Abstract
Phi29 DNA polymerase is a small DNA-dependent DNA polymerase that belongs to eukaryotic B-type DNA polymerases. Despite the small size, the polymerase is a multifunctional proofreading-proficient enzyme. It catalyzes two synthetic reactions (polymerization and deoxynucleotidylation of Phi29 terminal protein) and possesses two degradative activities (pyrophosphorolytic and 3′→5′ DNA exonucleolytic activities). Here we report that Phi29 DNA polymerase exonucleolyticaly degrades ssRNA. The RNase activity acts in a 3′ to 5′ polarity. Alanine replacements in conserved exonucleolytic site (D12A/D66A) inactivated RNase activity of the enzyme, suggesting that a single active site is responsible for cleavage of both substrates: DNA and RNA. However, the efficiency of RNA hydrolysis is ∼10-fold lower than for DNA. Phi29 DNA polymerase is widely used in rolling circle amplification (RCA) experiments. We demonstrate that exoribonuclease activity of the enzyme can be used for the target RNA conversion into a primer for RCA, thus expanding application potential of this multifunctional enzyme and opening new opportunities for RNA detection.
Keywords: Phi29 DNA polymerase, exoribonuclease, RNase, rolling circle amplification, RCA
INTRODUCTION
Phi29 DNA polymerase is a small (∼68 kDa) DNA-dependent DNA polymerase that belongs to eukaryotic B-type DNA polymerases (UniProtKB/TrEMBL: Q38545). Despite the small size, the polymerase is a multifunctional enzyme. It catalyzes two distinguishable synthetic reactions: polymerization reaction, characteristic for any other DNA-dependent polymerase, and Phi29 terminal protein (TP) deoxynucleotidylation (formation of covalent TP-dAMP complex) that enables initiation of DNA replication. In addition to catalysis of two synthetic reactions, Phi29 DNA polymerase also has two degradative activities: pyrophosphorolysis (the reverse reaction of polymerization) and DNA 3′→5′ exonucleolytical activity, responsible for the proofreading function of the enzyme (Blanco and Salas 1996). The biochemical and structural characterization of Phi29 DNA polymerase indicates that the first three activities are located in the C-terminal part of the enzyme, while the 3′→5′ degradative activity is in the N-terminal part of the enzyme (Bernad et al. 1989, Blanco and Salas 1996, Meijer et al. 2001; Kamtekar et al. 2004).
The exonuclease domains are conserved across proofreading-proficient DNA polymerases (Bernad et al. 1989; Blanco et al. 1991; Zuo and Deutscher 2001). They share a common catalytic mechanism of two metal ions, which are bound to the enzyme via four carboxylate groups (Derbyshire et al. 1991; Beese and Steitz 1991; Brautigam and Steitz 1998). Mutational analysis of Phi29 DNA polymerase confirmed that three N-terminally located conserved motifs, ExoI (D12X2ET), ExoII (H61NX2FD), and ExoIII (Y165X3D), form the 3′→5′ exonuclease active site, where D12, E14, D66, Y165, and D169 are involved in metal ion binding and catalysis (Bernad et al. 1989; Esteban et al. 1994) and T15, H61, N62, and F65 contact ssDNA (de Vega et al. 1996, 1998, 2000).
In addition to earlier observations that DNA exonuclease domain of Phi29 DNA polymerase possesses DNA 3′→5′ exonucleolytic activity (Garmendia et al. 1992; Esteban et al. 1992), we show here that in the presence of divalent metal ions as cofactors the same enzyme exonucleolytically degrades ssRNA. The RNase activity acts in a 3′ to 5′ polarity. This observation has also a practical implication: we show that the exoribonuclease activity of Phi29 DNA polymerase could be used to convert the target RNA into a primer for rolling circle amplification (RCA). This opens new opportunities for the RNA detection.
RESULTS
The comparative analysis of Phi29 DNA polymerase capacity to excise a single 3′ terminal ribonucleotide versus deoxyribonucleotide revealed that the enzyme is able to degrade both single nucleotides at the 3′ terminus of the DNA oligonucleotide with similar efficiency (Bonnin et al. 1999). This indicates that the enzyme does not discriminate against the 2′-OH group of the 3′-terminal nucleotide, and in both cases the enzyme cleaves DNA junctions in the same manner. We wanted to investigate if and how efficiently this enzyme would cleave RNA junctions.
RNase activity of Phi29 DNA polymerase
To test whether Phi29 DNA polymerase has an RNase activity, the initial RNA cleavage experiments were carried out using [3H] poly(A) RNA as a substrate. The RNA degradation activity could be detected by the formation of acid soluble products. We found that Phi29 DNA polymerase exhibited a distinctive RNase activity: the incubation of labeled RNA substrate with the enzyme yielded ∼30% of acid soluble products while only ∼5% of such products were observed in the control sample (data not shown). To explore if the 3′→5′ exonucleolytic center is also responsible for the RNase activity, the same experiments were carried out with Phi29 DNA polymerase double mutant (D12A/D66A). The specific DNA exonucleolytic activity of this mutant was found to be about three orders of magnitude lower than that of the wild-type DNA polymerase (Bernad et al. 1989). Our results demonstrate that the RNase activity of the double mutant is abolished (data not shown), thereby supporting the presumption that RNase activity of Phi29 DNA polymerase is linked to the 3′→5′ exonucleolytic center.
Phi29 DNA polymerase RNase activity was partially characterized. It was shown that the enzyme was catalytically active in the presence of bivalent metal ions, such as Mg2+ and Mn2+, as cofactors, and had a marginal activity in the presence of Co2+ ions (data not shown). Depending on pH and/or the composition of reaction buffer, the cofactor effect on RNase activity differed. At pH 7.9, in Tris–acetate buffer, Phi29 DNA polymerase degraded RNA more efficiently in the presence of 10 mM Mg2+ ions (the yield of acid soluble products in the presence of Mg2+ and Mn2+ ions exceeded corresponding background levels by ∼25% and ∼15%, respectively), while at pH 9.0, in Gly-KOH buffer, the enzyme preferred 10 mM Mn2+ ions (the yield of acid soluble products in the presence of Mg2+ and Mn2+ ions exceeded corresponding background levels by ∼10% and ∼20%, respectively). When both buffers were supplemented with 10 mM Co2+ ions, the yields of acid soluble products were only higher by ∼5% than the background levels. No Phi29 DNA polymerase RNase activity was observed at pH 6.0 in Mes-KOH buffer.
The type and polarity of RNA hydrolysis by Phi29 DNA polymerase
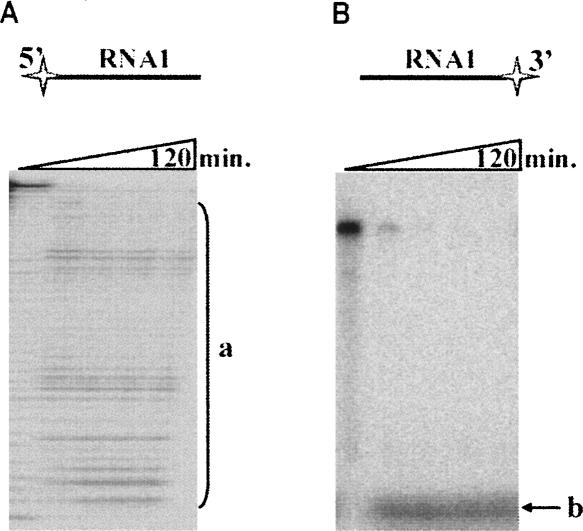
To determine the type (endo- or exo-) and polarity (3′→5′ or 5′→3′) of RNA cleavage by Phi29 DNA polymerase, we examined the ability of the enzyme to hydrolyze different RNA substrates. The synthetic RNA oligonucleotide RNA1, radioactively labeled at either the 5′ or 3′ end, was hydrolyzed by Phi29 DNA polymerase, and the accumulation of hydrolysis products was monitored (see Fig. 1A,B, respectively). The data show that when the substrate was the 5′-end labeled RNA molecule the truncated RNA oligonucleotide retained the label (Fig. 1A, products a), while the degradation of the 3′-end-labeled RNA molecule effectively removed the label, which was observed as a fast migrating band in the bottom of the gel (Fig. 1B, products b). Additional analysis of the reaction mix by HPLC using 5′-AMP, 5′-CMP, 5′-GMP, and 5′-UMP as the standards confirmed that liberated reaction products were mononucleotides (data not shown). These results indicate that Phi29 DNA polymerase degrades RNA by gradually releasing mononucleotides, nucleoside 5′-monophosphates (see below), from the 3′ end.
