Abstract
RNA-binding proteins regulate every aspect of RNA metabolism, including pre-mRNA splicing, mRNA trafficking, stability, and translation. This review summarizes the available information on molecular mechanisms of translational repression by RNA-binding proteins. By using a specific set of well-defined examples, we also describe how regulation can be reversed.
Keywords: translation, RNA-binding proteins, hnRNP K, SXL, Maskin, Cup, IRP, GAIT, CPEB, Smaug
INTRODUCTION
Translational regulation plays an important role in numerous biological situations. In conditions of amino acid starvation, apoptosis, or viral infection, a global response modifies the translational efficiency of most mRNAs in the cell. In other circumstances (e.g., embryonic pattern formation, sex determination, neuronal plasticity) the translation of specific mRNAs is regulated, leaving most cellular transcripts unaffected. Misregulation of global or mRNA-specific translation contributes to disease (Mamane et al. 2006; Kozma et al. 2007; Wiemer 2007). For example, Fragile X syndrome results from mutations in FMRP, a protein involved in translational regulation at synapses (Zalfa et al. 2006). While global regulation is normally driven by phosphorylation or proteolysis of key general translation initiation factors, mRNA-specific regulation is exerted by proteins (or microRNAs) that recognize sequence elements usually located in the untranslated regions (UTRs) of the transcript. Most often, regulation involves repression by proteins that bind to the 3′ UTR. Translational repression can be reversed, and this is generally achieved by removal of the repressor from its binding site on the mRNA or by remodeling of the repressor complex. In this review we briefly summarize the current knowledge on mechanisms of translational repression by RNA-binding proteins, as well as how regulation can be reversed. More extensive reviews concerning various aspects of translational regulation can be found elsewhere (de Moor et al. 2005; Piccioni et al. 2005; Wilhelm and Smibert 2005; Hentze et al. 2007; Jackson and Standart 2007; Thompson et al. 2007).
The majority of known translation regulatory proteins target the initiation step. Initiation in eukaryotes is a very elaborated process that requires the action of about 30 polypeptides in addition to the ribosomal proteins (Jackson 2005; Pestova et al. 2007). Initiation can be divided into three substeps: (1) recruitment of the small (40S) ribosomal subunit to the 5′ end of the mRNA, (2) scanning along the 5′ UTR and initiator AUG recognition, and (3) large (60S) ribosomal subunit joining. Examples of regulation at each of these steps are discussed below.
CAP-DEPENDENT MECHANISMS
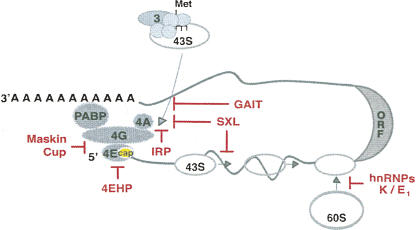
During their synthesis in the nucleus, mRNAs are endowed with a m7GpppN (m7G) cap structure at their 5′ end. In addition to promoting splicing, nuclear export, and stability, the cap is required for translation of nearly all mRNAs. Some transcripts, however, can initiate translation in a cap-independent fashion making use of RNA structures called internal ribosome entry sites (IRESs) (Stoneley and Willis 2004; Jackson 2005). The cap is recognized by the cap-binding complex eIF4F, composed of the eukaryotic translation initiation factors (eIFs) eIF4E, eIF4G, and eIF4A (Fig. 1). eIF4E directly binds to the cap, while eIF4G serves as a scaffolding protein that binds to the other components of the complex and, at least in higher eukaryotes, to eIF3. Because eIF3, in turn, binds to the small ribosomal subunit, these set of interactions are thought to bridge the 43S complex (an assembly of the small ribosomal subunit with eIF3 and other initiation factors) to the mRNA (Fig. 1). eIF4G also binds to poly(A) binding protein (PABP), promoting the formation of a cap–eIF4E–eIF4G–PABP–poly(A) complex that results in mRNA pseudo-circularization and is thought to underline the translational synergy between the cap and the poly(A) tail (for review, see Kahvejian et al. 2001). The formation of an mRNA “closed loop” provides a physical framework for the action of 3′ UTR binding proteins on translation initiation at the 5′ end. Indeed, such a closed loop seems to be required for the function of some inhibitory complexes that bind to the 3′ UTR, including micro-ribonucleoprotein particles (miRNPs) (Mazumder et al. 2001; Humphreys et al. 2005; Wang et al. 2006; Wakiyama et al. 2007).
FIGURE 1.
