Abstract
The pattern of microRNA (miRNA) expression is associated with the degree of tumor cell differentiation in human prostate cancer. MiRNAs bind complementarily to either oncogenes or tumor suppressor genes, which are consequently silenced, resulting in alterations of tumorigenecity. We have detected eight down-regulated and three up-regulated known miRNAs in androgen-independent human prostate cancer cells compared to those in androgen-dependent cells, using miRNA microarray analyses. These identified miRNAs showed the same expression patterns in hormone-refractory prostate carcinomas (HRPC) compared to androgen-sensitive noncancerous prostate epithelium as determined by fluorescent in situ hybridization assays in human prostate cancer tissue arrays. One of the eight down-regulated miRNAs, mir-146a, was selected and constitutively expressed to examine its effects on suppression of prostate cancer transformation from androgen-dependent to -independent cells as determined by in vitro tumorigenecity assays. Transfection of mir-146a, which perpetually express the miRNA, suppressed >82% of the expression of the targeted protein-coding gene, ROCK1, in androgen-independent PC3 cells, consequently markedly reducing cell proliferation, invasion, and metastasis to human bone marrow endothelial cell monolayers. Given that ROCK1 is one of the key kinases for the activation of hyaluronan (HA)-mediated HRPC transformation in vivo and in PC3 cells, mir-146a may function as a tumor-suppressor gene in modulating HA/ROCK1-mediated tumorigenecity in androgen-dependent prostate cancer.
Keywords: mir-146a, mir-184, microRNA (miRNA), hormone-refractory prostate cancer (HRPC), extracellular matrix (ECM), hyaluronan (HA), Rho-activated protein kinase (ROCK), microarray, tissue array
INTRODUCTION
Hormone-refractory prostate cancer (HRPC) is a leading cause of aging-related cancer death in men. Normal prostatic cell growth is controlled by androgens, whereas prostate cancer often occurs when this control is disturbed as one ages. The mainstay treatment of androgen-dependent prostate cancer is to remove androgen stimulation by surgery or hormonal therapy, which, although temporarily effective, eventually results in a deadly androgen-independent cancer transformation. To understand the mechanism by which this transformation mechanism takes place, several microarray studies were performed to profile expression patterns of specific genes and microRNAs (miRNAs) in relation to the progression of HRPC (Reis et al. 2004; Volinia et al. 2006; Porkka et al. 2007). As a result, there is a high correlation between the expression of miRNAs and the degree of tumor differentiation. However, the functional significance of these microarray-identified miRNAs has not been validated experimentally due to the lack of tissue-specific patterning and functional assays.
MiRNA is an abundant class of small noncoding RNAs, ∼17–25 nucleotides (nt) in length, capable of degrading mRNA or suppressing translation of the targeted genes with high sequence complementarity. These small RNAs are involved in suppression of oncogenes or tumor suppressor genes via complementary binding to their targeted gene transcripts, resulting in gene silencing and thus changes of cellular tumorigenecity in various cancers (Esquela-Kerscher and Slack 2006). Given that the progression of cancer is a multistep process involving changes of the expression of numerous oncogenes and tumor suppressor genes, it is difficult to define the function of a certain miRNA in modulating the tumorigenetic changes because most miRNAs have several to over 50 targets. Moreover, miRNAs bind the target genes with different affinity; some are stronger than others. Although current computer programs provide a means to predict the potential targets of a miRNA, as well as possible base pairs for binding between the miRNA and its target, the information based on the sequence complementarity is insufficient to pinpoint the exact functional interaction between the miRNA and its target(s). We present here a new approach for tissue- and stage-specific patterning of the microarray-identified miRNAs, which consists of two steps to ascertain the function: first, fluorescent in situ hybridization (FISH) in human prostate cancer tissue arrays in vivo; second, vector-based gain-of-function expression of the miRNA in human prostate cancer cells in vitro. This approach not only correlates the tissue-specific expression pattern of the miRNA to the stage of cancer, but also provides a functional assay for testing the role the identified miRNA plays in cancer progression.
Previous miRNA microarray studies have generated abundant information about differentially expressed miRNAs between HRPC and benign prostatic hyperplasia (BPH). However, the results varied probably due to limited patient samples used or the selection of cancer samples in different studies. We have recently analyzed differentially expressed miRNAs between human HRPC cell lines (i.e., LNCaP C4-2B and PC3) and their androgen-dependent counterparts (i.e., LNCaP and PC3-AR9) because these cell lines have high similarities to certain stages of human prostate cancers. Subsequently, a simple experimental procedure for correlating the in vitro identified miRNAs to their functions in vivo was established. We reported here that excessive expression of miRNA-146a was exclusively found in androgen-dependent LNCaP and PC3-AR9 cell lines, while the gain-of-function expression of miRNA-146a in androgen-independent PC3 cells markedly reduced the tumorigenecity as determined by cell proliferation, invasion, and metastasis to human bone marrow endothelial cell monolayers.