FIGURE 1.
The polarity of Phi29 DNA polymerase exoribonuclease activity. RNA hydrolysis studies were carried out under the conditions described in “Materials and Methods,” using 5′-end labeled (A) or 3′-end labeled (B) RNA1 oligonucleotides as substrates. The incubation mixtures, containing labeled RNA1 and Phi29 DNA polymerase, were incubated at 37°C. Aliquots were removed in timed intervals and analyzed by electrophoresis through denaturing 8% polyacrylamide gels. RNA1 oligonucleotides are shown above the gels with radioactive label indicated as a star. “a” represents the hydrolysis products of 5′-end labeled RNA1 (A), “b” represents the hydrolysis products of 3′-end labeled RNA1 (B).
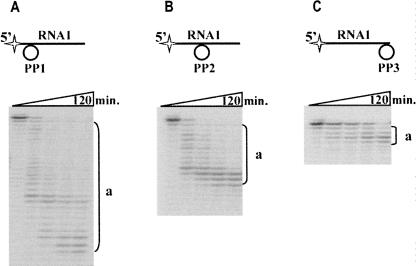
Further, we tested Phi29 DNA polymerase action on the RNA–DNA hybrids. In order to prevent DNA degradation by Phi29 DNA polymerase 3′→5′ exodeoxyribonuclease activity, circular DNA molecules were chosen for hybrid formation. Kinetic experiments were carried out with several hybrids composed of the RNA oligonucleotide RNA1 and preformed circular single stranded DNA molecules PP1, PP2, and PP3, which were complementary to the 5′ end, center part, or 3′ end of RNA1 molecule, respectively (Fig. 2). The results revealed that, similar to the hydrolysis of free RNA (Fig. 1), in the RNA–DNA hybrids Phi29 DNA polymerase hydrolyzed RNA in 3′→5′ direction as evidenced by the appearance of 5′-end-labeled truncated RNA1 products (Fig. 2, products a). Interestingly enough, the size of the prevailing RNA hydrolysis products depended on the location of the RNA–DNA hybridization area: the closer it was to the labeled 5′ end, the shorter were the RNA hydrolysis products formed (Fig. 2).
FIGURE 2.
Phi29 DNA polymerase 3′→5′ exoribonuclease activity on the RNA–DNA hybrids. The RNA–DNA hybrid hydrolysis studies were carried out under the conditions described in “Materials and Methods,” using 5′-end-labeled RNA–DNA hybrids. The DNA moiety of the hybrids was a preformed circular single stranded DNA molecule PP1, PP2, or PP3, complementary to the 5′ end (A), center part (B), or 3′ end (C) of the RNA1 molecule. The incubation mixtures, containing various RNA1–PP hybrids and Phi29 DNA polymerase, were incubated at 37°C. Aliquots were removed in timed intervals and analyzed by electrophoresis through denaturing 8% polyacrylamide gel. The labeled RNA1–PP hybrid substrates are shown above the gels. The radioactive label is indicated by the star. “a” represents the hydrolysis products of 5′-end-labeled RNA1-PP hybrids.
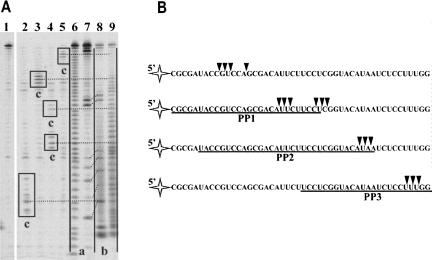
The final prevailing RNA1 hydrolysis products were characterized. The nature of their 3′ end (3′-OH or 3′-P) was determined by polyacrylamide gel electrophoresis under denaturing conditions, as previously described (Brown and Bevilacqua 2005). RNA1 alkaline hydrolysis and RNaseT1 cleavage (specificity: G) products were used as marker fragments having terminal 3′ phosphates (Fig. 3A, lanes 6,7, products a) or (after treatment with T4 PNK) 3′ hydroxyl groups (Fig. 3A, lanes 8,9, products b). We found that the migration of final prevailing RNA1 cleavage products generated by Phi29 DNA polymerase exonuclease activity (Fig. 3A, lanes 2–5, products c) was identical to that of corresponding T4 PNK treated oligoribonucleotides (Fig. 3A, lanes 8,9, products b), indicating that the enzyme hydrolyzed RNA1 to terminal 3′-OH groups containing oligonucleotides and 5′-NMPs. The alignment of prevailing degradation products to the hybridization areas (Fig. 3B) confirmed that Phi29 DNA polymerase stops RNA degradation at the RNA–DNA hybrid boundary. The enzyme acting as exoribonuclease paused either at the beginning of the double stranded structure or slightly inside the hybridization area. These data were supported by the observation that RNA degradation in the RNA–DNA hybrid area was evidently a much slower process in comparison to the degradation of free RNA (see Fig. 2C and Fig. 1A, respectively). Therefore, we conclude that Phi29 DNA polymerase is a 3′→5′ exoribonuclease, which prefers single-stranded RNA as a substrate, as opposed to the RNA–DNA hybrid. A similar trend was previously observed for this enzyme when the cleavage efficiency of a single-stranded and a double-stranded DNA was compared (Bonnin et al. 1999).
FIGURE 3.
(A) Sequencing of Phi29 DNA polymerase 3′→5′ exoribonuclecleolytic degradation products. The experiments were performed under the conditions described in “Materials and Methods.” The RNA degradation products of 5′-end-labeled free RNA1 or RNA1–PP hybrid substrates, generated by Phi29 DNA polymerase, were analyzed by electrophoresis through sequencing 8% polyacrylamide gel in the presence of RNA ladders. In lane 1 is a control sample of free RNA1. Lanes 2–5 represent the hydrolysis samples of RNA1 in different substrates by Phi29 DNA polymerase (lane 2, free RNA1; lane 3, RNA1-PP2; lane 4, RNA1–PP1; lane 5, RNA1–PP3 hybrids). Lanes 6 and 9 represent RNA1 alkaline hydrolysis ladders before and after the treatment with T4 PNK, respectively. Lanes 7 and 8 represent RNA1 digestion by RNaseT1 (specificity: G) fragments before and after the treatment with T4 PNK, respectively. The products of RNA1 degradation by Phi29 DNA polymerase (lanes 2–5, products c) were aligned with the fragments generated by alkaline hydrolysis and RNaseT1 treatment, and having 3′-phosphate (lanes 6,7, products a) or 3′-OH (lanes 8,9, products b) groups at their 3′ ends. The prevailing RNA1 degradation products are framed. Dotted lines indicate the assignments of bands of RNA1 degradation by Phi29 DNA polymerase to the corresponding bands of alkaline hydrolysis ladder treated with T4 PNK. Oblique dotted lines indicate the assignments of RNA1/RNaseT1 ladder fragments to the same fragments after T4 PNK treatment. (B) Mapping of prevailing RNA degradation products. The underlined sequences indicate areas of target RNA1 hybridization with respective circular DNA probes in RNA–DNA hybrids. Triangles indicate 3′-end points of prevailing RNA oligonucleotides generated by Phi29 DNA polymerase exonuclease activity.