Translation initiation and its regulation by RNA-binding proteins. The translation initiation pathway and factors are depicted in gray (see text for details). The term eIF has been omitted for simplicity. Only the translation factors mentioned in this review are shown. The regulators and the steps that they inhibit are shown in red. Cap-dependent mechanisms of translational control target the association of eIF4E with the cap structure (yellow oval), the interaction of eIF4E with eIF4G, the interaction of eIF4G with eIF3, or sterically hinder 43S ribosomal recruitment. Cap-independent mechanisms can block 43S recruitment, ribosomal scanning, or 60S subunit joining.
Given the relevance of the cap structure in translation initiation, it is not surprising that a large number of translation regulatory mechanisms target the cap or its associated factors. A variety of strategies are used to interfere with cap function, including blocking the access of eIF4E to the cap, preventing the eIF4E–eIF4G interaction, interfering with the eIF4G–eIF3 interaction, and sterically hindering ribosome recruitment (Fig. 1).
A regulator that blocks the access of the eIF4E to the cap is Drosophila 4EHP (4E homologous protein). 4EHP binds to the cap but cannot interact with eIF4G because it lacks the corresponding interaction site and, thus, acts as a decoy eIF4E (Fig. 1). Alone, 4EHP shows a low affinity for the cap structure and does not repress translation (Zuberek et al. 2007). However, in conjunction with the 3′ UTR binding protein Bicoid, 4EHP inhibits the translation of caudal mRNA at the anterior pole of early Drosophila embryos (Cho et al. 2005). 4EHP is also necessary for the optimal translational repression of maternal hunchback mRNA at the posterior pole. In this case, the complex Pumilio/Nanos/Brain tumor (Pum/Nos/Brat) binds to the 3′ UTR of hunchback, recruiting 4EHP via interactions with Brat (Cho et al. 2006). These translational repression events create opposing gradients of Caudal and Hunchback that are critical to activate axis-specific patterns of gene expression. In the case of hunchback mRNA, an additional mechanism contributes to translational silencing through mRNA deadenylation mediated by Nos (Wreden et al. 1997). Nos and Pum can interact with different subunits of the deadenylase CCR4/NOT, although it is not known whether this deadenylase functions in hunchback repression (Goldstrohm et al. 2006; Kadyrova et al. 2007). The use of multiple mechanisms to ensure appropriate translational regulation of a given mRNA is a recurrent feature in development and highlights the complexity of translation regulatory modes (see below).
Interference with the access of eIF4E to the cap has also been proposed as a mechanism for miRNP-mediated repression. The miRNP component Ago2 contains a cap-binding motif that is required for translational repression when Ago2 is artificially tethered to the 3′ UTR (Kiriakidou et al. 2007). Recently developed in vitro assays are consistent with miRNPs inhibiting translation initiation in a cap- and poly(A)-dependent fashion. However, the true mechanism of translational repression by miRNPs remains controversial, as some studies performed in vivo support a role of miRNPs in repression of post-initiation steps. These controversies have been extensively reviewed in two recent reports and will not be further discussed here (Jackson and Standart 2007; Standart and Jackson 2007, and references therein).
Another strategy to inhibit cap-dependent translation is to block the eIF4E–eIF4G interaction. This approach is used by the so-called 4E-binding proteins (4E-BPs), which bind to eIF4E in the same region that is recognized by eIF4G, preventing the formation of the cap-binding complex. General 4E-BPs, such as 4E-BP1, bind to eIF4E depending on their phosphorylation state and play a pivotal role in the global control of mRNA translation under mitogenic stimulation or stress conditions (Dann and Thomas 2006). Message-specific 4E-BPs have also been described, which, as for 4EHP, are recruited to specific transcripts via 3′ UTR binding proteins. The proteins Cup and Maskin are examples of this type of 4E-BPs (Fig. 1).
Cup represses the translation of oskar and nanos mRNAs, two transcripts that are transported to and localized at the posterior pole of Drosophila oocytes and embryos and that must be silenced prior to their localization (Wilhelm et al. 2003; Nakamura et al. 2004; Nelson et al. 2004; Zappavigna et al. 2004). Cup is recruited to oskar and nanos mRNAs by the 3′ UTR binding proteins Bruno and Smaug, respectively. As for hunchback, additional mechanisms contribute to translational repression of these transcripts, including Bruno-driven mRNA oligomerization and Smaug-dependent deadenylation (Chekulaeva et al. 2006; Jeske et al. 2006; Zaessinger et al. 2006). The examples of hunchback and nanos mRNAs illustrate that deadenylation often cooperates with mechanisms of translational control involving direct interference with initiation factors. The regulation of cyclin B1 mRNA also follows these principles. The 4E-BP protein Maskin represses the translation of cyclin B1 mRNA during late Xenopus oogenesis and is recruited to this transcript by CPEB (cytoplasmic polyadenylation element binding protein), a major regulator of mRNA polyadenylation and translation in vertebrates (Richter 2007). In addition to Maskin, CPEB recruits the deadenylase PARN, which contributes to mRNA silencing by keeping the poly(A) tail short (Fig. 2A; Kim and Richter 2006). Intriguingly, a protein complex containing CPEB, the repressor helicase p54, the vertebrate Cup homolog 4E-T (4E-transporter), and the eIF4E family member 4E1b, among other proteins, has been detected in early Xenopus oocytes (Minshall et al. 2007). As described for 4EHP, 4E1b shows low affinity for the cap structure and for eIF4G, suggesting that mechanisms combining specific 4E-BPs with decoy isoforms of eIF4E may operate for translational repression.