The miRNA family of mir-146a and mir-146b was first found in mouse, which shared ∼91% homology in a length of 22 nt (Lagos-Quintana et al. 2002). The identical sequences were also identified in human, indicating that this miRNA family might target the same genes conserved in humans (http://microrna.sanger.ac.uk). One of the major target genes of mir-146a, ROCK1, was recently reported to be highly involved in the transformation of HRPC and metastasis in vivo and in HRPC-derived PC3 cells (Lin et al. 2007). In >70% of advanced prostate cancer patients, hyaluronan (HA—an extracellular disaccharide matrix polymer) frequently bound to its receptors (CD168 and CD44) and then stimulated the activation of Rho-activated protein kinase (ROCK)-mediated signal transduction pathways. Active ROCK had two tumorigenetic functions. First, it increased myosin light chain (MLC) phosphorylation and actin/myosin-coupled contraction, which might enhance cancer migration and metastasis (Bourguignon et al. 2003; Somlyo et al. 2003). Second, HA-activated ROCK could phosphorylate its linker molecule, Gab1, and promoted the membrane localization of both Gab1 and CD168/CD44 for the activation of phosphatidylinositol-(3,4,5)P3 kinases (PI3K). This activation of PI3K further converted phosphatidylinositol (PtdIns)-(4,5)P2 to PtdIns-(3,4,5)P3 (IP3), subsequently leading to the activation of Akt/mTOR/eIF4E signal transduction (Lin et al. 2007). Activation of the Akt/mTOR/eIF4E signaling has been often reported to increase malignant transformation and drug resistance in HRPC (Rodriguez-Viciana et al. 1996; De Benedetti and Graff 2004). Thus, we examined the correlation between loss of mir-146a expression and elevation of HA/ROCK-associated tumorigenecity in the HRPC-derived PC3 cells. This report is the first observation of the mechanism by which mir-146a, as a tumor suppressor, modulates the transformation of prostate cancer.
RESULTS
Identification of differentially expressed miRNAs in HRPC-derived versus androgen-dependent human prostate cancer cell lines
We have consistently detected the same eight down-regulated and three up-regulated known miRNAs in androgen-independent human prostate cancer cell lines, including LNCaP-C81, LNCaP C4-2B, and PC3, as compared to those in androgen-dependent cell lines, such as LNCaP and PC3-AR9, using the miRNA microarray analysis (Fig. 1). These cell lines have different androgen responsiveness, which reflects in the mechanism by which androgen receptor (AR) mediates the tumorigenecity in androgen-dependent cancer but not in HRPC. For instance, a mutant-type AR, T877A, was highly expressed in LNCaP cells, which showed a several-fold higher sensitivity to androgen stimulation than that of the wild-type AR (Grossmann et al. 2001). Conversely, C4-2B cells, derived from LNCaP cells, possessed the same T877A AR but lost some downstream components of the AR signaling pathway and consequently became androgen independent (Grossmann et al. 2001). PC3 cells, a type of HRPC cells, possessed all the AR signaling components except AR itself (Litvinov et al. 2006). When the wild-type AR was transgenically expressed in PC3 cells (i.e., PC3-AR9 cell line), almost all AR responses to androgen stimulation were restored and resulted in about sixfold less HA retaining on the cell surface (Litvinov et al. 2006).
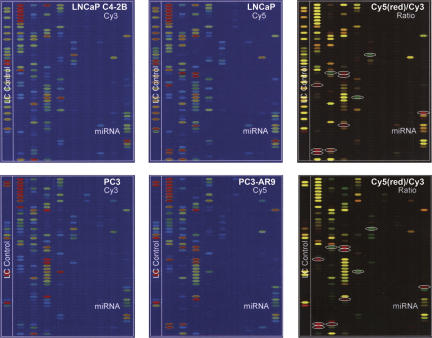
FIGURE 1.
MicroRNA microarray analyses of androgen-independent human prostate cancer cell lines, such as LNCaP C4-2B and PC3 (labeled by Cy3), compared to androgen-dependent cell lines, such as LNCaP and PC3-AR9 (labeled by Cy5), respectively (P <0.01, n = 3). Comparison of differentially expressed miRNAs in LNCaP versus C4-2B cells (upper panels) and PC3-AR9 versus PC3 cells (lower panels) shows 11 consistent miRNA alterations (white circles in the right two panels), including up-regulated mir-184, mir-361, and mir-424 (green dots) and down-regulated mir-19b, mir-29b, mir-128b, mir-146a, mir-146b, mir-221, mir-222, and mir-663 (red dots). Each of the four blue panels (the left and middle columns) represented the individual miRNA expression pattern in each cell line, showing miRNA expression from low abundant (blue–green) to high abundant (red) levels.