Comparative analysis of Phi29 DNA polymerase nuclease activities on RNA and DNA substrates
Phi29 DNA polymerase excises a deoxyribonucleotide and single ribonucleotide at the 3′-end of single stranded DNA with the same efficiency (Bonnin et al. 1999). However, in both cases the enzyme cleaves DNA junctions. To compare the Phi29 DNA polymerase's ability to cleave DNA versus RNA junctions, we performed experiments with 16-mer DNA and RNA oligonucleotides. The exonucleolytic degradation was expressed as a rate constant of phosphodiesteric bond cleavage.
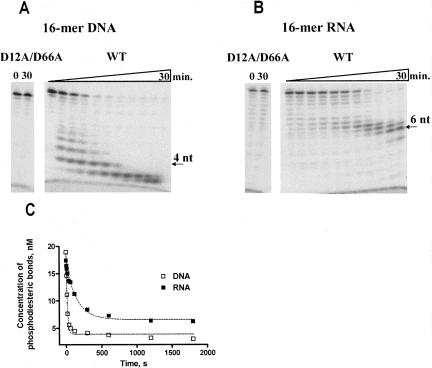
As expected, Phi29 DNA polymerase exo-mutant (D12A/D66A) had no nuclease activity (Fig. 4), while the wild-type (wt) enzyme was able to degrade 3′→5′ exonucleolytically both substrates, DNA and RNA (see Fig. 4A,B, respectively). We observed at least two differences in DNA and RNA hydrolyzes. First, we found that Phi29 DNA polymerase degrades DNA and RNA, leaving final products of different length. After incubation for 30 min the prevailing DNA hydrolysis products were 2–3 nucleotides (nt) long (Fig. 4A), while in the case of RNA they were 5–7 nt long (Fig. 4B). Second, we demonstrated that the efficiency of RNA hydrolysis was ∼10-fold lower than for DNA (Fig. 4C). The best fits to single exponentials gave rate constants of 4.01 ± 0.39 min−1 for DNA and 0.394 ± 0.045 min−1 for RNA.
FIGURE 4.
3′→5′ Exonucleolytic activity of Phi29 DNA polymerase on RNA and DNA substrates. The experiments were performed under the conditions described in “Materials and Methods,” using 5′-end-labeled 16-mer DNA (A) or RNA (B) oligonucleotides as substrates. Samples were incubated at 25°C for different time periods (5 s, 10 s, 10 s, 20 s, 40 s, 1 min, 2 min, 5 min, 10 min, 20 min, 30 min) and quenched by adding equal volume of 2× Loading dye solution. In the cases of Phi29 DNA polymerase exo− mutant (D12A/D66A), the samples were incubated for 30 min. Later the samples were analyzed by electrophoresis in 8% (w/v) denaturing polyacrylamide gel (29:1 [w/w] acrylamide/bisacrylamide, 7 M urea, 1× TBE buffer). The exonucleolytic degradation (C) was quantified by densitometry of autoradiographs and expressed as the rate constants of phosphodiesteric bond cleavage. The rates of cleavage were fit to single exponentials. The best fits (broken lines) gave rate constants of 4.01 ± 0.39 min−1 for DNA and 0.394 ± 0.045 min−1 for RNA.
RCA from target RNA
An extension of single-ribonucleotide containing primer terminus by Phi29 DNA polymerase was shown previously (Bonnin et al. 1999). Moreover, direct detection of RNA molecules in situ by RCA, using Phi29 DNA polymerase and RNA target, as a primer for RCA, was previously described for miRNA (Jonstrup et al. 2006). The RNA target–circular DNA probe duplex described in that publication was perfectly matched at the 3′ end of miRNA.
The fact that Phi29 polymerase exhibits RNase activity on [3H] poly(A) RNA substrate even in the presence of dNTPs (data not shown) allowed us to anticipate that the enzyme's 3′→5′ exoribonucleolytic activity could be used to convert the target RNA into a primer for RCA. The RNase activity of the enzyme should degrade the single stranded RNA up to the double stranded probe hybridization region, rendering RNA into a primer. Later in the presence of dNTPs this newly generated primer could be extended as a DNA strand complementary to the circular DNA probe.
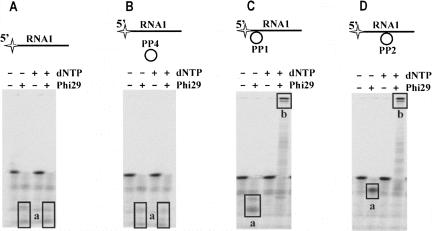
We tested the possibility of the 3′→5′ exoribonuclease activity of Phi29 DNA polymerase to convert the target RNA into the primer by using circular DNA probes PP1 and PP2 hybridized to the 5′ end or to the middle of the target RNA1, respectively (Fig. 5C,D). To verify reaction specificity, the control samples of free RNA1 or its mixture with circular noncomplementary DNA probe PP4 were used (see Fig. 5A,B, respectively).
FIGURE 5.
RCA from target RNA. The experiments were performed under the conditions described in “Materials and Methods.” The 5′-end-labeled RNA1 with no DNA (A), in the presence of nonspecific preformed circular DNA probe PP4 (B), and RNA1-DNA hybrids (C, D) were incubated with Phi29 DNA polymerase at 37°C with or without 1 mM dNTPs. RNA degradation and RCA product synthesis were analyzed by electrophoresis through denaturing 8% polyacrylamide gel. Different RNA1 substrates are shown above the gels. The radioactive label is indicated by the star. Prevailing reaction products are framed and represent 5′-end-labeled RNA1 hydrolysis products (“a” in A–D) and 5′-end-labeled RCA products (“b” in C, D).
As expected, in the control experiments, regardless of the presence of dNTPs, only the products of RNA degradation were observed (Fig. 5A,B, products a). When Phi29 DNA polymerase was added to the RNA–DNA hybrids in the absence of dNTPs, we observed the accumulation of truncated RNA fragments (Fig. 5C,D, products a). Similarly to the observations depicted in Figure 3, the length of the prevailing RNA fragments correlated with the hybrid structure location on RNA1 (see Fig. 5, C and D, respectively). The presence of dNTPs in the reaction mixtures induced the formation of long RCA products that comprised labeled RNA fragments at their 5′ ends (Fig. 5C,D, products b).
The target RNA conversion into a primer for RCA
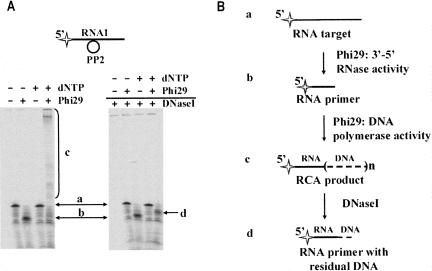
To verify that Phi29 DNA polymerase performs polymerization of the RCA product after RNA digestion, the above described experimental methodology was modified: after reaction the products were treated with DNaseI (Fig. 6A). The experimental data showed that DNaseI transforms the 5′-end-labeled RCA product (Fig. 6A, product c) into the fragment (Fig. 6A, product d) slightly longer than the truncated RNA target after the degradation by Phi29 DNA polymerase (Fig. 6A, product b). It is well known that DNaseI has a preferential specificity for sequence and structure (Potter et al. 1958), so the discrepancy in sizes of products b and d (Fig. 6) could be due to the specificity of the endonuclease action. Our data suggest that Phi29 DNA polymerase performed polymerization of RCA product after the digestion of target RNA1, when it was converted to a primer for polymerization (Fig. 6b).
FIGURE 6.