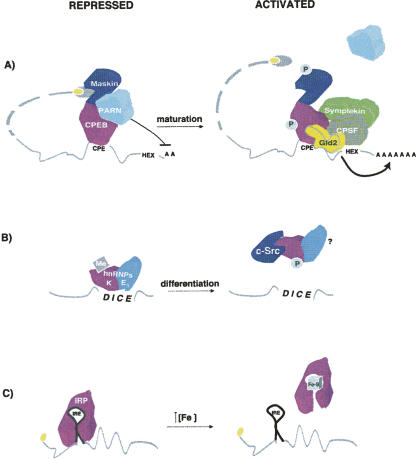
FIGURE 2.
Reversion of translational repression. mRNAs in their repressed and activated states are shown, as well as the signals leading to activation. (A) Translational activation by remodeling of the repressed RNP. In immature Xenopus oocytes, CPEB recruits a repressor complex composed of the 4E-BP Maskin and the deadenylase PARN. Upon progesterone stimulation, phosphorylation of CPEB allows the dissociation of PARN and the productive polyadenylation by a complex composed of symplekin, CPSF and Gld2. The poly(A) tail then recruits PABP, which binds to eIF4G, resulting in Maskin displacement. Maskin phosphorylation also contributes to de-repression. For simplicity, the polyadenylation complex is depicted bound to the mRNA only in maturing oocytes (see text for details). (B) Removal of the repressor by phosphorylation. hnRNP K is phosphorylated by c-Src, leading to its dissociation from the DICE element in LOX 3′ UTR. Methylation (Me) of hnRNP K inhibits its association with c-Src. (C) Binding of IRP to the small molecule cofactor [4Fe–4S] prevents its interaction with the IRE element in the 5′ UTR of ferritin mRNA.
The interaction of eIF4G with eIF3 can also be a target of regulation. Ceruloplasmin mRNA translation is repressed during inflammation by the binding of the GAIT (IFN-γ activated inhibitor of translation) complex to its 3′ UTR. GAIT is composed of four proteins: GAPDH, NSAP1, GluProRS, and the large ribosomal subunit protein L13a (Sampath et al. 2004). Translational repression seems to require the formation of a transcript closed loop in order to place the GAIT complex in proximity to the 5′ end (Mazumder et al. 2001). L13a then binds to eIF4G, impeding its interaction with eIF3 and blocking 43S complex recruitment (Kapasi et al. 2007).
The iron regulatory proteins (IRPs) use an alternative mechanism to prevent 43S recruitment without affecting the association of the eIF4F complex with the mRNA. Under conditions of low iron, IRP binds to a cap-proximal stem–loop in the 5′ UTR of ferritin mRNA and sterically hinders 43S recruitment (Muckenthaler et al. 1998). The finding that RNA-binding proteins with no role in translation can become repressors when bound to structures located within ∼40 nucleotides of the cap supports a steric mode of inhibition (Stripecke et al. 1994).
CAP-INDEPENDENT MECHANISMS
Cap-independent mechanisms of translational control are those that occur efficiently in the absence of the m7G cap structure. In this section, we will describe mechanisms that target translation initiation steps after eIF4F formation. We will not discuss regulation of IRES- dependent translation by UTR-binding proteins, as this type of translational control is mechanistically poorly understood.
Translational repression of msl-2 mRNA by Sex-lethal (SXL) is an essential regulatory step of X-chromosome dosage compensation in Drosophila. SXL binds to both the 5′ and 3′ UTRs of msl-2 and inhibits translation by a double-block mechanism: 3′-bound SXL inhibits the recruitment of the 43S ribosomal complex while 5′-bound SXL blocks the scanning of those complexes that presumably have escaped the 3′ UTR-mediated control (Fig. 1; Beckmann et al. 2005). Translational repression requires that SXL recruits the protein UNR to the 3′ UTR of msl-2 (Abaza et al. 2006; Duncan et al. 2006) and is equally efficient when the canonical m7GpppN cap at the 5′ end of the mRNA is substituted by an ApppN cap, which does not support eIF4E binding (Gebauer et al. 2003). How inhibition of 43S recruitment and scanning occurs is an open question, but the fact that a m7G cap is not essential suggests that eIF4E is not a target.