When these four cell lines were compared using miRNA microarray analyses, mir-184, mir-361, and mir-424 were significantly up-regulated, and mir-19b, mir-29b, mir-128b, mir-146a, mir-146b, mir-221, mir-222, and mir-663 were down-regulated in HRPC-related LNCaP C4-2B and PC3 cells as compared to androgen-dependent LNCaP and PC3-AR9 cells. Only mir-184 has been proposed to possess oncogenic activities in prostate cancer (Jagla et al. 2007). A recent study demonstrated that an aberrant splicing variant of androgen receptor, AR23, contained 69 nt of the intron 2 sequence, which separated the two AR zinc fingers required for nuclear entry (Jagla et al. 2007). It was suggested that the expression of mir-184 might silence its targeted splicing factor 1 (SF1) gene and cause aberrant splicing of the ARs, which prevents it from entering the nucleus. Consequently, the AR variants were not responsive to dihydrotestosterone (DHT) stimulation and became androgen insensitive with cancer transformation and elevated tumorigenecity. For these reasons, we examined tissue-specific expression patterns of mir-184 and mir-146a (probably mir-146b as well) in human prostate cancer tissue arrays to correlate miRNA expression to the progression of prostate cancer in vivo.
Confirmation of microarray-identified miRNA expressions in human prostate cancer tissue arrays in vivo
Prostatic carcinoma often shows a heterogeneous and multifocal incidence with diverse clinical and morphologic manifestations. Knowledge of the molecular basis for such heterogeneity is limited. Prostate cancer tissue samples were divided into four groups based on the Gleason scores and metastasis status: noncancerous prostatic tissues and prostate carcinomas with Gleason scores of 5–6, androgen-independent 7–8, and metastatic 9–10. The last group was selected from HRPC patients. Based on this pathological category, we correlated the expression patterns of two microarray-identified miRNAs, mir-184 and mir-146a, with the stages of prostate cancer progression in vivo, using FISH in human prostate cancer tissue arrays containing 60 patient samples.
Both mir-184 and loss of mir-146a were detected in high-grade HRPC tissues but not in androgen-sensitive noncancerous prostate epithelium, which was consistent with the miRNA microarray data (Fig. 2). The expression of mir-184 was weak in noncancerous tissues and intensified in advanced prostate cancers, particularly in metastatic HRPC, whereas mir-146a was expressed strongly in noncancerous prostatic epithelium but moderately in stromal cells, which gradually disappeared with cancer progression. Over 75% of the FISH-stained tissue array samples corresponded perfectly to the miRNA microarray-identified results, indicating that the concurrent increase of mir-184 and loss of mir-146a expression in HRPC was a prevalent incidence. It is possible that these specific miRNA profiles can be used as cancer stage signatures for predicting the progression of prostate cancer.
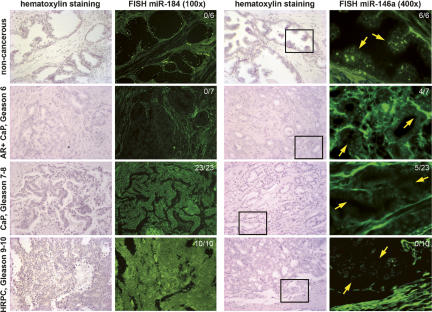
FIGURE 2.
Fluorescent in situ hybridization analyses of mir-184 and mir-146a expression in human prostate cancer (CaP) tissue arrays in vivo. In view of prostatic epithelium, mir-184 expression was gradually increased consistent with cancer progression, while mir-146a expression (yellow arrows) was greatly diminished in most of the advanced prostatic carcinomas with Gleason scores over 7. Ratios (upper right corner) showed the number of positive samples versus the total patient numbers from each stage of prostate cancer. Simultaneous elevation of mir-184 and loss of mir-146a expression was observed in >75% of advanced prostate caner epithelium (P <0.001; n = 4), suggesting the significance of miRNA-mediated cancer transformation in vivo.
Mir-146a-mediated silencing of ROCK1 oncogene and inhibition of HA/ROCK1-mediated cell proliferation
The effect of mir-146a on cancer cell growth was examined with a vector-based miRNA knock-in approach instead of antisense oligonucleotide-mediated miRNA knockout methods to prevent any potential cytotoxicity and transfection inconsistency. This miRNA knock-in approach used the Pol-II-mediated intronic miRNA expression system, which had been successfully demonstrated in many vertebrate models with special RNAi activities (Lin et al. 2006, 2007). Several transgenic loss-of-gene-function zebrafish, chicken, and mice have been established for studying various human diseases and neuropathological disorders, using this intronic miRNA expression system (Lin et al. 2006). The intronic miRNA biogenesis was regulated by intracellular Pol-II transcription, RNA splicing, and nonsense-mediated RNA decay (NMD) mechanisms; therefore, gene silencing achieved with this system was more specific, controllable, and effective than that with antisense oligonucleotide (Lin and Ying 2008). Given that loss of mir-146a was shown in PC3 cells that possessed very high HA/ROCK-mediated HRPC tumorigenecity (Lin et al. 2007), the gain-of-function assay of mir-146a was conducted in PC3 cells. The vector-based transgenic expression of the native mir-146a precursor significantly knocked down ROCK1 expression (> 82% ± 3%) in PC3 cells, while other off-target genes, such as integrin β1 (ITGb1) and GAPDH, were not affected (Fig. 3A). In addition, we showed that ITGb1 was an effective target for mir-507 rather than mir-146a. Based on the database of the miRBase∷Sequences program (http://microrna.sanger.ac.uk), mature mir-146a could bind to two highly complementary target sequences located in nucleotides 678–699 and 1504–1525 of the ROCK1 gene transcript, indicating that ROCK1 mRNA was a strong target for mir-146a. Our results validated that mir-146a triggered a strong and specific silencing effect on the targeted ROCK1 gene as predicted.