The target RNA conversion into a primer for RCA. The experiments (A) were performed under the conditions described in “Materials and Methods.” The 5′-end-labeled RNA1–DNA hybrids were incubated with Phi29 DNA polymerase at 37°C in the presence or absence of 1 mM dNTPs. After incubation the samples were divided into two equal parts, one of which was treated with the excess of DNaseI. The reaction products were analyzed by electrophoresis through denaturing 8% polyacrylamide gels. RNA1 substrate is shown above the gels. The radioactive label is indicated by the star. The reaction scheme (B) represents the conversions of target RNA. Reaction substrate and products are labeled as “a” for the 5′-end-labeled RNA1 substrate, “b” for the 5′-end-labeled RNA1 hydrolysis product serving as an RNA primer for RCA, “c” for the 5′-end labeled RCA product, and “d” for the 5′-end labeled RCA product after DNaseI treatment comprising the RNA primer with residual DNA.
Additional evidence for RCA induction by target RNA conversion into a primer was obtained using another RNA target—a 111-nt-long RNA transcript RNA2 hybridized to a circular DNA probe PP2 (data not shown). It was shown that the Phi29 DNA polymerase synthesized RCA product from RNA target after its 3′→5′ exonucleolytical degradation.
The induction of RCA by hybridization of the 3′ end of target RNA
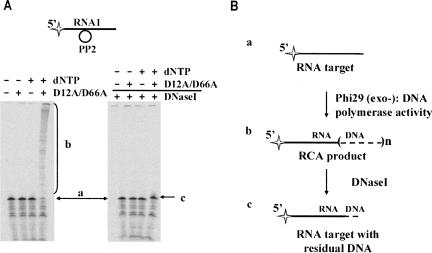
Is Phi29 DNA polymerase activity on RNA–DNA hybrids associated with its exonucleolytic domain and does it differ in their exo+ and exo− versions? To answer this question we tested Phi29 DNA polymerase exo− mutant (D12A/D66A), lacking 3′→5′ exonucleolytical activity (Fig. 7A). Surprisingly, it was shown that the exo− enzyme could produce RCA product (Fig. 7, products b) with efficiency comparable to that of the wt enzyme, and the yields of labeled RCA product were ∼85% and ∼65%, respectively (Table 1). However, contrary to previously described observation with wt Phi29 DNA polymerase (Fig. 6A), the digestion of RCA product with DNaseI produced a product almost as long as the initial target RNA (see Fig. 7A, products c,a, respectively). These data indicate that the exo− mutant polymerase could perform DNA synthesis without digesting the target RNA (Fig. 7B), most likely due to hybridization of the 3′ end of the RNA molecule to the DNA template. Scrutinized sequence of the padlock probe revealed that there was a possibility for hybridization of RNA1 3′-terminal guanine(s) to the closest cytosine(s) of circular DNA probe PP2.
FIGURE 7.
The induction of RCA by hybridization of the 3′ end of target RNA. The experiments (A) were performed under the conditions described in “Materials and Methods.” The 5′-end-labeled RNA1–DNA hybrids were incubated with Phi29 DNA polymerase exo− mutant (D12A/D66A) at 37°C in the presence or absence of 1 mM dNTPs. After incubation the samples were divided into two equal parts, one of which was treated with the excess of DNaseI. The reaction products were analyzed by electrophoresis through denaturing 8% polyacrylamide gels. RNA1 substrate is shown above the gels. The radioactive label is indicated by the star. The reaction scheme (B) represents the conversions of target RNA. Reaction substrate and products are labeled as “a” for the 5′-end-labeled RNA1 target serving as an RNA primer for RCA, “b” for the 5′-endlabeled RCA product, and “c” for the 5′-end-labeled RCA product after DNaseI treatment comprising the RNA target with residual DNA.
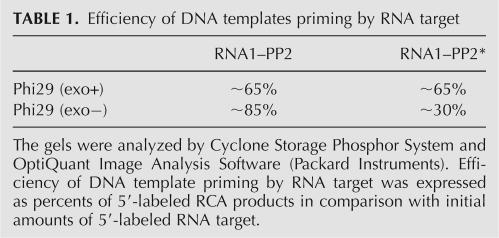
TABLE 1.
Efficiency of DNA templates priming by RNA target
To ascertain that the RNA mispriming on the DNA template is a result of enzyme action under “provocative” conditions, the circular DNA probe was locally changed. To avoid the hybridization of terminal guanine(s) at the 3′ end of target RNA1 to the DNA template, a few cytosines in circular DNA probe (PP2), closest to 3′ end of RNA1, were replaced by guanines (PP2*). It was shown that when PP2* served as a template (data not shown), the efficiency of DNA template priming by RNA with Phi29 DNA polymerase exo− mutant was ∼2.8-fold lower (∼30%) in comparison to PP2 template (∼85%) (Table 1). As expected, using Phi29 DNA polymerase the efficiency of both DNA templates priming by RNA remained the same (∼65%) (Table 1). These data confirmed our presumption that, using Phi29 DNA polymerase exo− mutant in RCA, the efficiency of DNA priming by RNA depends on RNA's ability to misprime on the DNA template.
DISCUSSION
We have demonstrated that B-type Phi29 DNA polymerase possesses a divalent metal dependent 3′→5′ exoribonucleolytic activity on ssRNA. Given that the mutant of Phi29 exonucleolytic center, D12A + D66A, was catalytically inactive against RNA, our expectations based on the predictions that the DNA and RNA substrates are cleaved via the same active site were experimentally confirmed.
The 3′→5′ DNA exonuclease active site is conserved in A- and B-type DNA polymerases (Bernad et al. 1989; Blanco et al. 1991; Zuo and Deutscher 2001). They share a common catalytic mechanism of two divalent metal ions. The conserved sequence motifs, termed ExoI, ExoII, and ExoIII, are clustered around of the active site and contain four negatively charged residues (D, E, D, D) that serve as ligands for two divalent metal ions required for catalysis, as well as catalytically active tyrosine from the ExoIII motif (Beese and Steitz 1991; Derbyshire et al. 1991; Brautigam and Steitz 1998).
The C-type DNA polymerases and some of the 3′→5′ DNA or RNA exonucleases were also predicted to have a structure and mechanism of action similar to the proofreading domain of A- and B-type DNA polymerases (Koonin and Deutscher 1993; Moser et al. 1997; Ito and Braithwaite 1998; Zuo and Deutscher 2001, 2002a). In the presumed catalytic mechanism four conserved carboxylate residues (D, E, D, D) are responsible for the divalent metal ion complexation and catalysis, and the conserved His residue, which is present in the position adjacent to Tyr residue in the ExoIII motif sequence and structure, may play a similar role as proposed for the Tyr residue in the proofreading domain of A- and B-type DNA polymerases (Zuo and Deutscher 2002b). Based on additional residues, His or Tyr, that are conserved in ExoIII motif, the DEDD exonuclease family can be divided into two groups: DEDDh and DEDDy (Zuo and Deutscher 2001). Despite the fact that the DEDD exonuclease family includes ribonucleases and deoxyribonucleases, only a few exonucleases have been previously reported to have a duality of polynucleotide substrate.