LOX (15-lipoxygenase) mRNA translation is repressed in erythroid precursor cells by the binding of hnRNPs K and E1 to its 3′ UTR. Sucrose gradient and toe-print analysis showed that hnRNP K/E1 inhibit the joining of the 60S ribosomal subunit to the 40S subunit placed at the initiation codon (Fig. 1; Ostareck et al. 2001). Repression occurs efficiently when translation is driven by the EMCV (encephalomyocarditis virus) or CSFV (classical swine fever virus) IRESs, indicating that LOX mRNA is repressed in a cap-independent fashion (Ostareck et al. 2001). The specific factors targeted for inhibition are unknown.
REVERSIBLE TRANSLATIONAL REPRESSION
Translational regulation is often regarded as a rapid and “reversible” way to control gene expression. However, although reversibility is an essential component of many translational regulatory mechanisms, not all mechanisms are reversible. For example, msl-2 translational repression is thought to persist throughout life in every tissue of female flies.
Relatively little is known about mechanisms that reverse translational repression. In most available examples, relief from translation inhibition is achieved by remodeling of the repressed RNP or, more often, by removal of the repressor from the target mRNA (Fig. 2). Strategies for removal include phosphorylation of the repressor and binding of the regulator to small molecules or to activator factors.
One of the best examples of de-repression by RNP remodeling is that of Xenopus cyclin B1 mRNA. As mentioned above, translation of this transcript is repressed by a complex nucleated by CPEB on the 3′ UTR that contains Maskin and PARN. Two phosphorylation events are thought to be responsible for the full translational activation of cyclin B1 during oocyte maturation. On one hand, phosphorylation of CPEB by Aurora A causes the dissociation of PARN, allowing productive polyadenylation by a cytoplasmic complex composed of the scaffolding protein symplekin, CPSF (cleavage and polyadenylation specificity factor), and the cytoplasmic poly(A) polymerase Gld2 (Fig. 2A; Barnard et al. 2004; Kim and Richter 2006). Basal levels of this complex can be found associated to CPEB in immature oocytes, but association increases upon maturation (Mendez et al. 2000; Barnard et al. 2004). Importantly, CPEB is a necessary factor for polyadenylation (Hake and Richter 1994). Elongation of the poly(A) tail then allows recruitment of PABP, which binds to eIF4G and displaces Maskin from eIF4E (Cao and Richter 2002). On the other hand, phosphorylation of Maskin by cdk1 contributes to its dissociation from eIF4E (Barnard et al. 2005). Cycles of phosphorylation and dephosphorylation of Maskin by cdk1 and calcineurin, respectively, combined with phosphorylation and dephosphorylation of CPEB direct the cyclic production of cyclin B1 in extracts that mimic the mitotic cell cycles of the early Xenopus embryo (Groisman et al. 2002; Cao et al. 2006). Thus, CPEB is an example of a regulator that can behave as a repressor or an activator depending on its phosphorylation status, revealing yet another level of complexity in translational regulation.
Phosphorylation also plays an important role in other forms of de-repression. In order to prevent LOX mRNA translation, hnRNP K binds to the 3′ UTR-located differentiation control element (DICE) through its KH domain 3. A tyrosine residue within this domain (Y458) is critical for DICE binding (Messias et al. 2006). Phosphorylation of Y458 by c-Src impairs DICE binding and translation inhibition in vitro and in Hela cells (Fig. 2B; Ostareck-Lederer et al. 2002; Messias et al. 2006). Activation of c-Src is mediated by direct binding to hnRNP K (Adolph et al. 2007). Thus, hnRNP K is both an activator and a substrate of c-Src in these systems. Interestingly, arginine dimethylation of hnRNP K by PRMT1 prevents its interaction with c-Src, suggesting that this modification could be critical to regulate the timing of LOX mRNA de-repression (Ostareck-Lederer et al. 2006). Deciphering whether this mechanism operates during red blood cell differentiation, in addition to Hela cells and reticulocyte lysates, awaits the establishment of the appropriate experimental system.