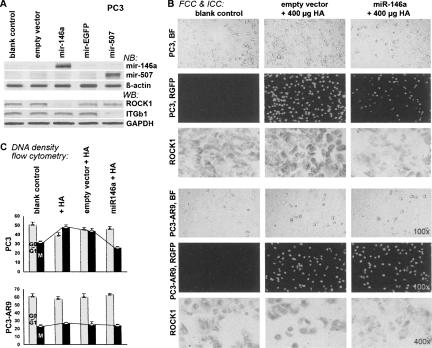
FIGURE 3.
Mir-146a-mediated ROCK suppression in androgen-independent human prostate cancer PC3 cells. (A) Northern blotting (NB) of mir-146a and Western blotting (WB) of ROCK1 were concurrently performed. Transfection of mir-146a suppressed >82% of ROCK1 expression, while other off-target genes, such as integrin β1 (ITGb1) and GAPDH, were not affected (P <0.001; n = 4). (B) Immunocytochemical staining (ICC) of ROCK1 protein in fluorescent-labeled PC3-mir146a cells (FCC). Mir-146a-transfected PC3 cells coexpressed a fluorescent marker protein, RGFP (middle). The ROCK1 protein was markedly reduced in RGFP-positive PC3-mir146a cells, while the PC3 cells transfected with an empty vector showed elevated levels of active ROCK1 after HA stimulation. Cell proliferation rates were changed in response to these ROCK1 alterations, which displayed a much faster cell growth in HA-stimulated PC3 cells than those in PC3-mir146a cells, as shown in C. No significant change was found in PC3-AR9 cells with the same treatments. (C) Flow cytometry analysis of HA-stimulated cell proliferation (P <0.05; n = 4). Bar charts indicated the ratio of different cell populations (x axis) versus different treatments (y axis), including untreated control cells (Ctl), cells treated with 400 μg/mL HA (+ HA), cells treated with empty vector and then HA (empty vector+HA), and cells treated with mir-146a-expressing vector and then HA (mir-146a+HA). The white bar refers to the cell population resting in the G0/G1 nondividing phase, whereas the black bar represents the mitotic (M) cell population.
Cell counting and DNA-density flow cytometry analyses were carried out to determine the cell proliferation rates and mitotic cell populations in mir-146a-mediated, ROCK1-depleted PC3 cells, namely PC3-mir146a. Figure 3B showed that PC3-mir146a cells grew much slower than the original PC3 cells with or without empty vector transfection, even in the presence of HA stimulation. The rate of cell number duplication in PC3-mir146a was very similar to that of PC3-AR9 cells; however, the growth rate of PC3-AR9 cells was not affected by the mir-146a transfection. Since abundant mir-146a expression was observed in PC3-AR9 cells as determined by miRNA microarray analyses, it is highly possible that mir-146a plays an important role in regulating PC3 cell growth. Further, HA-stimulated cell proliferation in PC3-mir146a cells showed a 50% reduction, whereas PC3 cells with empty-vector transfection had only 6% reduction as determined by cell-cycle analyses using DNA-density flow cytometry (Fig. 3C). In the presence of HA, the mitotic cell population (the M phase) decreased from 48.5% ± 3.1% to 24.4% ± 1.6% (50% loss) in PC3 cells, whereas that of PC3-AR9 cells was not changed (23.7% ± 1.3% to 21.7% ± 1.7%), suggesting that mir-146a plays an inhibitory role in PC3 cell growth via silencing ROCK1.
Tumor invasion and metastasis assays in PC3-mir146a cells
The tumor-suppressor function of mir-146a in prostate cancer invasion and metastasis was ascertained by examining matrigel cell invasion and PC3-mir146a cell adhesion to human bone marrow endothelial cell (hBMEC) monolayers, respectively. HA stimulation elevated the invasive cell population in PC3 cells from 6.7% ± 1.0% to 15.4% ± 1.1% (130% enhancement), whereas ROCK1-depleted PC3-mir146a cells treated with HA showed an inhibitory effect (34% reduction) in matrigel cell invasion assays (Fig. 4A). We have previously demonstrated that HA markedly promoted the invasiveness and metastasis of HRPC-derived PC3 cells mainly through the CD168–ROCK1 signaling pathway; thus, blockade of ROCK1 function in PC3-mir146a cells prevented the tumorigenecity of PC3. Figure 4B indicated that a significant increase of PC3 cell adhesion to hBMEC was detected after HA stimulation, whereas PC3-mir146a showed no response to the same HA treatment. Over 29% of HA-stimulated PC3 cells were adhered to hBMEC as compared to 17% in blank controls (70% enhancement) within 50 min of HA stimulation. In contrast, PC3-mir146a cells responded to HA stimulation with only 7% adhesion to hBMEC, which was a 59% reduction from the blank controls. Interestingly, the mir-146a-expressing PC3-AR9 cells showed neither invasiveness nor metastasis activities, suggesting that mir-146a may be responsible for preventing the HA/ROCK-mediated invasiveness and metastasis in androgen-dependent prostate cancer cells. Therefore, loss of mir-146a regulation in androgen-independent PC3 cells indeed caused HRPC-associated cancer invasiveness and metastasis.