One of them is RNaseT, a member of the DEDDh group of enzymes. Similarly to Phi29 DNA polymerase, RNaseT exhibited a duality for polynucleotide substrates. Initially RNaseT was characterized as an enzyme responsible for the 3′-end turnover of tRNA in Escherichia coli (Deutscher and Marlor 1985). Later it was demonstrated that E. coli RNaseT has divalent metal ions dependent 3′→5′ exonucleolytic activity that degrades both ssRNA and ssDNA substrates (Zuo and Deutscher 1999, 2002c), and achieved optimal activity on both substrates under comparable conditions (Viswanathan et al. 1998). First, the DNase and RNase activities of E. coli RNaseT exhibited similar requirements for divalent metal ions. Mg2+ and Mn2+ ions served as effective cofactors for the RNA and DNA cleavage, and Ba2+, Ca2+, Cd2+, Cu2+, Ni2+, and Zn2+ ions could not serve as cofactors for either of the substrates (Deutscher and Marlor 1985; Viswanathan et al. 1998). Co2+ ions supported partial RNase activity (Deutscher and Marlor 1985), but they were unable to substitute the cofactors for DNA cleavage (Viswanathan et al. 1998). Second, E. coli RNaseT showed optimal activity on both substrates (relative activity: 80%–100%) at a pH range from 8.0 to 9.0 (Deutscher and Marlor 1985; Viswanathan et al. 1998). The activity of RNaseT was strongly reduced in lower pH—more than 95% decrease in activity was noted at pH 6.0.
We found that optimal conditions for RNase activity of Phi29 DNA polymerase resembled those for E. coli RNaseT activity, including optimal pH and the role of different divalent metal cations when tested as cofactors. Similar to E. coli RNaseT, Phi29 DNA polymerase has a distinctive RNase activity at pH 7.9 and pH 9.0 and was completely inactive at pH 6.0. Also, for Phi29 DNA polymerase, Mg2+ and Mn2+ ions served as the cofactors for RNA cleavage, and Co2+ ions supported only marginal RNase activity. Similar results in metal ion requirements for Phi29 DNA polymerase were previously described in DNA cleavage (Esteban et al. 1992). The electrophoretic analysis of the Phi29 DNA polymerase DNA cleavage efficiency on the 5′-end-labeled DNA oligonucleotide as substrate showed that the enzyme preferred Mg2+ and Mn2+ ions as the cofactors (∼90% and 100% activities, correspondingly), whereas Co2+ ions yielded ∼20% of Mn2+-activated reaction.
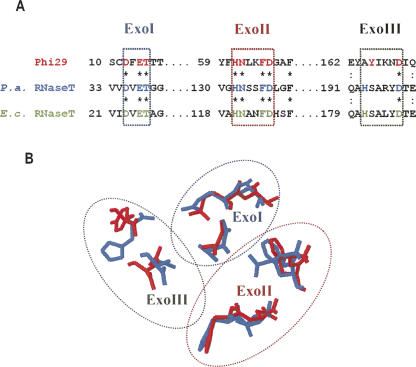
The intriguing similarity between Phi29 DNA polymerase (member of DEDDy group) and E. coli RNaseT (member of DEDDh group) in requirement for cofactors and the resemblance in the 3′→5′ exonucleolytic type of activity on both substrates most likely could be explained by similar organization of the active site. The primary sequence of the N-terminal part of Phi29 DNA polymerase containing ExoI, ExoII, and ExoIII motifs, was similar to the RNaseT orthologs from E. coli and Pseudomonas aeruginosa (Fig. 8A). The mutational analysis of E. coli RNaseT (Zuo and Deutscher 2002a) and the X-ray structure of P. aeruginosa RNaseT (PDB: 2F96) showed that these three conserved regions form an exonucleolytic active site. The superposition of Phi29 DNA polymerase 3′→5′ active site, including Tyr residue (PDB: 1XHX) and P. aeruginosa RNaseT active site, including His residue (PDB: 2F96) confirmed that these enzymes are well structurally conserved (Fig. 8B). These observations are in good agreement with the conception that the duality of polynucleotide substrates in both enzymes is related to similar active site organization and catalytic mechanism.
FIGURE 8.
Sequence comparison and active sites superposition of Phi29 DNA polymerase and RNaseT orthologs. (A) The sequence analysis of Phi29 DNA polymerase (UniProtKB/TrEMBL: Q38545), Escherichia coli RNaseT (UniProtKB/TrEMBL: Q1RBE3), and Pseudomonas aeruginosa RNaseT (UniProtKB/TrEMBL: Q9HY82) sequences were performed using the CLUSTALW program (Thompson et al. 1994) at PôleBio-Informatique Lyonnais (http://pbil.univ-lyon1.fr). Asterisks (*) indicate identical amino acid residues; double dots (:) stand for similar amino acid residues. The conserved active site motifs (ExoI, ExoII, ExoIII) are framed. The amino acid residues that play key role in catalysis are presented in red (Phi29 DNA polymerase), blue (P. aeruginosa RNaseT), or green (E. coli RNaseT). (B) Superposition of Phi29 DNA polymerase 3′→5′ exonucleolytic (PDB: 1XHX) and P. aeruginosa RNaseT (PDB: 2F96) active sites was performed using the Swiss_PdbVieWer program (Guex and Peitsch 1997). The amino acids of Phi29 DNA polymerase are shown in red; P. aeruginosa RNaseT is in blue.
The novel 3′→5′ exoribonucleolytic activity of Phi29 DNA polymerase could be successfully used for direct target RNA detection and analysis by the RCA technique, sometimes termed the rolling circle replication (RCR) technique (Baner et al. 1998; Lizardi et al. 1998), in which Phi29 DNA polymerase is mostly preferred due to high processivity and efficient strand-displacement activity. Usually, the detection and quantity measurements of RNA molecules by RCA are performed routinely using immuno-RCA (Zhou et al. 2001), by DNA circularization on RNA (Christian et al. 2001; Nilsson et al. 2001), or cDNA analysis with padlock probes (Baner et al. 2003, 2005). However, the disadvantage of these methods is that they are indirect and require DNA primers to initiate RCA. Since RCA is a very sensitive reaction and often is used in multiparallel analysis of different targets in the same sample, the addition of exogenous RCA primers adds a further process step and may decrease reaction specificity due to nonspecific primer hybridization to other padlock probes.
Direct detection of RNA molecules by RCA, using target RNA as a primer for RCA, was previously described for miRNA (Jonstrup et al. 2006). However, the target RNA–circular DNA probe duplexes described in that publication were perfectly matched at the 3′ end of miRNA, and the 3′→5′ exonucleolytic shortening of target RNA was not necessary to initiate RCA. The observation that Phi29 DNA polymerase exhibits RNase activity in the presence of dNTPs prompted us to use the enzyme's 3′→5′ exoribonucleolytic activity in the conversion of target RNA into a primer for RCA. We showed that the enzyme's RNase activity degraded the RNA up to the double stranded region of probe hybridization, transforming target RNA into the primer, from which synthesis of DNA, complementary to the DNA template, begins in the presence of dNTPs. This opens new opportunities for the RNA detection by RCA.
MATERIALS AND METHODS
Proteins, buffers, and reagents
BSA, glycogen, DNaseI, and DNaseI buffer with MgCl2 (10×), DEPC-treated water, Phi29 DNA polymerase, Phi29 DNA polymerase exo− mutant (D12A + D66A), Ribolock ribonuclease inhibitor, RNaseT1, T4 polynucleotide kinase (T4 PNK), T4 DNA ligase, T4 RNA ligase, Tango buffer (10×), TBE buffer (10×), and RNA Loading Dye Solution (2×) were products of Fermentas UAB. RNA labeling KinaseMax kit was purchased from Ambion; yeast tRNA was obtained from Serva.