The regulator IRP leads a double life. Under conditions of low iron, it behaves as a RNA-binding protein that recognizes specific stem–loops called iron responsive elements (IREs) in the 5′ or 3′ UTRs of mRNAs encoding factors involved in iron homeostasis (Wallander et al. 2006). As explained above, binding of IRP to the 5′ IRE of ferritin mRNA represses translation. Under conditions of high iron, IRP binds to a cubic [4Fe–4S] cluster and shows cytoplasmic aconitase activity (Fig. 2C). The two functions of IRP, translational repression and aconitase, are mutually exclusive. Cysteine 437 of IRP is particularly critical for RNA binding and is essential to anchor the iron–sulphur cluster (Hirling et al. 1994). Consistently, binding of IRP to the [4Fe–4S] cluster results in the loss of affinity for RNA and the subsequent translation of ferritin mRNA (Hirling et al. 1994).
Reversible translational repression is a common theme in mRNA localization. In order to achieve localized expression, the mRNA is silenced after transcription and is only expressed when it reaches its final cellular destiny. A mechanism for local translational activation has been recently proposed (Zaessinger et al. 2006). Localized expression of Nos is accomplished by the translational repression of the ∼96% nanos mRNA present in the bulk cytoplasm of the early Drosophila embryo versus the translational activation of the ∼4% of posteriorly localized nanos mRNA. Translational repression is carried out by Smaug, which binds to the 3′ UTR of nanos and recruits both the 4E-BP protein Cup and the deadenylase CCR4/NOT (see above). At the posterior pole, translation de-repression requires the posteriorly localized factor Oskar. This protein has been shown to interact with the RNA-binding domain of Smaug in yeast two-hybrid and GST pull-down assays (Dahanukar et al. 1999). In addition, overexpression of Oskar prevents nanos deadenylation and decreases the interaction of nanos with Smaug (Zaessinger et al. 2006). The simplest explanation for these results is that nanos translational de-repression is achieved by Oskar directly inhibiting Smaug binding. Further experiments will be required to test whether this mechanism indeed operates in vivo.
CONCLUDING REMARKS
Recent progress on translational control highlights the complexity and versatility of regulation by RNA-binding proteins. Multistep overlapping mechanisms are often used to keep the mRNA silenced. In addition, a single regulator can exploit different modes of control, sometimes with opposing outcomes, which depend on the binding context and the composition of the ribonucleoprotein particle at the time of binding. Microarray analyses have revealed that regulators bind to multiple mRNAs, usually covering >10% of the transcriptome, leading to networks of post-transcriptional regulation that often include factors involved in the same biological pathway and that constitute genuine post-transcriptional operons (Keene 2007). New small RNA molecules have been uncovered that regulate both the translation and stability of the mRNA. Despite these discoveries, much remains to be learned. Mechanistic understanding on how regulators can affect ribosomal scanning, subunit joining, elongation, or termination is lacking. Except for a few examples, how translational repression is reversed and which are the signaling cascades that connect mRNA-specific translational control with general cellular function are unknown. Ultimately, research on these aspects will help us understand not only how the translation process itself occurs, but also how it is altered in disease.
ACKNOWLEDGMENTS
We thank Nancy Standart for communication of results prior to publication and Robin Wharton for discussions. We also thank Raúl Méndez, Josep Vilardell, Juan Valcárcel, and members of F. Gebauer laboratory for critical reading of this manuscript. Work in our laboratory is supported by grants BFU2006-01874 from the Spanish Ministry of Education and Science (MEC) and 2005SGR00669 from the Department of Universities, Information and Sciences of the Generalitat of Catalunya (DURSI).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.848208.