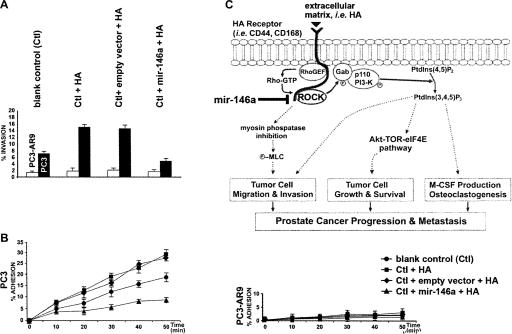
FIGURE 4.
Effects of mir-146a-suppressed ROCK expression on HA-dependent cancer cell invasion and metastasis to hBMEC in androgen-independent PC3 and androgen-dependent PC3-AR9 cells. (A) Functional analysis of mir-146a-suppressed tumor invasion in matrigel chambers (P <0.05; n = 4). Black and white bars referred to PC3 and PC3-AR9 cells, respectively. In the presence of high HA density, a significant increase of tumor invasion was detected in PC3 cells (Ctl+HA), whereas mir-146a-mediated ROCK1 suppression in PC3-mir146a cells (Ctl+mir-146a+HA) completely reversed this tumorigenetic effect. Transfection of empty vector in PC3 cells (Ctl+empty vector+HA) did not provide the suppression effect on cancer cell invasion. No significant change was observed in PC3-AR9 cells with the treatment. (B) Comparison of cell adhesion to the hBMEC monolayer between PC3 and PC3-AR9 cells (P <0.05; n = 6). The x and y axes refer to time duration after treatments and cell population, respectively. Within 50 min, >29% of PC3 cell population adhered to the hBMEC monolayer after HA treatment (square mark) as compared with a 17% adhesion rate in PC3 cells without any treatment (circle mark). Mir-146a-mediated ROCK1 suppression in PC3-mir146a cells (triangle mark) greatly reduced the HA-stimulated cell adhesion rate from 29% to 7% (76% reduction). PC3-AR9 cells responded weakly to all treatments and barely showed any adhesion to hBMEC (right chart). (C) Schematic mechanisms by which mir-146a is involved in the HA-mediated HRPC transformation, including cancer cell proliferation (anti-apoptosis), invasion (migration), and metastasis to bone marrow tissues. In all three pathways, mir-146a, as a tumor suppressor gene, directly suppresses ROCK1 oncogene expression to prevent the tumorigenetic effects of HA-mediated ROCK signaling in prostate cancer.
DISCUSSION
While the oncogenic mechanism of mir-184 has been recently proposed (Jagla et al. 2007), the tumor-suppressor role of mir-146a was unclear. Our results provided the first insight into the function of mir-146a (and probably mir-146b) in regulating prostate cancer progression. ROCK1 was one of the predicted mir-146a target genes, which has been identified to be highly involved in HA-mediated HRPC transformation and metastasis in vivo (Lin et al. 2007). Figure 4C shows three oncogenic mechanisms for the HA-stimulated ROCK1 signaling in prostate cancer (Lin et al. 2007), including direct MLC phosphorylation to increase cell migration and invasion (route 1), activation of the PI3K-mediated Akt/TOR/eIF4E signaling pathway that supports cancer cell proliferation and anti-apoptosis (route 2), and augmentation of M-CSF cytokine production to facilitate osteolytic metastasis (route 3). The elevated HA content in many solid tumors in vivo could be as high as >100 μg/mL (Paszek et al. 2005). Thus, in the presence of high HA density, mir-146a-mediated ROCK1 silencing may play an important role in suppressing HRPC-associated tumorigenecity, such as cancer cell proliferation (anti-apoptosis), invasion, and metastasis.
Our previous findings have established that the HA-stimulated ROCK1 signaling pathway was responsible for >70% of HRPC transformation in vivo and in androgen-independent PC3 cells, resulting in dramatic elevation of androgen-independent cancer cell proliferation, invasion, and metastasis. The present study provided more evidence in the deregulation of HA-stimulated ROCK1 signaling by loss of mir-146a function in androgen-independent prostate cancer cells. The loss of mir-146a frequently occurred in androgen-independent PC3 and LNCaP C4-2B cell lines as well as in >75% of advanced and metastatic prostate carcinomas in vivo. Transfection of mir-146a into PC3 cells restored the mir-146a-mediated ROCK1 gene silencing and thus significantly decreased the HA/ROCK-dependent tumorigenecity, in terms of markedly reduced cancer cell proliferation, invasion, and adhesion to hBMEC monolayers. These findings indicated a tumor suppressor role of mir-146a in preventing HA/ROCK-mediated cancer transformation in human prostate epithelium. Further, more identified miRNAs were concurrently and consistently involved in the processes of prostate cancer transformation. Thus, a thorough investigation of all microarray-identified miRNA functions might be crucial for a better understanding of the miRNA-mediated mechanisms of HRPC transformation.
