extent Mmp1, in axon guidance of motor neurons as they migrate to their target muscles. Motor axons bundle together and migrate as nerves, but individual axons must separate from the nerve when they reach their appropriate target. Although it had been previously thought that MMPs might be involved in clearing a path for axons during pathfinding [28], the Drosophila MMP phenotypes tell a very different story. In Mmp2 null mutants, motor neuron axons fail to remain bundled together (fasciculated), and splinter off inappropriately before they reach their targets. This phenotype is evident in many nerves, including the ISNb and the SNa. Mmp1 has similar but weaker phenotypes [27]. In the ISNb, the double mutant phenotype is very similar to that of the Mmp2 single mutant phenotype, suggesting that Mmp1 does not contribute much to the guidance of this nerve.
Interestingly, in the SNa nerve the double mutant displays a phenotype that is stronger than both single mutants, indicating that Mmp1 and Mmp2 are partially redundant in guiding the path of these axons. Other genetic evidence also indicates that Mmp1 and Mmp2 share substrates that influence the developing motor neurons. When either Mmp1 or Mmp2 is misexpressed, axon bundling is much tighter and axons fail to separate at their appropriate choice points, indicating that the two MMPs can perform similar functions when similarly expressed. Also interesting is the fact that a dominant negative form of Mmp1 can mimic the Mmp2 phenotype in the ISNb, a nerve that does not display a strong Mmp1 loss-of-function phenotype. Thus it appears that the two MMPs share substrates during axon pathfinding, but have distinct roles because of their different regulation. Indeed, they are expressed in very different patterns, with Mmp2 expressed robustly in neurons and glia, whereas Mmp1 expression is much more limited [27].
How could a mutation in a protease result in motor neurons losing interaxonal adhesion? Vertebrate models posited that MMPs promoted outgrowth, and these models predicted that MMP mutants would display stalled axons – exactly the opposite of the fly phenotypes. One intriguing possibility is that MMPs cleave guidance molecules or their receptors. Guidance molecules can act as attractive or repulsive cues to axons. The semaphorin family of guidance molecules promotes inter-axonal repulsion, resulting in defasciculation; axons in Semaphorin-1a loss-of-function mutants fail to separate when they reach their appropriate target [29]. Importantly, decreasing a semaphorin gene dose by half (in a Semaphorin-1a heterozygote) suppresses the Mmp2 mutant phenotype. Although it is unclear whether Mmp2 acts directly (to cleave and inactivate) or indirectly on semaphorin or its receptor, clearly MMPs function in vivo in axon guidance. These Drosophila studies support the hypothesis that MMPs function in guiding vertebrate axons as well.
Drosophila MMPs in dendritic remodeling
Neuronal dendritic remodeling also requires MMPs in Drosophila. In mammals, dendrites are considered to be relatively plastic, altering their neuronal connections in response to activity and environment as well as in response to wounding and trauma [30]. Drosophila provides a model system for identifying the molecular and cellular changes underlying dendritic remodeling. Dendritic remodeling happens in a stereotyped manner during metamorphosis when neurons remodel their dendrites but maintain their axonal connections, effectively rewiring the developing adult brain. The larval dendrites are severed, the severed dendrites are then cleared, and new adult dendrites grow from the cell body. However, in weak mutants for Mmp1 or Mmp2, or in flies engineered to misexpress Timp during nervous system remodeling, the severed dendrites persist and are not cleared [26]. Thus both MMPs are required for dendrite remodeling, and this MMP function may be related to histolysis functions discussed above. It had been hypothesized that vertebrate MMPs might be involved in dendritic remodeling [31], and the fly data is strong support for this model. This anatomical study in flies complements recent vertebrate studies showing that MMP-9 is required for non-pathological synaptic function and plasticity [32].
MMPs contribute to Drosophila tumor invasiveness
Some of the most intensive work in understanding Drosophila MMPs has been examining how they contribute to tumor metastasis in fly tumor models. Single gene mutations in any of five Drosophila genes, including scribble, lethal giant larvae (lgl), and brain tumor (brat), can cause tissues to take on most of the hallmarks of cancer: loss of tissue architecture, overproliferation, defective differentiation, and invasiveness [33]. Genes with these mutant phenotypes are known as neoplastic tumor suppressors: scribble and lgl are known to regulate cell polarity, and brat is a translational repressor. There are two distinct models for Drosophila tumor invasion: the first is a simple transplantation of tissue from a tumorous fly mutant into a wild-type host, similar the classic approach of mouse peritoneal injection of tumor cells. The second is genetic induction of tumors initiated by gene disruption or misexpression in a single cell of a developing fly; this results in a clonal tumor in situ, surrounded by genetically distinct host tissue (Figure 3) [33]. Both tumor models harness the advantages of Drosophila – fast generation time, sophisticated genetics, excellent molecular tools – for understanding how conserved genetic pathways contribute to tumorigenesis.