Nucleotides and oligonucleotides
[γ33P]ATP (∼3000 Ci/mmol), [32P]pCp (∼3000 Ci/mmol), and poly[8-3H]adenylic acid (∼490 mCi/mmol) were purchased from Amersham Biosciences; unlabeled polyadenylic acid was obtained from Pharmacia. Unlabeled dNTPs were products of Fermentas UAB. DNA oligonucleotides PP1 (5′-GTATCGCGAAAATGTAAAGCAATGCTGCTGCTGTACTACGAGCGGTCTCCAAGGAATGCGCATTTAGAAGAATGTCGCTGGACG), PP2 (5′-ATGTCGCTGGACGGTAATGCTGCTGCTGTACTACGAGCGGTCTCCAGGAATGCGCATTTATGTACCGAGGAAGA), PP2* (5′-ATGTCGCTGGACGGTAATGCTGCTGCTGTACTACGAGGGGTGTGGAGGAATGGGGATTTATGTACCGAGGAAGA), PP3 (5′-TATGTACCGAGGAAGATGATGCTGCTGCTGTACTACGAGCGGTCTCCAGGAACGCATGAAACGCCAAAGGAGAT), PP4 (5′-GTTTTACAACGCCAAGCTTGCATGAATGCAGGTTAGGATCCAATGGTACCGAGCTCGAATTCACTGGCCGTC), PL1 (5′-CGTTTACATTTTCGCGATACCGTCCAGCGACATTCTTCCT), PL2 (5′-ACCGTCCAGCGACATTCTTCCTCGGTACAT), PL3 (5′-CTTCCTCGGTACATAATCTCCTTTGGCGTT), PL4 (5′-TTGGCGTTGTAAAACGACGGCCAGTGAATT), D1 (5′-CACTGGTCTTCAGCCG), and RNA oligonucleotides RNA1 (5′-CGGGAUACCGUCCAGCGACAUUCUUCCUCGGUACAUAAUCUCCUUUGG), and R1 (5′-CACUGGUCUUCAGCCG) were obtained from Metabion; RNA2 (5′-GGGAAAGCUUUACAUUUUCGCGAUACCGUCCAGCGACAUUCUUCCUCGGUACAUAAUCUCCUUUGGCGUUUCCCGAUGUCCGUCACGCACAUGGGAUCCCCGGGUACCGAG) was obtained from Fermentas UAB.
Phi29 DNA polymerase RNase activity assay
RNase activity was evaluated by measuring the amount (%) of acid soluble products.
Twenty-five microliters of reaction mixture (1× Tango buffer [33 mM Tris-HCl at pH 7.9, 10 mM Mg-acetate, 66 mM K-acetate, 0.1 mg/mL BSA], 24 μM poly[8-3H]adenylic acid [43 mCi/mmol]) was incubated with 25 U Phi29 DNA polymerase or 25 U Phi29 (exo−) DNA polymerase (final concentrations of the enzymes were ∼150 nM) for 3 h at 37°C. An identical mixture without the enzyme was set up as a control. After incubation, 25 μL of BSA (10 mg/mL) were added to the samples and reactions were terminated by addition of 50 μL of cold 10% (w/v) trichloroacetic acid (TCA). To evaluate total radioactivity of samples (100%), TCA in samples without the enzymes was replaced with DEPC-treated water. The samples were centrifuged at 13,000 rpm for 10 min. One hundred microliters of the supernatant were added to 5 mL of dioxane scintillation fluid. The amount of radioactive nucleotides released was determined by scintillation counting.
Phi29 DNA polymerase RNase activity assays at pH 6.0 and pH 9.0 were carried out as described above, only Mes-KOH buffer (10 mM Mes-KOH at pH 6.0, 10 mM MgCl2 100 mM KCl, 0.1 mg/mL BSA) and Gly-KOH buffer (10 mM Gly-KOH at pH 9.0, 10 mM MgCl2, 100 mM KCl, 0.1 mg/mL BSA) were used instead of 1× Tango buffer. In some experiments the reaction conditions were altered, replacing magnesium salts with 10 mM MnCl2 or 10 mM CoCl2.
DNA labeling
5′-end labeling of DNA was carried out using T4 PNK (Fermentas UAB) following the manufacturer's recommendations.
RNA labeling
5′-end labeling of RNA was carried out using a KinaseMax kit (Ambion) following the manufacturer's recommendations. 3′-end labeling of RNA was carried out with T4 RNA ligase and [32P]pCp as described previously (Uhlenbeck and Gumport 1982). 3′-terminal dephosphorylation of 3′-end labeled RNA was performed at a final concentration of 100 nM in 10 μL reaction mixture (1× Tango buffer, 10 U Ribolock ribonuclease inhibitor, 10 U T4 PNK) for 30 min at 37°C. Afterward reaction mixture was heated for 10 min at 70°C and placed on ice.
Preparation of RNA alkaline hydrolysis ladder
Two microliters of 5′-end labeled RNA1 (1 μM) were mixed with 3 μL of yeast tRNA in DEPC-treated water (1 mg/mL), followed by the addition of 10 μL of alkaline hydrolysis buffer (50 mM sodium carbonate at pH 9.2, 1 mM EDTA) to the ice-cold RNA mixture. The sample was heated at 95°C for 2 min and immediately placed on ice, where an equal volume of 2× Loading dye solution was added.
Preparation of RNA RNaseT1 sequencing ladder
Two microliters of 5′-end labeled RNA1 (1 μM) were mixed with 3 μL of yeast tRNA in DEPC-treated water (1 mg/mL), followed by the addition of 10 μL of RNA sequencing buffer (20 mM sodium citrate at pH 5.0, 7 M urea, 1 mM EDTA) to the ice-cold RNA mixture. After the mixture was heated at 50°C for 5 min and cooled to room temperature, 3 μL of RNaseT1 (0.2 U/μL) were added. The sample was incubated for 15 min at room temperature, followed by the phenol/chloroform (5:1 [v/v], pH 4.5) extraction. Afterward the sample was placed on ice and mixed with an equal volume of 2× Loading dye solution.
Preparation of RNA ladders with 3′-terminal hydroxyls
One-microliter aliquots of RNA1 alkaline hydrolysis and RNaseT1 sequencing ladders (with Loading dye) were mixed with 0.25 μL glycogen (20 mg/mL), 0.5 μL sodium acetate (3 M, pH 5.2), and 5.25 μL EtOH, chilled for few hours at −20°C, and centrifuged at 1600g for 15 min at 4°C. The pellet was washed with 70% EtOH and centrifuged at 16,000g for 15 min at 4°C, dried for 10 min at room temperature, and dissolved in 5 μL of DEPC-treated water. The sample was diluted to 6 μL of reaction mixture (1× DNaseI buffer with MgCl2, 6 U Ribolock inhibitor, 6 U T4 PNK) and incubated for 30 min at 37°C, then placed on ice and mixed with an equal volume of 2× Loading dye solution.
Preparation of circular single stranded DNA probes
PP oligonucleotides (PP1, PP2 or PP2*, PP3, PP4) were 5′-phosphorylated at a final concentration of 10 μM in 10 μL of reaction mixture (1× Tango buffer, 50 μM ATP, 5 U T4 PNK) for 30 min at 37°C. After incubation, samples were heated for 10 min at 70°C and placed on ice. The phosphorylated PP oligonucleotides were hybridized to appropriate complementary PL oligonucleotides (PL1, PL2, PL3, PL4) in 1× Tango buffer at final concentrations of 500 nM of each oligonucleotides by heating the sample in a boiling water bath for a few minutes and slow cooling to room temperature for 2 h. The ligation reactions were performed in 500 μL of reaction mixtures (1× Tango buffer, 200 nM PP-PL oligoduplexes, 1 mM ATP, 75 U T4 DNA ligase) for 1 h at 37°C. Afterward the samples were heated for 10 min at 70°C and placed on ice. After circularization, linear DNA oligonucleotides were degraded with 100 U T7 DNA polymerase for 1 h at 37°C. Then the sample was heated for 10 min at 70°C and placed on ice.
Preparation of labeled RNA–DNA hybrids
5′-end-labeled RNA1 hybrids with circularized PP oligonucleotides were prepared in 15 μL of reaction mixture (1× Tango buffer, 4 nM 5′-end-labeled RNA1, 40 nM circularized PP oligonucleotide) by heating for 3 min at 65°C and cooling to room temperature for 10 min. After cooling the samples were placed on ice.