MATERIALS AND METHODS
Cell culture and treatments
Human prostate cancer cell lines, LNCaP, and PC3 cells were obtained from the American Type Culture Collection, and C4-2B and PC3-AR9 cell lines were kindly provided by Dr. Gerhard Coetzee at the University of Southern California and Dr. Chawnshang Chang at the University of Rochester Medical Center, respectively. All cell lines were grown in phenol red-free DMEM with 10% charcoal-stripped FBS and 1% gentamycin as reported previously (Lin et al. 2007). In addition, a mir-146a-mediated, ROCK1-depleted PC3 cell line, namely, PC3-mir146a, was generated using a cytomegalovirus (CMV) promoter-driven intronic miRNA expression vector system as reported previously (Lin et al. 2003; Lin and Ying 2005). To inhibit ROCK1 expression in PC3 cells, a precursor has-mir-146a sequence, encoding 5′-CCGAUGUGUAUCCUCAGCUUUGAGAACUGAAUUCCAUGGGUUGUGUCAGUGUCAGACCUCUGAAAUUCAGUUCUUCAGCUGGGAUAUCUCUGUCAUCGU-3′, was transfectively expressed. This sequence targeted against the ROCK1 gene nucleotide 1504–1525 region (accession number NM005406), which contained a Rho-binding domain required for ROCK kinase activation. For miRNA knock-in assays, the designed vectors were liposomally encapsulated in a FuGENE reagent (Roche) and applied to cell cultures at 40% confluency. Positively transfected cells were isolated and collected 24 h later for subculturing, using flow cytometry selection with anti-RGFP monoclonal antibody (Clontech) as reported (Lin et al. 2007). The efficacy of ROCK1-knockdown was determined by Northern and Western blot analyses (Lin et al. 2007). For further HA stimulation, low molecular weight HA at 400 μg/mL was added to the cells at ∼40% confluency (Lin et al. 2007).
MicroRNA microarray analysis
Human prostate cancer cell lines, LNCaP, LNCaP C4-2B, PC3, and PC3-AR9, were prepared as described above. At 70% confluency, small RNAs from each cell line were isolated, using the mirVana miRNA isolation kit (Ambion, Inc.) following the manufacturer's suggestion. The purity and quantity of the isolated small RNAs were assessed using 1% formaldehyde-agarose gel electrophoresis and spectrophotometer measurement (Bio-Rad) and then submitted to LC Sciences for miRNA microarray analysis. Each microarray chip was a hybridized single sample labeled with either Cy3 or Cy5 or a pair of samples labeled with Cy3 and Cy5, respectively. Background subtraction and normalization were performed. For a dual sample assay, a P-value calculation was performed and a list of differentially expressed transcripts more than threefold was produced.
Fluorescent in situ hybridization in tissue arrays
Human prostate cancer tissue arrays were obtained from Imgenex. FISH kits were purchased from Ambion, Inc., and performed according to the manufacturer's suggestions. We used synthetic locked nucleic acid (LNA) probes (Sigma-Genosys) directed against the mature mir-184 and mir-146a sequences, respectively. The LNA-modified DNA oligonucleotide was a high-affinity RNA analog with a bicyclic furanose unit locked in an RNA-mimicking sugar conformation, which provided strong hybridization affinity toward complementary single-stranded RNA molecules. In a series of experiments, tissue arrays were dewaxed in xylene, rehydrated through an ethanol series (100%, 95%, 90%, 80%, 70%, 50%, 30%) and postfixed in 4% paraformaldehyde for 30 min. Then, the arrays were digested with proteinase K (10 μg/mL; Roche) for 5 min, refixed with 4% paraformaldehyde, and washed in Tris/glycine buffer. After that, the arrays were hybridized overnight at 60°C within cloverslip chambers in in situ hybridization buffer (40% formamide, 5× SSC, 1× Denhard's solution, 100 μg/mL salmon testis DNA, 100 μg/mL tRNA), containing 1 ng/μL of fluorescin-labeled LNA probes. After post-hybridization washes once with 5× SSC and once with 0.5× SSC at 25°C for 1 h, positive results were observed under a 100× microscope with whole field scanning and recorded at 100× and 400× magnification (Nikon 80i microscopic quantitation system).