REFERENCES
- Abaza, I., Coll, O., Patalano, S., Gebauer, F. Drosophila UNR is required for translational repression of male-specific lethal 2 mRNA during regulation of X-chromosome dosage compensation. Genes & Dev. 2006;20:380–389. doi: 10.1101/gad.371906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolph, D., Flach, N., Mueller, K., Ostareck, D.H., Ostareck-Lederer, A. Deciphering the cross-talk between hnRNP K and c-Src: The c-Src activation domain in hnRNP K is distinct from a second interaction site. Mol. Cell. Biol. 2007;27:1758–1770. doi: 10.1128/MCB.02014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard, D.C., Ryan, K., Manley, J.L., Richter, J.D. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Barnard, D.C., Cao, Q., Richter, J.D. Differential phosphorylation controls Maskin association with eukaryotic translation initiation factor 4E and localization on the mitotic apparatus. Mol. Cell. Biol. 2005;25:7605–7615. doi: 10.1128/MCB.25.17.7605-7615.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann, K., Grskovic, M., Gebauer, F., Hentze, M.W. A dual inhibitory mechanism restricts msl-2 mRNA translation for dosage compensation in Drosophila . Cell. 2005;122:529–540. doi: 10.1016/j.cell.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Cao, Q., Richter, J.D. Dissolution of the maskin-eIF4E complex by cytoplasmic polyadenylation and poly(A)-binding protein controls cyclin B1 mRNA translation and oocyte maturation. EMBO J. 2002;21:3852–3862. doi: 10.1093/emboj/cdf353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Q., Kim, J.H., Richter, J.D. CDK1 and calcineurin regulate Maskin association with eIF4E and translational control of cell cycle progression. Nat. Struct. Mol. Biol. 2006;13:1128–1134. doi: 10.1038/nsmb1169. [DOI] [PubMed] [Google Scholar]
- Chekulaeva, M., Hentze, M.W., Ephrussi, A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124:521–533. doi: 10.1016/j.cell.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Cho, P.F., Poulin, F., Cho-Park, Y.A., Cho-Park, I.B., Chicoine, J.D., Lasko, P., Sonenberg, N. A new paradigm for translational control: Inhibition via 5′–3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell. 2005;121:411–423. doi: 10.1016/j.cell.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Cho, P.F., Gamberi, C., Cho-Park, Y.A., Cho-Park, I.B., Lasko, P., Sonenberg, N. Cap-dependent translational inhibition establishes two opposing morphogen gradients in Drosophila embryos. Curr. Biol. 2006;16:2035–2041. doi: 10.1016/j.cub.2006.08.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahanukar, A., Walker, J.A., Wharton, R.P. Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila . Mol. Cell. 1999;4:209–218. doi: 10.1016/s1097-2765(00)80368-8. [DOI] [PubMed] [Google Scholar]
- Dann, S.G., Thomas, G. The amino acid sensitive TOR pathway from yeast to mammals. FEBS Lett. 2006;580:2821–2829. doi: 10.1016/j.febslet.2006.04.068. [DOI] [PubMed] [Google Scholar]
- de Moor, C.H., Meijer, H., Lissenden, S. Mechanisms of translational control by the 3′ UTR in Development and differentiation. Semin. Cell Dev. Biol. 2005;16:49–58. doi: 10.1016/j.semcdb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Duncan, K., Grskovic, M., Strein, C., Beckmann, K., Niggeweg, R., Abaza, I., Gebauer, F., Wilm, M., Hentze, M.W. Sex-lethal imparts a sex-specific function to UNR by recruiting it to the msl-2 mRNA 3′ UTR: Translational repression for dosage compensation. Genes & Dev. 2006;20:368–379. doi: 10.1101/gad.371406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer, F., Gskovic, M., Hentze, M. Drosophila Sex-lethal inhibits the stable association if the 40S ribosomal subunit with msl-2 mRNA. Mol. Cell. 2003;11:1397–1404. doi: 10.1016/s1097-2765(03)00176-x. [DOI] [PubMed] [Google Scholar]
- Goldstrohm, A.C., Hook, B.A., Seay, D.J., Wickens, M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- Groisman, I., Jung, M.Y., Sarkissian, M., Cao, Q., Richter, J.D. Translational control of the embryonic cell cycle. Cell. 2002;109:473–483. doi: 10.1016/s0092-8674(02)00733-x. [DOI] [PubMed] [Google Scholar]
- Hake, L.E., Richter, J.D. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell. 1994;79:617–627. doi: 10.1016/0092-8674(94)90547-9. [DOI] [PubMed] [Google Scholar]
- Hentze, M.W., Gebauer, F., Preiss, T. Cis-regulatory sequences and trans-acting factors in translational control. In: Mathews M.B., editor. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 269–295. [Google Scholar]