Hydrolysis of free RNA and RNA–DNA hybrid
For cleavage reaction 5′- or 3′-end-labeled free RNA1 or 5′-end-labeled RNA1–PP hybrids were diluted with 1× Tango buffer, containing Ribolock (1 U/μL), to a final concentration of 1 nM. Fifty microliters of reaction mixture were prewarmed at 37°C. Before the addition of an enzyme an 8-μL aliquot was removed, placed on ice, and mixed with 8 μl of 2× Loading dye solution to be used as a control. The remaining reaction mixture was supplemented with 6 U Phi29 DNA polymerase (final concentration was ∼20 nM) and incubated at 37°C. Aliquots were removed at various time points (0.5, 2, 10, 120 min), placed on ice, and mixed with 8 μL of 2× Loading dye solution. Before electrophoresis the samples were heated for 10 min at 70°C and cooled on ice, then loaded onto the 8% (w/v) denaturing polyacrylamide gel (29:1 [w/w] acrylamide/bisacrylamide, 7 M urea, 1× TBE buffer). After electrophoresis the RNA degradation products were detected and analyzed by Cyclone Storage Phosphor System and OptiQuant Image Analysis Software (Packard Instruments).
HPLC analysis of RNA degradation products
The chromatography experiments were performed at 25°C on an AKTA HPLC system using a 5C18 column (Amersham-Pharmacia-Biotech) equilibrated with 100 mM triethylamonium-acetate at pH 7.0). Ten microliters of reaction mixture (1× Tango buffer, 0.1 mM RNA1 oligonucleotide) were incubated with 10 U Phi29 DNA polymerase (final concentration was ∼150 nM) for 3 h at 37°C. Afterward the sample was subjected to column chromatography using acetolnitril gradient (0%–60%). Elution of minimal nucleic acid degradation products was followed spectrophotometrically, monitoring the absorbance at 260 nm. Individual 5′-AMP, 5′-CMP, 5′-GMP, or 5′-UMP ribonucleotides were used as markers.
Determination of Phi29 DNA polymerase 3′→5′ exoribonucleolytic degradation products
RNA1 cleavage by Phi29 DNA polymerase was carried out for 2 h at 37°C as described in section “Hydrolysis of free RNA and RNA–DNA hybrid.”
The RNA hydrolysis products, generated by Phi29 DNA polymerase, were analyzed by 8% (w/v) sequencing polyacrylamide gel (29:1 [w/w] acrylamide/bisacrylamide, 7 M urea, 1× TBE buffer) electrophoresis. The RNA oligonucleotide degradation products were detected and analyzed by Cyclone Storage Phosphor System and OptiQuant Image Analysis Software (Packard Instruments). Alkaline hydrolysis and RNaseT1 digestion products of the same RNA oligonucleotide, containing 3′-phosphate or 3′-OH groups at their terminal 3′ ends, were used as markers for establishing RNA degradation products.
3′→5′ Exonucleolytic activity of Phi29 DNA polymerase on RNA and DNA substrates
3′→5′ exonuclease activity assay was carried as described previously (Bonnin et al. 1999; de Vega et al. 1998). Incubation mixture (12.5 μL) contained 1× Tango buffer, Ribolock (1 U/μL), 1.2 nM 5′-labeled 16-mer DNA or RNA oligonucleotides (D1 or R1), and 1.67 U of Phi29 DNA polymerase (∼20 nM). Samples were incubated at 25°C for different time periods (5 s, 10 s, 10 s, 20 s, 40 s, 1 min, 2 min, 5 min, 10 min, 20 min, 30 min) and quenched by adding an equal volume of 2× Loading dye solution. In the case of Phi29 DNA polymerase exo− mutant (D12A/D66A), the samples were incubated for 30 min. Afterward the samples were analyzed by 8% (w/v) denaturing polyacrylamide gel (29:1 [w/w] acrylamide/bisacrylamide, 7 M urea, 1× TBE buffer) electrophoresis. The exonucleolytic degradation was quantified by densitometry of autoradiographs and expressed as rate constants of phosphodiesteric bond cleavage.
RCA from target RNA
The reactions were performed for 2 h at 37°C in 20 μL of reaction mixture containing 1× Tango buffer, 20 U Ribolock ribonuclease inhibitor, and 3 U Phi29 (exo+/−) DNA polymerase (final concentration was ∼20 nM), and complemented with 1 nM labeled free RNA, or its mixture with nonspecific PP, or RNA-PP hybrids (final concentrations of RNA and circular PP oligonucleotide were 1 nM and 10 nM, respectively), in the absence or presence of 1 mM dNTPs. Free RNA and its mixture with circular PP oligonucleotide having no complementary sequence to RNA1 were used as controls of reaction specificity. Following incubation 20 μL of 2×, Loading dye solution was added to stop the reaction. The samples were heated for 10 min at 70°C and placed on ice before loading onto denaturing polyacrylamide gel. The gels were analyzed by the Cyclone Storage Phosphor System and OptiQuant Image Analysis Software (Packard Instruments). Efficiency of DNA template priming by RNA target was expressed as percents of 5′-labeled RCA products in comparison with initial amounts of 5′-labeled RNA target.
In the case of DNaseI treatment of reaction products, the experimental procedures were modified. Following incubation the reaction mixtures were diluted with an equal volume of miliQ water. The samples were heated for 10 min at 70°C and supplemented with 1 mM CaCl2 and DNaseI (final concentration was 0.3 U/μL). The DNaseI digestion reactions were performed for 20 min at 37°C. Afterward an equal volume of 2× Loading dye solution was added to stop the reaction. The samples were heated for 10 min at 70°C and placed on ice before loading onto denaturing polyacrylamide gel.
AKNOWLEDGMENTS
This work was partially funded by an EU FP6 Integrated Project “MolTools” (http://www.moltools.org/) award. We are grateful to Roberta Kunceviciene and Tadas Jarasunas for HPLC analysis. Also, we express our appreciation to Dr. Egle Merkiene for discussion and Dr. Egle Cesnaviciene, Dr. Lolita Zaliauskiene, and Dr. Ramune Leipuviene for help in preparation of the manuscript.
Footnotes
Abbreviations: BSA, bovine serum albumin; RCA, rolling circle amplification; T4 PNK, T4 polynucleotide kinase; TCA, trichloroacetic acid.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.622108.