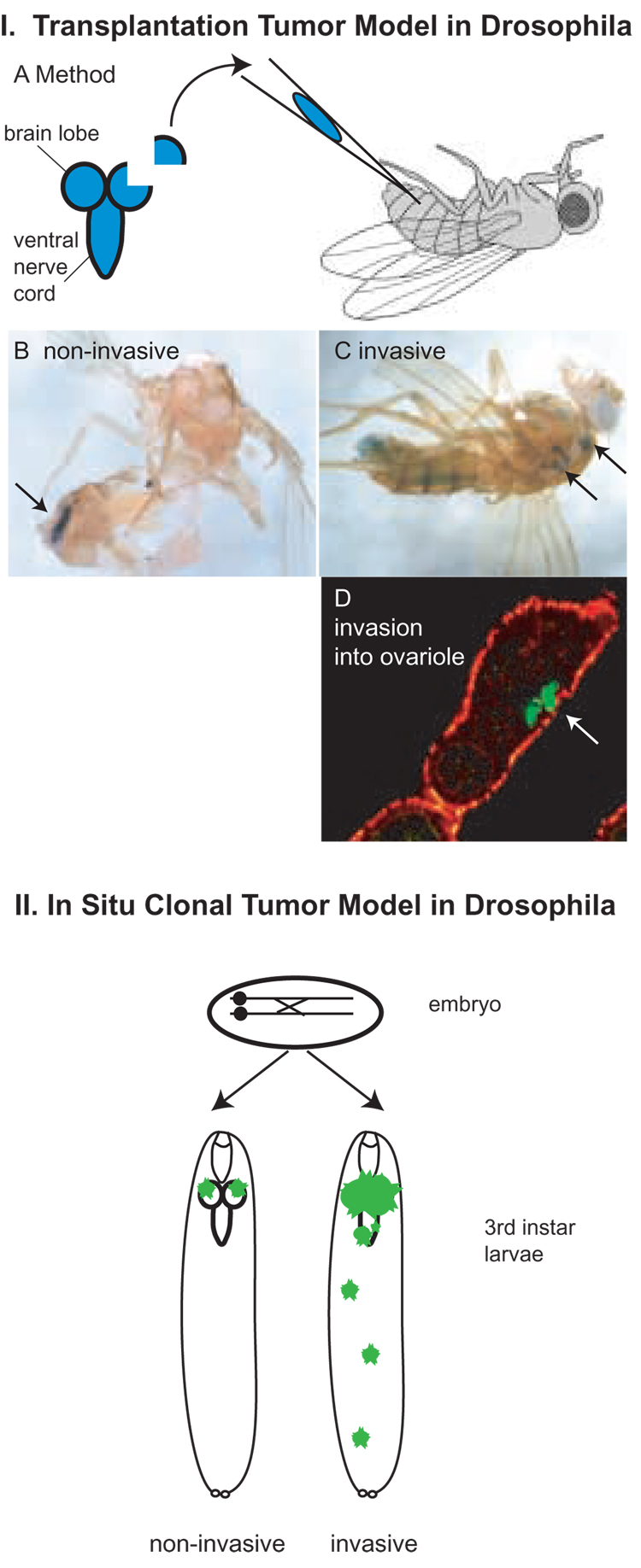
Figure 3. Drosophila Models of Tumor Invasion.
In the transplantation tumor model (I) a fragment of a brain lobe or imaginal disc is injected into the abdomen of an adult fly. The donor fragment is genetically marked with lacZ or another marker. IB. Fragments of wild-type tissue do not invade host tissue after many days and are identified as a discreet tissue after lacZ staining (arrow). IC. Fragments of tissue from tumor suppressor mutants invade host tissues, invading through basement membranes. Secondary tumor sites can be identified as blue lacZ staining throughout the animal (some labeled with arrows). ID. (adapted from [14]) lgl tumor that has invaded through two basement membranes to enter ovariole, labeled with antibodies against lacZ (green, arrow); phalloidin staining in red.
In the in situ clonal tumor model (II), mitotic recombination is induced in an embryo heterozygous for a tumor suppressor, generating mutant cells that give rise to clones of mutant tissue in a phenotypically wild-type background or host. The clones have also lost a repressor that controls the expression of GFP, so that they can be identified as fluorescent green tissue. Wild-type clones or mutants that are non-invasive remain in the tissue where they were generated (left). Invasive tumor clones invade surrounding and distant tissues (right). For a clone to develop aggressive invasive capacity, it must lose a tumor suppressor and also gain activity of an oncogene (under the same genetic control as GFP so that it is expressed only in the clone.)
In the tumor transplantation model, tumor tissue (a brain lobe or imaginal disc fragment) from genetically marked homozygous neoplastic mutants is transplanted into the abdomen of a wild-type host. Wild-type tissue exhibits limited growth in a host abdomen and remain distinct from the host tissues (Figure 3B); in contrast, tissue from tumor mutants proliferates wildly, loses its tissue architecture, and invades into host tissues, killing the host (Figure 3C). Importantly, in the transplantation model metastasis is assayed as the presence of micrometastases (identified as marked tumor cells) observed in host ovarioles (reproductive tissue) because tumor cells found in ovarioles have not only migrated but also invaded through two layers of basement membrane (Figure 3D) [34].
Recently it was established that Drosophila MMPs contribute significantly to the invasive nature of neoplastic tumors (lgl and brat) in the transplantation model. Mmp1 but not Mmp2 is upregulated 10-fold in lgl mutant brains compared to wild-type brains; neither MMP is upregulated in brat mutant brains. Mmp1 is critical for the invasiveness of lgl tumors: tumors from lgl Mmp1 double mutant animals establish significantly fewer micrometastases than tumors from lgl single mutants. Interestingly, although brat tumors do not upregulate Mmp1, hosts injected with brat tumors upregulate Mmp1 20-fold in the host tissue. One advantage of this tumor model is that it is straightforward to manipulate both the genotype of the tumor and also the genotype of the host, which provides the tumor microenvironment. To test for a function of this upregulated Mmp1 expression in the host tissue, the MMP inhibitor Timp was expressed in the host. The rate of micrometastases from both brat and lgl tumors decreased significantly. Thus for both neoplastic mutants, Mmp1 is a crucial factor that allows tumor invasion – in one case Mmp1 is expressed in the tumor; in the other case, Mmp1 expression is induced in the host tissue by the tumor [14].
MMPs in Drosophila clonal tumors
Similar results about the role of Mmp1 in tumor invasion were found using a very different Drosophila tumor model. With the advent of controlled mitotic recombination utilizing the FLP/FRT system, loss of heterozygosity can be induced at will [35]. Thus in situ clonally-derived tumors can be induced in host tissue heterozygous for tumor suppressor genes. The genetic origin of these tumors closely mimics human tumors, which also develop clonally from genetic alterations often involving loss of heterozygosity [36]. In addition to the neoplastic tumor suppressor genes, other mutations can cause clones to overgrow, including gain-of-function mutations in oncogenes like ras (rasact). In the context of wild-type tissue architecture, neoplastic tumor suppressor clones such as scribble display limited aggressiveness because the mutant cells undergo apoptosis [37–40]. It appears that in order for tumors to metastasize to secondary sites and threaten the life of the fly, two mutations are required; for example, the combination of scribble loss of function and ras gain of function (scrib−/− rasact) causes tumors to invade surrounding tissues [37, 41]. Invasion can be easily monitored, as the tumors are engineered to express GFP only in the clonally derived tumor cells (via the MARCM system [42]). When these GFP-expressing scrib−/− rasact tumors are induced in eye/antennal disc tissue, they proliferate wildly and invade the ventral nerve cord (Figure 3, lower panel) [37].