- Hirling, H., Henderson, B.R., Kuhn, L.C. Mutational analysis of the [4Fe–4S]-cluster converting iron regulatory factor from its RNA-binding form to cytoplasmic aconitase. EMBO J. 1994;13:453–461. doi: 10.1002/j.1460-2075.1994.tb06280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys, D.T., Westman, B.J., Martin, D.I., Preiss, T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc. Natl. Acad. Sci. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, R.J. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem. Soc. Trans. 2005;33:1231–1241. doi: 10.1042/BST0331231. [DOI] [PubMed] [Google Scholar]
- Jackson, R.J., Standart, N. How do microRNAs regulate gene expression? Sci. STKE. 2007;367:re1. doi: 10.1126/stke.3672007re1. [DOI] [PubMed] [Google Scholar]
- Jeske, M., Meyer, S., Temme, C., Freudenreich, D., Wahle, E. Rapid ATP-dependent deadenylation of Nanos mRNA in a cell-free system from Drosophila embryos. J. Biol. Chem. 2006;281:25124–25133. doi: 10.1074/jbc.M604802200. [DOI] [PubMed] [Google Scholar]
- Kadyrova, L.Y., Habara, Y., Lee, T.H., Wharton, R.P. Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development. 2007;134:1519–1527. doi: 10.1242/dev.002212. [DOI] [PubMed] [Google Scholar]
- Kahvejian, A., Roy, G., Sonenberg, N. The mRNA closed-loop model: The function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harb. Symp. Quant. Biol. 2001;66:293–300. doi: 10.1101/sqb.2001.66.293. [DOI] [PubMed] [Google Scholar]
- Kapasi, P., Chaudhuri, S., Vyas, K., Baus, D., Komar, A.A., Fox, P.L., Merrick, W.C., Mazumder, B. L13a blocks 48S assembly: Role of a general initiation factor in mRNA-specific translational control. Mol. Cell. 2007;25:113–126. doi: 10.1016/j.molcel.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene, J.D. RNA regulons: Coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Kim, J.H., Richter, J.D. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell. 2006;24:173–183. doi: 10.1016/j.molcel.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Kiriakidou, M., Tan, G.S., Lamprinaki, S., de Planell-Saguer, M., Nelson, P.T., Mourelatos, Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Kozma, S.C., Um, S.H., Thomas, G. Translational control in metabolic diseases: The role of mTOR signaling in obesity and diabetes. In: Mathews M.B., et al., editors. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 459–483. [Google Scholar]
- Mamane, Y., Petroulakis, E., LeBacquer, O., Sonenberg, N. mTOR, translation initiation and cancer. Oncogene. 2006;25:6416–6422. doi: 10.1038/sj.onc.1209888. [DOI] [PubMed] [Google Scholar]
- Mazumder, B., Seshadri, V., Imataka, H., Sonenberg, N., Fox, P.L. Translational silencing of ceruloplasmin requires the essential elements of mRNA circularization: Poly(A) tail, poly(A)-binding protein, and eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 2001;21:6440–6449. doi: 10.1128/MCB.21.19.6440-6449.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez, R., Murthy, K.G., Ryan, K., Manley, J.L., Richter, J.D. Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol. Cell. 2000;6:1253–1259. doi: 10.1016/s1097-2765(00)00121-0. [DOI] [PubMed] [Google Scholar]
- Messias, A.C., Harnisch, C., Ostareck-Lederer, A., Sattler, M., Ostareck, D.H. The DICE-binding activity of KH domain 3 of hnRNP K is affected by c-Src-mediated tyrosine phosphorylation. J. Mol. Biol. 2006;361:470–481. doi: 10.1016/j.jmb.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Minshall, N., Reiter, M.H., Weil, D., Standart, N. CPEB interacts with an ovary-specific eIF4E and 4E-T in early Xenopus oocytes. J. Biol. Chem. 2007;282:37389–37401. doi: 10.1074/jbc.M704629200. [DOI] [PubMed] [Google Scholar]
- Muckenthaler, M., Gray, N.K., Hentze, M.W. IRP-1 binding to ferritin mRNA prevents the recruitment of the small ribosomal subunit by the cap-binding complex eIF4F. Mol. Cell. 1998;2:383–388. doi: 10.1016/s1097-2765(00)80282-8. [DOI] [PubMed] [Google Scholar]
- Nakamura, A., Sato, K., Hanyu-Nakamura, K. Drosophila Cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev. Cell. 2004;6:69–78. doi: 10.1016/s1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]
- Nelson, M.R., Leidal, A.M., Smibert, C.A. Drosophila cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression. EMBO J. 2004;23:150–159. doi: 10.1038/sj.emboj.7600026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck, D.H., Ostareck-Lederer, A., Shatsky, I.N., Hentze, M.W. Lipoxygenase mRNA silencing in erythroid differentiation: The 3′ UTR regulatory complex controls 60S ribosomal subunit joining. Cell. 2001;104:281–290. doi: 10.1016/s0092-8674(01)00212-4. [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer, A., Ostareck, D.H., Cans, C., Neubauer, G., Bomsztyk, K., Superti-Furga, G., Hentze, M.W. c-Src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Mol. Cell. Biol. 2002;22:4535–4543. doi: 10.1128/MCB.22.13.4535-4543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck-Lederer, A., Ostareck, D.H., Rucknagel, K.P., Schierhorn, A., Moritz, B., Huttelmaier, S., Flach, N., Handoko, L., Wahle, E. Asymmetric arginine dimethylation of heterogeneous nuclear ribonucleoprotein K by protein–arginine methyltransferase 1 inhibits its interaction with c-Src. J. Biol. Chem. 2006;281:11115–11125. doi: 10.1074/jbc.M513053200. [DOI] [PubMed] [Google Scholar]