Fluorescent cytochemical and immunocytochemical staining (FCC and ICC)
The successful transfection of the intronic miRNA expression vectors coexpressed a red fluorescent RGFP marker protein with the desired miRNA at a 1:1 ratio so that the RGFP could be observed directly under a fluorescent microscopic system (Lin et al. 2003; Lin and Ying 2005). For ICC, primary antibodies and immunohistochemical staining kits were obtained from Imgenex. The active form of ROCK proteins was immunostained as reported (Lin et al. 2007). The Thr-286 site of the ROCK kinase domain was usually masked by an inhibitory Cys/His-rich pleckstrin homology (PH) domain located in the carboxyl (C) terminus of inactive ROCK. Rho, a small GTPase bound and removed the PH domain of the inactive ROCK, allowing the detection of active ROCK proteins by the ROCK1(thr286) antibody. Since the ROCK1(thr286) antibody was prelabeled with biotin, alkaline phosphatase-conjugated sheep streptavidin-Fab fragment antibody (Roche) was used as the secondary antibody. Then, the bound antibody was detected with Fast Red staining (Roche) for 2 min. All positive results were examined under a 100× microscope with whole field scanning and measured at 400× magnification for quantitative analysis by a Metamorph Imaging program (Nikon 80i and TE2000 microscopic quantitation systems).
DNA-density flow cytometry
Cells were trypsinized, pelleted, and fixed by resuspension in 1 mL of prechilled 70% methanol in PBS for 1 h at −20°C. The cells were pelleted and washed once with 1 mL of PBS. The cells were pelleted again and resuspended in 1 mL of 1 mg/mL propidium iodide, 0.5 μg/mL RNase in PBS for 30 min at 37°C. Approximately 15,000 cells were then analyzed on a BD FACSCalibur. Cell doublets were excluded by plotting pulse width versus pulse area and gating on the single cells. The collected data were analyzed using the software package Flowjo using the “Watson Pragmatic” algorithm.
Tumor cell invasion assay
Chamber inserts (12-μm pore size; Chemicon) were coated with 200 μg/mL of matrigel alone or supplemented with 400 μg/mL of HA in phenol red-free-DMEM with 1% L-glutamine and dried overnight under sterile conditions. Cells were harvested, washed, and resuspended in phenol red-free-DMEM to give a final cell density of 1 × 106 cells/mL. Then, 500 μL of the resulting cell suspension were dispensed into the top chamber, whereas DMEM-conditioned medium (1.5 mL) was added to the bottom chamber to create a chemotactic gradient. Invasion was measured after overnight incubation at 37°C for 16 h. Top chambers were wiped with cotton wool, and invading cells on the underside of the membrane were fixed in 100% methanol for 10 min, air dried, stained in cresyl violet for 20 min, and gently rinsed in water. When dry, the cresyl violet stain on membranes was eluted using a 100% ethanol/0.2 M NaCitrate (1:1) wash for 20 min and absorbance read at 570 nm using a Precision Microplate Reader (Molecular Dynamics). The percentage of invading cells was calculated by comparison of absorbance in test samples against absorbance determined on membrane inserts that were not wiped (total cells).
Tumor cell adhesion assay
Cells were trypsinized, washed in adhesion media (RPMI 1640/0.1% BSA/20 mM HEPES at pH7.4) and sterile saline once, and resuspended at 1 × 106 cells/mL in PBS with 10 μM fura-4 acetoxymethyl ester (fluorescent probe; Sigma) for 1 h at 37°C in the dark. Cells were then pelleted, washed in serum-free medium containing 1% (v/v) of probenecid (100 mM), and incubated for 20 min in adhesion media at 37°C in the dark to activate the intracellular fluorescent probe. Cells (3 × 105cells/mL) were resuspended in adhesion medium and protected from the light until experimentation. Human bone marrow endothelial cells (hBMEC) were seeded at a density of 1 × 105 cells/mL in 96-well plates and washed with adhesion media before assays as reported (Lin et al. 2007). Cancer cells were added (300 μL cell suspension/well) to the confluent hBMEC monolayers and incubated for specific times up to 50 min at 37°C in the presence of 400 μg/mL HA. Nonadherent cells were removed using 2 × 250 μL washes of adhesion medium. Finally, plates were read in a fluorescent plate reader (Molecular Dynamics) at 37°C using an excitation wavelength of 485 nm and an emission wavelength of 530 nm.
Statistical analysis
Results were presented as mean±SE. Statistical analysis of the data was performed by one-way ANOVA. When main effects were significant, the Dunnett's post hoc test was used to identify the groups that differed significantly from the controls. For pairwise comparison between two treatment groups, the two-tailed Student's t-test was used. For experiments involving more than two treatment groups, an ANOVA was performed followed by a post hoc multiple range test. Probability values of P <0.05 were considered significant. All P values were determined from two-tailed tests.
ACKNOWLEDGMENTS
This study was supported by NIH/NCI Grant CA-85722, Wright Foundation Award #291, and Prostate Cancer Foundation Award 2006.