Mmp1 contributes significantly to the invasive capacity of these in situ tumors. Mmp1 is highly expressed in clonally induced scrib−/− rasact invading tumor tissue [19, 40]. When these clones are also engineered to express the MMP inhibitor Timp, however, the clones lose invasiveness, even though they proliferate as much as control scrib−/− rasact clones [19, 40]. Expression of the MMP negative regulator RECK can further reduce invasiveness [19]. However, when the tumorous clones are engineered to express dsRNA knocking down Mmp1 function [40], or are induced in a weak Mmp1 mutant background [19], only partial inhibition of invasion is achieved. Using RNAi to knock down both Mmp2 and Mmp1 at the same time completely reproduces the non-invasive phenotype observed with overexpressing Timp [40]. Thus induced tumor clones in Drosophila express high levels of Mmp1, but use both Mmp1 and Mmp2 to mediate their invasion into surrounding tissue.
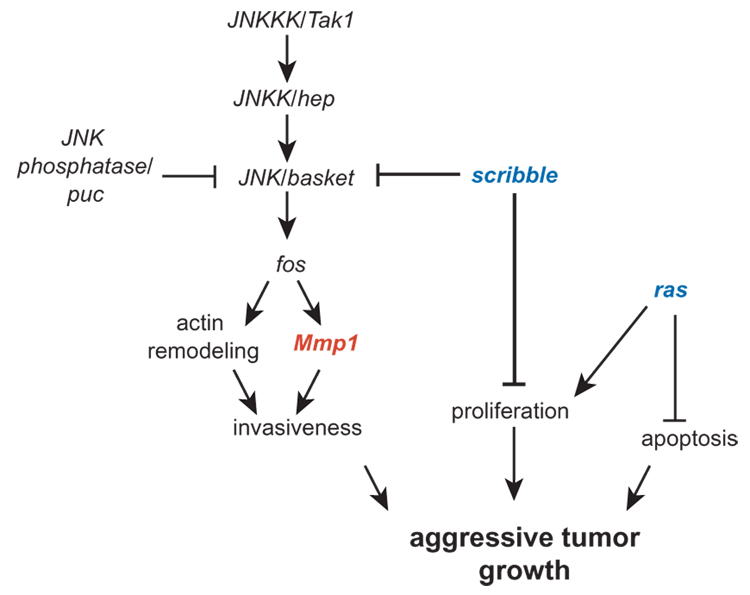
Significant progress has been made in understanding the regulation of Mmp1 in these in situ tumors thanks to the precise genetic manipulations available in Drosophila (Figure 4). Mmp1 upregulation in scrib−/− rasact tumors is caused by the loss of scrib and not by rasact, as clones mutant only for scrib also express Mmp1, although they are not able to metastasize without rasact [40]. In scrib− clones, the Jun N-terminal Kinase (JNK) pathway is activated, and the JNK pathway is in turn responsible for the activation of Mmp1. This JNK-regulated activation of Mmp1 is mediated by the JNK transcriptional effector fos, a member of the AP-1 family, as knocking down fos in scrib−/− mutant clones ablates Mmp1 expression [40]. Indeed, ectopic JNK pathway activation is also sufficient to upregulate Mmp1 [40]. Importantly, the JNK pathway is required for scrib−/− rasact tumors to invade into nearby tissue; when the JNK pathway is inactivated in tumor cells, eye-antennal disc tumors no longer invade the nearby ventral nerve cord as do tumors with a functional JNK pathway[40]. Tumors carrying an inactivating mutation in the JNK pathway also do not upregulate Mmp1, which probably accounts for their non-invasive character [19, 40]. JNK activation is sufficient to allow invasion of clones that are otherwise non-invasive, those expressing rasact [40]. These data have led to the model that the reason two mutations are required for metastatic tumor invasion is that one (such as rasact) is required to promote cell proliferation and inhibit apoptosis; the other (such as scribble) is required to promote MMP-mediated invasion [40].
Figure 4. A Molecular Mechanism for Drosophila Tumor Invasion.
Mmp1 is regulated by the JNK pathway, which is in turn repressed by the polarity gene scribble. In scribble rasact clones, invasiveness and proliferation are activated while apoptosis is downregulated. The JNK pathway also promotes apoptosis (not shown), although rasact epistatically inactivates apoptosis in scribble rasact clones. Model based on [19, 40, 41, 69].
The tumor transplantation experiments demonstrate that both the tumor and the surrounding host tissue contribute to the metastatic potential of the tumor. The role of host tissue can also be assessed in the in situ tumor model by examining clones of different sizes where the ratio of mutant and wild-type tissue is altered; or by examining cells that are close to the clone border. When the Src-inhibitor csk is knocked down throughout the tissue of a fly eye or wing leaving little wild-type tissue, the organ overgrows because of excessive cell proliferation and insufficient apoptosis [43]. However, when small clones that knock down csk are induced in epithelia, the csk cells invade through the basement membrane rather than divide. Interestingly, this invasion appears to require Mmp2, as either reducing the amount of Mmp2 by half (in a heterozygote) or overexpressing Timp eliminates the cell invasive behavior [43]. Mmp1 has a similar function in promoting invasion in csk clones (Vidal and Cagan, personal communication). csk invasive cells are found at the boundary of the csk clone, within 3–4 cell diameters from the edge, indicating that the interplay of wild-type and mutant tissue causes the MMP-dependent invasive behavior. The invasive cells eventually die by apoptosis [43].
Clearly MMPs are required to mediate tumor invasion in Drosophila. Two different models of metastasis highlight the importance of MMPs: transplanted tumors and in situ clonal tumors. Invasive tissue with four distinct genetic origins (lgl, brat, scrib−rasact, and csk) all utilize MMPs to mediate invasion. Mammalian MMPs contribute to tumor progression at many stages, and they contribute to late-stage metastasis by promoting invasion, migration, and adhesive changes [44]. The fly results highlight the essential and conserved functions of MMPs in promoting metastasis, probably in part by directly degrading basement membrane. More importantly, because of the genetic tractability of Drosophila, these studies have unambiguously identified the JNK pathway as a regulatory pathway functioning in vivo, in tumors, to upregulate Mmp1 expression. Although it was clear the JNK pathway regulates MMPs in some cultured cells [45–49], the significance of this regulation was unclear in tumors. Further insight into the in vivo regulation of MMPs is certain to come from future studies. MMPs are considered potentially important but difficult pharmaceutical targets for cancer therapies [50, 51], and understanding the genetic regulation of MMPs may provide additional paradigms for therapeutic inhibition.