REFERENCES
- Baner, J., Nilsson, M., Mendel-Hartvik, M., Landegren, U. Signal amplification of padlock probes by rolling circle replication. Nucleic Acids Res. 1998;26:5073–5078. doi: 10.1093/nar/26.22.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baner, J., Isaksson, A., Waldenstrom, E., Jarvius, J., Landegren, U., Nilsson, M. Parallel gene analysis with allele-specific padlock probes and tag microarrays. Nucleic Acids Res. 2003;31:e103. doi: 10.1093/nar/gng104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baner, J., Marits, P., Nilsson, M., Winqvist, O., Landegren, U. Analysis of T-cell receptor V β gene repertoires after immune stimulation and in malignancy by use of padlock probes and microarrays. Clin. Chem. 2005;51:768–775. doi: 10.1373/clinchem.2004.047266. [DOI] [PubMed] [Google Scholar]
- Beese, L.S., Steitz, T.A. Structural basis for the 3′→5′ exonuclease activity of Escherichia coli DNA polymerase I: A two metal ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad, A., Blanco, L., Lazaro, J.M., Martin, G., Salas, M. A conserved 3′→5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989;59:219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- Blanco, L., Salas, M. Relating structure to function in φ29 DNA polymerase. J. Biol. Chem. 1996;271:8509–8512. doi: 10.1074/jbc.271.15.8509. [DOI] [PubMed] [Google Scholar]
- Blanco, L., Bernad, A., Blasco, M.A., Salas, M. A general structure for DNA-dependent DNA polymerases. Gene. 1991;100:27–38. doi: 10.1016/0378-1119(91)90346-d. [DOI] [PubMed] [Google Scholar]
- Bonnin, A., Lázaro, J.M., Blanco, L., Salas, M. A single tyrosine prevents insertion of ribonucleotides in the eukaryotic-type φ29 DNA polymerase. J. Mol. Biol. 1999;290:241–251. doi: 10.1006/jmbi.1999.2900. [DOI] [PubMed] [Google Scholar]
- Brautigam, C.A., Steitz, T.A. Structural principles for the inhibition of the 3′→5′ exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates. J. Mol. Biol. 1998;277:363–377. doi: 10.1006/jmbi.1997.1586. [DOI] [PubMed] [Google Scholar]
- Brown, T.S., Bevilacqua, P.C. Method for assigning double-stranded RNA structures. Biotechniques. 2005;38:368–372. doi: 10.2144/05383BM04. [DOI] [PubMed] [Google Scholar]
- Christian, A.T., Pattee, M.S., Attix, C.M., Reed, B.E., Sorensen, K.J., Tucker, J.D. Detection of DNA point mutations and mRNA expression levels by rolling circle amplification in individual cells. Proc. Natl. Acad. Sci. 2001;98:14238–14243. doi: 10.1073/pnas.251383598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vega, M., Lazaro, J.M., Salas, M., Blanco, L. Primer-terminus stabilization at the 3′→5′ exonuclease active site of φ29 DNA polymerase. Involvement of two amino acid residues highly conserved in proofreading DNA polymerases. EMBO J. 1996;15:1182–1192. [PMC free article] [PubMed] [Google Scholar]
- de Vega, M., Lazaro, J.M., Salas, M., Blanco, L. Mutational analysis of φ29 DNA polymerase residues acting as ssDNA ligands for 3′→5′ exonucleolysis. J. Mol. Biol. 1998;279:807–822. doi: 10.1006/jmbi.1998.1805. [DOI] [PubMed] [Google Scholar]
- de Vega, M., Lazaro, J.M., Salas, M. Phage φ29 DNA polymerase residues involved in the proper stabilisation of the primer-terminus at the 3′→5′ exonuclease active site. J. Mol. Biol. 2000;304:1–9. doi: 10.1006/jmbi.2000.4178. [DOI] [PubMed] [Google Scholar]
- Derbyshire, V., Grindley, N.D., Joyce, C.M. The 3′→5′ exonuclease of DNA polymerase I of Escherichia coli: Contribution of each amino acid at the active site to the reaction. EMBO J. 1991;10:17–24. doi: 10.1002/j.1460-2075.1991.tb07916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher, M.P., Marlor, C.W. Purification and characterization of Escherichia coli RNase T. J. Biol. Chem. 1985;260:7067–7071. [PubMed] [Google Scholar]
- Esteban, J.A., Bernad, A., Salas, M., Blanco, L. Metal activation of synthetic and degradative activities of φ29 DNA polymerase, a model enzyme for protein-primed DNA replication. Biochemistry. 1992;31:350–359. doi: 10.1021/bi00117a006. [DOI] [PubMed] [Google Scholar]
- Esteban, J.A., Soengas, M.S., Salas, M., Blanco, L. 3′→5′ exonuclease active site of φ29 DNA polymerase. Evidence favoring a metal-ion-assisted reaction mechanism. J. Biol. Chem. 1994;269:31946–31954. [PubMed] [Google Scholar]
- Garmendia, C., Bernad, A., Esteban, J.A., Blanco, L., Salas, M. The bacteriophage φ29 DNA polymerase, a proofreading enzyme. J. Biol. Chem. 1992;267:2594–2599. [PubMed] [Google Scholar]
- Guex, N., Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Ito, J., Braithwaite, D.K. RNase T is related to dnaQ and family C DNA polymerases. Mol. Microbiol. 1998;27:235–237. doi: 10.1046/j.1365-2958.1998.00673.x. [DOI] [PubMed] [Google Scholar]
- Jonstrup, S.P., Koch, J., Kjems, J. A microRNA detection system based on padlock probes and rolling circle amplification. RNA. 2006;12:1747–1752. doi: 10.1261/rna.110706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamtekar, S., Berman, A.J., Wang, J., Lazaro, J.M., de Vega, M., Blanco, L., Salas, M., Steitz, T.A. Insights into strand displacement and processivity from the crystal structure of the protein-primed DNA polymerase of bacteriophage φ29. Mol. Cell. 2004;16:609–618. doi: 10.1016/j.molcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Koonin, E.V., Deutscher, M.P. RNase T shares conserved sequence motifs with DNA proofreading exonucleases. Nucleic Acids Res. 1993;21:2521–2522. doi: 10.1093/nar/21.10.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizardi, P.M., Huang, X., Zhu, Z., Bray-Ward, P., Thomas, D.C., Ward, D.C. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- Meijer, W.J.J., Horcajadas, J.A., Salas, M. φ29 Family of phages. Microbiol. Mol. Biol. Rev. 2001;65:261–287. doi: 10.1128/MMBR.65.2.261-287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser, M.J., Holley, W.R., Chatterjee, A., Mian, I.S. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res. 1997;25:5110–5118. doi: 10.1093/nar/25.24.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson, M., Antson, D.-O., Barbany, G., Landegren, U. RNA-templated DNA ligation for transcript analysis. Nucleic Acids Res. 2001;29:578–581. doi: 10.1093/nar/29.2.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter, J.L., Laurila, U.-R., Laskowski, M. Studies of specificity of Deoxyribonuclease I. J. Biol. Chem. 1958;233:915–916. [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck, O.C., Gumport, R.I. T4 RNA ligase. In: Boyer P.D., editor. The Enzymes. vol. XV. Academic Press; New York: 1982. pp. 31–58. part B, [Google Scholar]
- Viswanathan, M., Dower, K.W., Lovett, S.T. Identification of a potent DNase activity associated with RNase T of Escherichia coli . J. Biol. Chem. 1998;273:35126–35131. doi: 10.1074/jbc.273.52.35126. [DOI] [PubMed] [Google Scholar]
- Zhou, Y., Calciano, M., Hamann, S., Leamon, J.H., Strugnell, T., Cristian, M.W., Lizardi, P.M. In situ detection of messenger RNA using digoxigenin-labeled oligonucleotides and rolling circle amplification. Exp. Mol. Pathol. 2001;70:281–288. doi: 10.1006/exmp.2001.2365. [DOI] [PubMed] [Google Scholar]
- Zuo, Y., Deutscher, M.P. The DNase activity of RNase T and its application to DNA cloning. Nucleic Acids Res. 1999;27:4077–4082. doi: 10.1093/nar/27.20.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, Y., Deutscher, M.P. Exoribonuclease superfamilies: Structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, Y., Deutscher, M.P. Mechanism of action of RNase T. I. Identification of residues required for catalysis, substrate binding, and dimerization. J. Biol. Chem. 2002a;277:50155–50159. doi: 10.1074/jbc.M207706200. [DOI] [PubMed] [Google Scholar]
- Zuo, Y., Deutscher, M.P. Mechanism of action of RNase T. II. A structural and functional model of the enzyme. J. Biol. Chem. 2002b;277:50160–50164. doi: 10.1074/jbc.M207707200. [DOI] [PubMed] [Google Scholar]
- Zuo, Y., Deutscher, M.P. The physiological role of RNase T can be explained by its unusual substrate specificity. J. Biol. Chem. 2002c;277:29654–29661. doi: 10.1074/jbc.M204252200. [DOI] [PubMed] [Google Scholar]