Footnotes
Abbreviations: miRNA, microRNA; HRPC, hormone-refractory prostate cancer; HA, hyaluronan; CD168 (RHAMM), receptor for hyaluronan-mediated motility; ROCK, Rho-activated protein kinase; PI3K, phosphatidylinositol-(3,4,5)P3 kinase; eIF4E, cap-dependent eukaryotic initiation factor 4E; FISH, fluorescent in situ hybridization; hBMEC, human bone marrow endothelial cell.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.874808.
REFERENCES
- Bourguignon, L.Y.W., Singleton, P.A., Zho, H., Diedrich, F. Hyaluronan-mediated CD44 interaction with RhoGEF and Rho kinase promotes Grb2-associated binder-1 phosphorylation and phosphatidylinositol 3-kinase signaling leading to cytokine (macrophage-colony stimulating factor) production and breast tumor progression. J. Biol. Chem. 2003;278:29420–29434. doi: 10.1074/jbc.M301885200. [DOI] [PubMed] [Google Scholar]
- De Benedetti, A., Graff, J.R. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23:3189–3199. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher, A., Slack, F.J. Oncomirs–MicroRNAs with a role in cancers. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Grossmann, M.E., Huang, H., Tindall, D.J. Androgen receptor signaling in androgen-refractory prostate cancer. J. Natl. Cancer Inst. 2001;93:1687–1697. doi: 10.1093/jnci/93.22.1687. [DOI] [PubMed] [Google Scholar]
- Jagla, M., Feve, M., Kessler, P., Lapouge, G., Erdmann, E., Serra, S., Bergerat, J.P., Ceraline, J. A splicing variant of the androgen receptor detected in a metastatic prostate cancer exhibits exclusively cytoplasmic actions. Endocrinology. 2007;148:4334–4343. doi: 10.1210/en.2007-0446. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., Tuschl, T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lin, S.L., Ying, S.Y. Gene silencing in vitro and in vivo using intronic microRNAs. In: Ying S.-Y., editor. MicroRNA protocols. Humana Press; Totowa, NJ: 2005. pp. 295–312. [Google Scholar]
- Lin, S.L., Ying, S.Y. Recent progress of the polymerase II-mediated intronic microRNA expression system. In: Gaur R.K., Rossi J., editors. Regulation of gene expression by small RNAs. CRC Press; Boca Raton, FL: 2008. [Google Scholar]
- Lin, S.L., Chang, D., Wu, D.Y., Ying, S.Y. A novel RNA splicing-mediated gene silencing mechanism potential for genome evolution. Biochem. Biophys. Res. Commun. 2003;310:754–760. doi: 10.1016/j.bbrc.2003.09.070. [DOI] [PubMed] [Google Scholar]
- Lin, S.L., Chang, S.J.E., Ying, S.Y. Transgene-like animal model using intronic microRNAs. Methods Mol. Biol. 2006;342:321–334. doi: 10.1385/1-59745-123-1:321. [DOI] [PubMed] [Google Scholar]
- Lin, S.L., Chang, D., Ying, S.Y. Hyaluronan stimulates transformation of androgen-independent prostate cancer. Carcinogenesis. 2007;28:310–320. doi: 10.1093/carcin/bgl134. [DOI] [PubMed] [Google Scholar]
- Litvinov, I.V., Antony, L., Dalrymple, R.B., Cheng, L., Isaacs, J.T. PC3, but not DU145, human prostate cancer cells retain the coregulators required for tumor suppressor ability of androgen receptor. Prostate. 2006;66:1329–1338. doi: 10.1002/pros.20483. [DOI] [PubMed] [Google Scholar]
- Paszek, M.J., Zahir, N., Johnson, K.R., Lakins, J.N., Rozenberg, G.I., Gefen, A., Reinhart-King, C.A., Margulies, S.S., Dembo, M., Boettiger, D., et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Porkka, K.P., Pfeiffer, M.J., Waltering, K.K., Vessella, R.L., Tammela, T.L., Visakorpi, T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- Reis, E.M., Nakaya, H.I., Louro, R., Canavez, F.C., Flatschart, A.V., Almeida, G.T., Egidio, C.M., Paquola, A.C., Machado, A.A., Festa, F., et al. Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer. Oncogene. 2004;23:6684–6692. doi: 10.1038/sj.onc.1207880. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana, P., Warne, P.H., Vanhaesbroeck, B., Waterfield, M.D., Downward, J. Activation of phosphoinositide 3-kinase by interaction with Ras and by point mutation. EMBO J. 1996;15:2442–2451. [PMC free article] [PubMed] [Google Scholar]
- Somlyo, A.V., Phelps, C., Dipierro, C., Eto, M., Read, P., Barrett, M., Gibson, J.J., Burnitz, M.C., Myers, C., Somlyo, A.P. Rho kinase and matrix metalloproteinase inhibitors cooperate to inhibit angiogenesis and growth of human prostate cancer xenotransplants. FASEB J. 2003;17:223–234. doi: 10.1096/fj.02-0655com. [DOI] [PubMed] [Google Scholar]
- Volinia, S., Calin, G.A., Liu, C.G., Ambs, S., Cimmino, A., Petrocca, F., Visone, R., Iorio, M., Roldo, C., Ferracin, M., et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]