Fly and Worm MMPs contribute to developmental tissue invasion
The invasiveness conferred by MMPs to tumor cells is similar to the normal function of the MMPs during Drosophila metamorphosis. Both MMPs are required during metamorphosis, as weak (hypomorphic) mutants for each gene display defects in the adult body plan (Figure 2). The adult head normally develops inside the body cavity and then everts midway through metamorphosis; weak Mmp1 mutants do not evert their heads, giving a headless or cryptocephalic phenotype [9]. Additionally, both Mmp1 and Mmp2 mutants display clefts in their notum (back of the thorax), and this defect, like the headless phenotype, is caused by a failure of disc eversion [9, 19]. Normally the imaginal discs, which develop inside the larva, evert and fuse to form the adult body at metamorphosis; during this process, the disc epithelium invades the larval epithelium requiring the degradation of two layers of basement membrane. In the weak MMP mutants, or animals engineered to misexpress Timp, the wing discs forming the notum do not evert, and the collagen IV-containing basement membrane is not degraded, resulting in clefted notums [19]. Similar phenotypes are observed in mutants of the JNK pathway [52]. In normal metamorphic development, as in tumor progression, Mmp1 is regulated by the JNK pathway: Mmp1 expression is ablated in JNK-pathway inactivating mutants, and ectopic activation of the JNK pathway by activating mutations is sufficient to induce ectopic basement membrane degradation and Mmp1 expression [19]. Basement membrane degradation is inhibited by Timp in this ectopic activation system, indicating that MMPs are likely responsible for its degradation [19]. Thus Mmp1 is activated by the JNK pathway normally during metamorphosis, pathologically during tumor invasion, and ectopically.
An MMP contributes to a developmentally important cell invasion event in the C. elegans nematode worm, and this invasion has molecular parallels to developmental invasion in flies. In worms, the anchor cell is a specialized uterine cell that invades into vulval epithelium. Initially, uterine and vulval cells develop independently, but a connection between them is essential for later egg-laying, and this connection is initiated by anchor cell invasion [53]. The anchor cell expresses the AP-1 transcription factor fos-1. fos-1 mutants cannot break down the basement membranes separating the epithelial layers, even though the mutant anchor cell processes display normal morphology. This phenotype demonstrates that fos-1 is required for local destruction of basement membrane that enables cell invasion. In vivo mutant analysis demonstrates that fos-1 is required for the expression of at least three different transcriptional targets: zmp-1, a GPI-anchored MMP; cdh-3, a Fat-like protocadherin; and him-4, or Hemicentrin, a member of the fibulin family of ECM proteins. At a subcellular level, all three proteins localize to the invasive membrane domain. To test the function of these fos-1 targets, anchor cell invasion was assayed in mutants carrying a null allele of each gene. None of the single mutants was able to recapitulate the defect in anchor cell invasion. However, triple mutants defective for all three genes showed delayed invasion in 25% of the animals, indicating that these fos-1 transcriptional targets function together to promote anchor cell invasion [54]. zmp-1 expression in the anchor cell is also dependent on the transcriptional repressor EGL-43, an ortholog of the EVI1 oncogene, which functions downstream of fos-1 and upstream of zmp-1 [55].
Thus in Drosophila metamorphic invasion and in C. elegans anchor cell invasion, MMPs play important roles in breaching the intervening basement membranes. Not all developmentally regulated invasion events require MMP function, however, as Drosophila border cell migration, and the basement membrane breakdown that precedes it, is not affected by misexpression of Timp or either MMP [56, 57]. It is interesting that in flies, MMP-mediated invasion is under control of the JNK-pathway, which works through AP-1 transcription factors; and in worms, MMP-mediated invasion is regulated by fos, an AP-1 component. It is likely that in both systems the JNK pathway is regulating MMP expression for its function in basement membrane invasion. Although MMPs were known to be regulated by the JNK pathway in mammalian cell culture, invertebrate studies have established the relevance of this regulation during developmental tissue invasion.
What proteins interact with Drosophila MMPs?
Drosophila and other invertebrate MMPs have been examined mostly in vivo through mutant analysis, and little work has been done in vitro or in cell culture to identify potential endogenous substrates. The genetic interactions between Mmp2 and a semaphorin indicates that these genes have opposing functions in axon guidance (above), but there is no evidence of a physical interaction [27]. Mmp1 degrades basement membrane as assessed by collagen IV staining in whole-mount tissues (above), but the precise targets for this degradation are unknown [19]. Two-hybrid screening with the hemopexin domain of Mmp1 has identified 7 interacting gene fragments, none of which encode the expected ECM proteins or known signaling molecules [58]. One of these two hybrid candidates encodes the protein Ninjurin A, a transmembrane protein named for its upregulation in injured nerves in rats [59]. Ninjurins are conserved two-pass transmembrane proteins, previously known to play a role in cell adhesion [59, 60].
Mmp1 and Ninjurin A colocalize in fly tissues, and microarrays indicate they are both subject to regulation by the Jun N-terminal Kinase (JNK) pathway [58, 61]. My lab has shown that in cultured insect cells, Mmp1 is responsible for activating the Ninjurin A signaling molecule by liberating it from the cell surface. Cells expressing both Drosophila Mmp1 and Ninjurin A condition their medium with an activity that causes these semi-adherent cells to release adhesion from their substratum. This conditioned medium is capable of inducing even wild-type cells to lose adhesion; conversely, when cells expressing both Ninjurin A and Mmp1 are washed in fresh media, they revert to adhesive behavior. Although the Ninjurin A-induced loss-of-adhesion phenotype requires functional Mmp1, when Ninjurin A is engineered so that its ectodomain is constitutively secreted from cells then Mmp1 function is no longer required. This data indicates that Mmp1 liberates the Ninjurin A ectodomain, perhaps via cleavage. Endogenous Drosophila Mmp1 and Ninjurin A co-immunoprecipitate from lysates of whole animals suggesting that if Mmp1 does cleave Ninjurin A, the cleavage is regulated rather than constitutive. The liberated Ninjurin A ectodomain acts as a signaling molecule, as it conveys information to wild-type cells instructing them to lose adhesion, indicating new roles for both MMPs and Ninjurins.
An MMP in Hydra regeneration
The small freshwater Hydra is one of the best model systems for understanding regeneration (see [62] for an excellent review). Hydra can regenerate its entire body from any body fragment so long as the piece contains more than a few hundred cells – representing only about 1/50th of its final size [63]. In Hydra, regeneration occurs in the absence of cell proliferation (termed morphallactic regeneration), so that existing cells and structures are reprogrammed to take on new identities. Thus regeneration in Hydra represents a striking example of tissue remodeling.