- Pestova, T.V., Lorsh, J.R., Hellen, C.U.T. The mechanism of translation initiation in eukaryotes. In: Mathews M.B., et al., editors. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 87–128. [Google Scholar]
- Piccioni, F., Zappavigna, V., Verrotti, A.C. Translational regulation during oogenesis and early development: The cap-poly(A) tail relationship. C. R. Biol. 2005;328:863–881. doi: 10.1016/j.crvi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Richter, J.D. CPEB: A life in translation. Trends Biochem. Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Sampath, P., Mazumder, B., Seshadri, V., Gerber, C.A., Chavatte, L., Kinter, M., Ting, S.M., Dignam, J.D., Kim, S., Driscoll, D.M., et al. Noncanonical function of glutamyl-prolyl-tRNA synthetase: Gene-specific silencing of translation. Cell. 2004;119:195–208. doi: 10.1016/j.cell.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Standart, N., Jackson, R.J. MicroRNAs repress translation of m7Gppp-capped target mRNAs in vitro by inhibiting initiation and promoting deadenylation. Genes & Dev. 2007;21:1975–1982. doi: 10.1101/gad.1591507. [DOI] [PubMed] [Google Scholar]
- Stoneley, M., Willis, A.E. Cellular internal ribosome entry segments: Structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- Stripecke, R., Oliveira, C.C., McCarthy, J.E., Hentze, M.W. Proteins binding to 5′ untranslated region sites: A general mechanism for translational regulation of mRNAs in human and yeast Cells. Mol. Cell. Biol. 1994;14:5898–5909. doi: 10.1128/mcb.14.9.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, B., Wickens, M., Kimble, J. Translational control in development. In: Mathews M.B., et al., editors. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 507–544. [Google Scholar]
- Wakiyama, M., Takimoto, K., Ohara, O., Yokoyama, S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes & Dev. 2007;21:1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallander, M.L., Leibold, E.A., Eisenstein, R.S. Molecular control of vertebrate iron homeostasis by iron regulatory proteins. Biochim. Biophys. Acta. 2006;1763:668–689. doi: 10.1016/j.bbamcr.2006.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B., Love, T.M., Call, M.E., Doench, J.G., Novina, C.D. Recapitulation of short RNA-directed translational gene silencing in vitro. Mol. Cell. 2006;22:553–560. doi: 10.1016/j.molcel.2006.03.034. [DOI] [PubMed] [Google Scholar]
- Wiemer, E.A. The role of microRNAs in cancer: No small matter. Eur. J. Cancer. 2007;43:1529–1544. doi: 10.1016/j.ejca.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Wilhelm, J.E., Smibert, C.A. Mechanisms of translational regulation in Drosophila . Biol. Cell. 2005;97:235–252. doi: 10.1042/BC20040097. [DOI] [PubMed] [Google Scholar]
- Wilhelm, J.E., Hilton, M., Amos, Q., Henzel, W.J. Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz. J. Cell Biol. 2003;163:1197–1204. doi: 10.1083/jcb.200309088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wreden, C., Verrotti, A.C., Schisa, J.A., Lieberfarb, M.E., Strickland, S. Nanos and Pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development. 1997;124:3015–3023. doi: 10.1242/dev.124.15.3015. [DOI] [PubMed] [Google Scholar]
- Zaessinger, S., Busseau, I., Simonelig, M. Oskar allows Nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development. 2006;133:4573–4583. doi: 10.1242/dev.02649. [DOI] [PubMed] [Google Scholar]
- Zalfa, F., Achsel, T., Bagni, C. mRNPs, polysomes or granules: FMRP in neuronal protein synthesis. Curr. Opin. Neurobiol. 2006;16:265–269. doi: 10.1016/j.conb.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Zappavigna, V., Piccioni, F., Villaescusa, J.C., Verrotti, A.C. Cup is a nucleocytoplasmic shuttling protein that interacts with the eukaryotic translation initiation factor 4E to modulate Drosophila ovary development. Proc. Natl. Acad. Sci. 2004;101:14800–14805. doi: 10.1073/pnas.0406451101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuberek, J., Kubacka, D., Jablonowska, A., Jemielity, J., Stepinski, J., Sonenberg, N., Darzynkiewicz, E. Weak binding affinity of human 4EHP for mRNA cap analogs. RNA. 2007;13:691–697. doi: 10.1261/rna.453107. [DOI] [PMC free article] [PubMed] [Google Scholar]