The Hydra MMP (HMMP) is required for regeneration: animals cannot regenerate either a head or a foot after amputation when HMMP is inactivated. HMMP inactivation was achieved by pharmacological inhibition with the broad-spectrum MMP inhibitor GM6001 and by using antisense technology [6, 7]. In the course of normal regeneration, ECM at the site of amputation is first lost and then rebuilt [7], and recombinant HMMP is able to cleave Hydra ECM in vitro [6]. It seems likely that HMMP is involved directly in reshaping the ECM at sites of regeneration. Interestingly, vertebrate MMPs are upregulated in newt limb regeneration, suggesting that they may have parallel functions in these two model systems of regeneration [64].
The expression pattern of HMMP suggests an additional function. As expected from its function in regeneration, HMMP is upregulated at sites of amputation. However, it is also expressed at high levels in the endoderm of the tentacles and foot near the poles of the animal [6]. What is the significance of that expression? Hydra has an unusual way of growing in that stem cells along the body column continually divide; this constant proliferation causes cells to migrate continually poleward, to the foot or the tentacles, where they transdifferentiate into specialized epithelial cells for those regions [62]. HMMP expression coincides with these areas of transdifferentiated specialized cells. When HMMP is inhibited in the foot region, the population of specialized basal disk cells is lost [6]. Thus HMMP appears to maintain the basal disk cell population, either by inhibiting their de-differentiation or by promoting their transdifferentiation from the body wall cells.
At this writing the genome of Hydra magnipapillata is not completed, but results from the EST project demonstrate that there are at least six MMPs in the Hydra genome and perhaps more. Functional information about the other Hydra MMPs should be a priority, especially as the advent of RNAi increases the feasibility of such studies.
The sea urchin genome contains many MMPs
The genome of the purple sea urchin Strongylocentrotus purpuratus was sequenced recently and found to contain at least 26 predicted MMP genes [8] (Table 1). Like the Drosophila MMPs, it is not possible to assign orthologs among the vertebrate MMPs and the sea urchin MMPs, as they appear to have duplicated independently [8]. At least nine membrane-type MMP (MT-MMP) family members are predicted [8], comparable to four MT-MMPs plus three GPI-anchored MMPs in vertebrates [1]. Of the 26 MMPs, 19 are expressed in embryonic stages of development [65]. Surprisingly, 10 TIMP genes are predicted in the sea urchin genome, more than twice the number found in vertebrates [8].
Table 1.
MMPs in the Sequenced Genomes of Invertebrates
| Organism | Number of MMPs | Predicted Secreted MMPs | Predicted Membrane-Associated MMPs# |
|---|---|---|---|
| Drosophila melanogaster fruitfly [9, 10] | 2 | Dm Mmp1 (experimentally confirmed) | Dm Mmp2 (experimentally confirmed) |
| C. elegans nematode worm (source: MEROPS, GenBank, TMHMM and DGPI) | 6 | H36L18.1 | ZMP-1 |
| C31B8.8 | |||
| H19M22.3 | |||
| T21D11.1 | |||
| W09D12.1 | |||
| Strongylocentrotus purpuratus sea urchin [8] | ≥26 | Sp-HE | Sp-MT-MMP-a |
| Sp-HE2 | Sp-MT-MMP-b | ||
| Sp-MMP-a | Sp-MT-MMP-c | ||
| Sp-MMP-b | Sp-MT-MMP-d | ||
| Sp-MMP-c | Sp-MT-MMP-e | ||
| Sp-MMP-d | Sp-MT-MMP-f | ||
| Sp-MMP-e | Sp-MT_MMP-g | ||
| Sp-MMP-f | Sp-MT-MMP-h | ||
| Sp-MMP-g | Sp-MT_MMP-i | ||
| Sp-MMP-h | |||
| Sp-MMP-i | |||
| Sp-MMP-j | |||
| Sp-MMP-k | |||
| Sp-MMP-l | |||
| Sp-MMP-m | |||
| Sp-MMP-n | |||
| Sp-MMP-o | |||
includes both transmembrane (sea urchin) and GPI-anchored MMPs
Two of these new sea urchin MMPs are highly related to the hatching enzyme from the another sea urchin, Paracentrotus lividus. Hatching enzyme, or envelysin, was first purified based on its activity of degrading the fertilization envelope; this activity allows the growing embryo to emerge from the ECM of the maternal egg [66]. Hatching enzyme is one of the first zygotically expressed genes in the genome, and its spatial and temporal expression is remarkably specific, confined only to the animal two-thirds of the embryo during the time right before hatching, when it appears to be transported from cytoplasmic puncta to the apical membrane where it degrades the egg ECM [67, 68]. Perhaps the sea urchin has evolved many other such highly specific MMPs, which could help explain the large number of MMP genes in this invertebrate.
Conclusions
The invertebrate MMPs appear to have functions that are similar to the vertebrate MMPs, most of which center on tissue remodeling. Some of these findings are straightforward confirmations of MMP functional conservation, such as the finding that Drosophila Mmp2 is required for tissue histolysis; or that Drosophila Mmp1 mediates basement membrane degradation; or that Drosophila MMPs contribute to tumor invasiveness. In contrast, many of the insights about invertebrate MMPs modify or clarify our understanding of vertebrate MMP functions. Some examples of this are the requirement for MMPs in axon guidance, the requirement for MMPs in dendritic remodeling, the requirement for MMPs in tissue regeneration, and the in vivo regulation of MMPs by the JNK pathway.
The advantage of invertebrate systems is that they have complementary strengths to vertebrates and cell culture: embryos develop outside the mother and are experimentally accessible; Drosophila and C. elegans have powerful genetics; many model invertebrates have short generation times; Hydra is a simple model for regeneration; all offer opportunities to study gene function in vivo; all are inexpensive compared to mice. The first functional study of a nematode MMP in 2005 may open the door for many more [54]. Certainly, many Drosophila laboratories began investigating MMPs after we reported the fly MMP mutants four years ago [9]. For Hydra, the sequencing of the genome and the advent of transgenesis and RNAi will probably enable more studies in molecular mechanisms of regeneration, which we know involve an MMP. Although the body of work on MMPs in invertebrates is still fairly small, it is expanding rapidly. Future invertebrate studies will likely move beyond confirmations and validations—to inform us about entirely new and surprising MMP functions and mechanisms.
Acknowledgements
I am grateful to Heather Broihier, Ross Cagan, and Ajay Srivastiva for discussions and sharing unpublished data; to Andrew Ewald, Eleanor Marion Ewald, Heather Broihier, and members of my laboratory for critically reading the manuscript and assistance; and to Michelle Beaucher and Shuning Zhang for help with figures. Research in my laboratory is funded by NIH grant R01GM073883 and by a Basil O’Connor Starter Scholars Award from the March of Dimes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007 Aug;:221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maidment JM, Moore D, Murphy GP, Murphy G, Clark IM. Matrix Metalloproteinase Homologues from Arabidopsis thaliana: Expression and Activity. J. Biol. Chem. 1999 274;:34706–34710. doi: 10.1074/jbc.274.49.34706. [DOI] [PubMed] [Google Scholar]
- 3.Golldack D, Popova OV, Dietz KJ. Mutation of the Matrix Metalloproteinase At2-MMP Inhibits Growth and Causes Late Flowering and Early Senescence in Arabidopsis. J. Biol. Chem. 2002 277;:5541–5547. doi: 10.1074/jbc.M106197200. [DOI] [PubMed] [Google Scholar]
- 4.Coates D, Siviter R, Isaac RE. Exploring the Caenorhabditis elegans and Drosophila melanogaster genomes to understand neuropeptide and peptidase function. Biochem Soc Trans. 2000 28;:464–469. [PubMed] [Google Scholar]
- 5.Wada K, Sato H, Kinoh H, Kajita M, Yamamoto H, Seiki M. Cloning of three Caenorhabditis elegans genes potentially encoding novel matrix metalloproteinases. Gene. 1998 211;:57–62. doi: 10.1016/s0378-1119(98)00076-6. [DOI] [PubMed] [Google Scholar]
- 6.Leontovich A, Zhang J, Shimokawa K, Nagase H, Sarras M. A novel hydra matrix metalloproteinase (HMMP) functions in extracellular matrix degradation, morphogenesis and the maintenance of differentiated cells in the foot process. Development. 2000 127;:907–920. doi: 10.1242/dev.127.4.907. [DOI] [PubMed] [Google Scholar]
- 7.Shimizu H, Zhang X, Zhang J, Leontovich A, Fei K, Yan L, et al. Epithelial morphogenesis in hydra requires de novo expression of extracellular matrix components and matrix metalloproteinases. Development. 2002 129;:1521–1532. doi: 10.1242/dev.129.6.1521. [DOI] [PubMed] [Google Scholar]
- 8.Angerer L, Hussain S, Wei Z, Livingston BT. Sea urchin metalloproteases: a genomic survey of the BMP-1/tolloid-like, MMP and ADAM families. Dev Biol. 2006 300;:267–281. doi: 10.1016/j.ydbio.2006.07.046. [DOI] [PubMed] [Google Scholar]
- 9.Page-McCaw A, Serano J, Sante JM, Rubin GM. Drosophila matrix metalloproteinases are required for tissue remodeling, but not embryonic development. Dev Cell. 2003 Apr;:95–106. doi: 10.1016/s1534-5807(02)00400-8. [DOI] [PubMed] [Google Scholar]
- 10.Llano E, Adam G, Pendas AM, Quesada V, Sanchez LM, Santamaria I, et al. Structural and Enzymatic Characterization of Drosophila Dm2-MMP, a Membrane-bound Matrix Metalloproteinase with Tissue-specific Expression. J. Biol. Chem. 2002 277;:23321–23329. doi: 10.1074/jbc.M200121200. [DOI] [PubMed] [Google Scholar]
- 11.Llano E, Pendas AM, Aza-Blanc P, Kornberg TB, Lopez-Otin C. Dm1-MMP, a Matrix Metalloproteinase from Drosophila with a Potential Role in Extracellular Matrix Remodeling during Neural Development. J. Biol. Chem. 2000 275;:35978–35985. doi: 10.1074/jbc.M006045200. [DOI] [PubMed] [Google Scholar]
- 12.Wei S, Xie Z, Filenova E, Brew K. Drosophila TIMP is a potent inhibitor of MMPs and TACE: similarities in structure and function to TIMP-3. Biochemistry. 2003 42;:12200–12207. doi: 10.1021/bi035358x. [DOI] [PubMed] [Google Scholar]
- 13.Beitel GJ, Krasnow MA. Genetic control of epithelial tube size in the Drosophila tracheal system. Development. 2000 127;:3271–3282. doi: 10.1242/dev.127.15.3271. [DOI] [PubMed] [Google Scholar]
- 14.Beaucher M, Hersperger E, Page-McCaw A, Shearn A. Metastatic ability of Drosophila tumors depends on MMP activity. Dev Biol. 2007 303;:625–634. doi: 10.1016/j.ydbio.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Lee CY, Cooksey BA, Baehrecke EH. Steroid regulation of midgut cell death during Drosophila development. Dev Biol. 2002 250;:101–111. doi: 10.1006/dbio.2002.0784. [DOI] [PubMed] [Google Scholar]
- 16.Lee CY, Baehrecke EH. Steroid regulation of autophagic programmed cell death during development. Development. 2001 128;:1443–1455. doi: 10.1242/dev.128.8.1443. [DOI] [PubMed] [Google Scholar]
- 17.Gross J, Lapiere CM. Collagenolytic activity in amphibian tissues: a tissue culture assay. Proc Natl Acad Sci U S A. 1962 47;:1014–1022. doi: 10.1073/pnas.48.6.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee CY, Clough EA, Yellon P, Teslovich TM, Stephan DA, Baehrecke EH. Genome-wide analyses of steroid- and radiation-triggered programmed cell death in Drosophila. Curr Biol. 2003 13;:350–357. doi: 10.1016/s0960-9822(03)00085-x. [DOI] [PubMed] [Google Scholar]
- 19.Srivastava A, Pastor-Pareja JC, Igaki T, Pagliarini R, Xu T. Basement membrane remodeling is essential for Drosophila disc eversion and tumor invasion. Proc Natl Acad Sci U S A. 2007 104;:2721–2726. doi: 10.1073/pnas.0611666104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiger JA, Jr., Natzle JE, Kimbrell DA, Paddy MR, Kleinhesselink K, Green MM. Tissue remodeling during maturation of the Drosophila wing. Dev Biol. 2007 301;:178–191. doi: 10.1016/j.ydbio.2006.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godenschwege TA, Pohar N, Buchner S, Buchner E. Inflated wings, tissue autolysis and early death in tissue inhibitor of metalloproteinases mutants of Drosophila. Eur J Cell Biol. 2000 79;:495–501. doi: 10.1078/0171-9335-00072. [DOI] [PubMed] [Google Scholar]
- 22.Brower DL, Jaffe SM. Requirement for integrins during Drosophila wing development. Nature. 1989 342;:285–287. doi: 10.1038/342285a0. [DOI] [PubMed] [Google Scholar]
- 23.Pohar N, Godenschwege TA, Buchner E. Invertebrate tissue inhibitor of metalloproteinase: structure and nested gene organization within the synapsin locus is conserved from Drosophila to human. Genomics. 1999 57;:293–296. doi: 10.1006/geno.1999.5776. [DOI] [PubMed] [Google Scholar]
- 24.Rahkonen OP, Koskivirta IM, Oksjoki SM, Jokinen E, Vuorio EI. Characterization of the murine Timp4 gene, localization within intron 5 of the synapsin 2 gene and tissue distribution of the mRNA. Biochim Biophys Acta. 2002 1577;:45–52. doi: 10.1016/s0167-4781(02)00404-9. [DOI] [PubMed] [Google Scholar]
- 25.Yu WP, Brenner S, Venkatesh B. Duplication, degeneration and subfunctionalization of the nested synapsin-Timp genes in Fugu. Trends Genet. 2003 19;:180–183. doi: 10.1016/S0168-9525(03)00048-9. [DOI] [PubMed] [Google Scholar]
- 26.Kuo CT, Jan LY, Jan YN. Dendrite-specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin-proteasome, and ecdysone signaling. Proc Natl Acad Sci U S A. 2005 102;:15230–15235. doi: 10.1073/pnas.0507393102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozora-Miller CM, Page-McCaw A, Broihier HT. Matrix metalloproteinases promote motor axon fasciculation in the Drosophila embryo. 2007 doi: 10.1242/dev.011072. Submitted. [DOI] [PubMed] [Google Scholar]
- 28.McFarlane S. Metalloproteases: carving out a role in axon guidance. Neuron. 2003 37;:559–562. doi: 10.1016/s0896-6273(03)00089-8. [DOI] [PubMed] [Google Scholar]
- 29.Yu HH, Huang AS, Kolodkin AL. Semaphorin-1a acts in concert with the cell adhesion molecules fasciclin II and connectin to regulate axon fasciculation in Drosophila. Genetics. 2000 156;:723–731. doi: 10.1093/genetics/156.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alvarez VA, Sabatini BL. Anatomical and Physiological Plasticity of Dendritic Spines. Annu Rev Neurosci. 2007 doi: 10.1146/annurev.neuro.30.051606.094222. [DOI] [PubMed] [Google Scholar]
- 31.Kaczmarek L, Lapinska-Dzwonek J, Szymczak S. Matrix metalloproteinases in the adult brain physiology: a link between c-Fos, AP-1 and remodeling of neuronal connections? Embo J. 2002 21;:6643–6648. doi: 10.1093/emboj/cdf676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bozdagi O, Nagy V, Kwei KT, Huntley GW. In vivo roles for matrix metalloproteinase-9 in mature hippocampal synaptic physiology and plasticity. J Neurophysiol. 2007 doi: 10.1152/jn.00202.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brumby AM, Richardson HE. Using Drosophila melanogaster to map human cancer pathways. Nat Rev Cancer. 2005 May;:626–639. doi: 10.1038/nrc1671. [DOI] [PubMed] [Google Scholar]
- 34.Woodhouse E, Hersperger E, Stetler-Stevenson WG, Liotta LA, Shearn A. Increased type IV collagenase in lgl-induced invasive tumors of Drosophila. Cell. Growth Differ. 1994 May;:151–159. [PubMed] [Google Scholar]
- 35.Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993 117;:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- 36.Godbout R, Dryja TP, Squire J, Gallie BL, Phillips RA. Somatic inactivation of genes on chromosome 13 is a common event in retinoblastoma. Nature. 1983 304;:451–453. doi: 10.1038/304451a0. [DOI] [PubMed] [Google Scholar]
- 37.Pagliarini RA, Xu T. A genetic screen in Drosophila for metastatic behavior. Science. 2003 302;:1227–1231. doi: 10.1126/science.1088474. [DOI] [PubMed] [Google Scholar]
- 38.Bilder D, Li M, Perrimon N. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science. 2000 289;:113–116. doi: 10.1126/science.289.5476.113. [DOI] [PubMed] [Google Scholar]
- 39.Uhlirova M, Jasper H, Bohmann D. Non-cell-autonomous induction of tissue overgrowth by JNK/Ras cooperation in a Drosophila tumor model. Proc Natl Acad Sci U S A. 2005 102;:13123–13128. doi: 10.1073/pnas.0504170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uhlirova M, Bohmann D. JNK- and Fos-regulated Mmp1 expression cooperates with Ras to induce invasive tumors in Drosophila. Embo J. 2006 25;:5294–5304. doi: 10.1038/sj.emboj.7601401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brumby AM, Richardson HE. scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila. Embo J. 2003 22;:5769–5779. doi: 10.1093/emboj/cdg548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee T, Winter C, Marticke SS, Lee A, Luo L. Essential roles of Drosophila RhoA in the regulation of neuroblast proliferation and dendritic but not axonal morphogenesis. Neuron. 2000 25;:307–316. doi: 10.1016/s0896-6273(00)80896-x. [DOI] [PubMed] [Google Scholar]
- 43.Vidal M, Larson DE, Cagan RL. Csk-deficient boundary cells are eliminated from normal Drosophila epithelia by exclusion, migration, and apoptosis. Dev Cell. 2006 Oct;:33–44. doi: 10.1016/j.devcel.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 44.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nature Rev Cancer. 2002 Feb;:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 45.Simon C, Simon M, Vucelic G, Hicks MJ, Plinkert PK, Koitschev A, et al. The p38 SAPK pathway regulates the expression of the MMP-9 collagenase via AP-1-dependent promoter activation. Exp Cell Res. 2001 271;:344–355. doi: 10.1006/excr.2001.5374. [DOI] [PubMed] [Google Scholar]
- 46.Sugioka Y, Watanabe T, Inagaki Y, Kushida M, Niioka M, Endo H, et al. c-Jun NH2-terminal kinase pathway is involved in constitutive matrix metalloproteinase-1 expression in a hepatocellular carcinoma-derived cell line. Int J Cancer. 2004 109;:867–874. doi: 10.1002/ijc.20095. [DOI] [PubMed] [Google Scholar]
- 47.Gum R, Wang H, Lengyel E, Juarez J, Boyd D. Regulation of 92 kDa type IV collagenase expression by the jun aminoterminal kinase- and the extracellular signal-regulated kinase-dependent signaling cascades. Oncogene. 1997 14;:1481–1493. doi: 10.1038/sj.onc.1200973. [DOI] [PubMed] [Google Scholar]
- 48.Cheung LW, Leung PC, Wong AS. Gonadotropin-releasing hormone promotes ovarian cancer cell invasiveness through c-Jun NH2-terminal kinase-mediated activation of matrix metalloproteinase (MMP)-2 and MMP-9. Cancer Res. 2006 66;:10902–10910. doi: 10.1158/0008-5472.CAN-06-2217. [DOI] [PubMed] [Google Scholar]
- 49.Byun HJ, Hong IK, Kim E, Jin YJ, Jeoung DI, Hahn JH, et al. A splice variant of CD99 increases motility and MMP-9 expression of human breast cancer cells through the AKT-, ERK-, and JNK-dependent AP-1 activation signaling pathways. J Biol Chem. 2006 281;:34833–34847. doi: 10.1074/jbc.M605483200. [DOI] [PubMed] [Google Scholar]
- 50.Overall CM, Kleifeld O. Tumour microenvironment - opinion: validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat Rev Cancer. 2006 Jun;:227–239. doi: 10.1038/nrc1821. [DOI] [PubMed] [Google Scholar]
- 51.Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002 295;:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 52.Zeitlinger J, Bohmann D. Thorax closure in Drosophila: involvement of Fos and the JNK pathway. Development. 1999 126;:3947–3956. doi: 10.1242/dev.126.17.3947. [DOI] [PubMed] [Google Scholar]
- 53.Sherwood DR, Sternberg PW. Anchor cell invasion into the vulval epithelium in C. elegans. Dev Cell. 2003 May;:21–31. doi: 10.1016/s1534-5807(03)00168-0. [DOI] [PubMed] [Google Scholar]
- 54.Sherwood DR, Butler JA, Kramer JM, Sternberg PW. FOS-1 promotes basement-membrane removal during anchor-cell invasion in C. elegans. Cell. 2005 121;:951–962. doi: 10.1016/j.cell.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 55.Hwang BJ, Meruelo AD, Sternberg PW. C. elegans EVI1 proto-oncogene, EGL-43, is necessary for Notch-mediated cell fate specification and regulates cell invasion. Development. 2007 134;:669–679. doi: 10.1242/dev.02769. [DOI] [PubMed] [Google Scholar]
- 56.Medioni C, Noselli S. Dynamics of the basement membrane in invasive epithelial clusters in Drosophila. Development. 2005 132;:3069–3077. doi: 10.1242/dev.01886. [DOI] [PubMed] [Google Scholar]
- 57.Starz-Gaiano M, Montell DJ. Genes that drive invasion and migration in Drosophila. Curr Opin Genet Dev. 2004 14;:86–91. doi: 10.1016/j.gde.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 58.Zhang S, Dailey GM, Kwan E, Glasheen BM, Sroga GE, Page-McCaw A. An MMP liberates the Ninjurin A ectodomain to signal a loss of cell adhesion. Genes Dev. 2006 20;:1899–1910. doi: 10.1101/gad.1426906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Araki T, Milbrandt J. Ninjurin, a novel adhesion molecule, is induced by nerve injury and promotes axonal growth. Neuron. 1996 17;:353–361. doi: 10.1016/s0896-6273(00)80166-x. [DOI] [PubMed] [Google Scholar]
- 60.Araki T, Milbrandt J. Ninjurin2, a novel homophilic adhesion molecule, is expressed in mature sensory and enteric neurons and promotes neurite outgrowth. J Neurosci. 2000 20;:187–195. doi: 10.1523/JNEUROSCI.20-01-00187.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boutros M, Agaisse H, Perrimon N. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev Cell. 2002 Mar;:711–722. doi: 10.1016/s1534-5807(02)00325-8. [DOI] [PubMed] [Google Scholar]
- 62.Bosch TC. Why polyps regenerate and we don't: towards a cellular and molecular framework for Hydra regeneration. Dev Biol. 2007 303;:421–433. doi: 10.1016/j.ydbio.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 63.Shimizu H, Sawada Y, Sugiyama T. Minimum tissue size required for hydra regeneration. Dev Biol. 1993 155;:287–296. doi: 10.1006/dbio.1993.1028. [DOI] [PubMed] [Google Scholar]
- 64.Miyazaki K, Uchiyama K, Imokawa Y, Yoshizato K. Cloning and characterization of cDNAs for matrix metalloproteinases of regenerating newt limbs. Proc Natl Acad Sci U S A. 1996 93;:6819–6824. doi: 10.1073/pnas.93.13.6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Samanta MP, Tongprasit W, Istrail S, Cameron RA, Tu Q, Davidson EH, et al. The transcriptome of the sea urchin embryo. Science. 2006 314;:960–962. doi: 10.1126/science.1131898. [DOI] [PubMed] [Google Scholar]
- 66.Lepage T, Gache C. Purification and characterization of the sea urchin embryo hatching enzyme. J Biol Chem. 1989 264;:4787–4793. [PubMed] [Google Scholar]
- 67.Lepage T, Gache C. Early expression of a collagenase-like hatching enzyme gene in the sea urchin embryo. Embo J. 1990 Sep;:3003–3012. doi: 10.1002/j.1460-2075.1990.tb07493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lepage T, Sardet C, Gache C. Spatial expression of the hatching enzyme gene in the sea urchin embryo. Dev Biol. 1992 150;:23–32. doi: 10.1016/0012-1606(92)90004-z. [DOI] [PubMed] [Google Scholar]
- 69.Igaki T, Pagliarini RA, Xu T. Loss of cell polarity drives tumor growth and invasion through JNK activation in Drosophila. Curr Biol. 2006 16;:1139–1146. doi: 10.1016/j.cub.2006.04.042. [DOI] [PubMed] [Google Scholar]