Abstract
Translational repression of male-specific-lethal 2 (msl-2) mRNA by Sex-lethal (SXL) is an essential regulatory step of X chromosome dosage compensation in Drosophila. Translation inhibition requires that SXL recruits the protein upstream of N-ras (UNR) to the 3′ UTR of msl-2 mRNA. UNR is a conserved, ubiquitous protein that contains five cold-shock domains (CSDs). Here, we dissect the domains of UNR required for translational repression and complex formation with SXL and msl-2 mRNA. Using gel-mobility shift assays, the domain involved in interactions with SXL and msl-2 was mapped specifically to the first CSD (CSD1). Indeed, excess of a peptide containing this domain derepressed msl-2 translation in vitro. The CSD1 of human UNR can also form a complex with SXL and msl-2. Comparative analyses of the CSDs of the Drosophila and human proteins together with site-directed mutagenesis experiments revealed that three exposed residues within CSD1 are required for complex formation. Tethering assays showed that CSD1 is not sufficient for translational repression, indicating that UNR binding to SXL and msl-2 can be distinguished from translation inhibition. Repression by tethered UNR requires residues from both the amino-terminal Q-rich stretch and the two first CSDs, indicating that the translational effector domain of UNR resides within the first 397 amino acids of the protein. Our results identify domains and residues required for UNR function in translational control.
Keywords: msl-2, SXL, UNR, dosage compensation, translational control
INTRODUCTION
Translational control is widely used in development to regulate processes such as embryonic patterning, cell differentiation, synaptic plasticity, sex determination, and dosage compensation (for review, see Kuersten and Goodwin 2003; Wilhelm and Smibert 2005; Thompson et al. 2007). Dosage compensation is the process that equalizes the expression of X-linked genes in those organisms in which sex determination relies on highly dimorphic sex chromosomes (for review, see Larsson and Meller 2006; Straub and Becker 2007). In Drosophila, dosage compensation is achieved by increasing the transcriptional output of the single male X chromosome by approximately twofold, as a result of the activity of a ribonucleoprotein assembly known as the dosage compensation complex (DCC) or male-specific-lethal (MSL) complex (Hamada et al. 2005; Straub et al. 2005; Mendjan and Akhtar 2007). The DCC fails to assemble in females because the expression of one of its subunits, the protein MSL2, is blocked (Kelley and Kuroda 1995). The female-specific RNA-binding protein Sex-lethal (SXL) prevents msl-2 expression via a dual mechanism that includes the inhibition of the splicing of a facultative intron in the 5′ UTR of msl-2 pre-mRNA, and the subsequent translational repression of the unspliced message (Bashaw and Baker 1997; Kelley et al. 1997; Gebauer et al. 1998). Translational repression requires SXL binding to specific U-rich sequences in both the 5′ and 3′ UTRs of msl-2 mRNA. SXL binding to the 3′ UTR is thought to inhibit the recruitment of the small ribosomal subunit to the mRNA, while SXL binding to the 5′ UTR blocks the scanning toward the AUG initiation codon of those subunits that presumably have escaped control through the 3′ UTR (Beckmann et al. 2005). How SXL inhibits these steps of translation initiation is unknown. Recently, a factor necessary for SXL-mediated translational repression has been identified as the protein upstream of N-ras (UNR) (Abaza et al. 2006; Duncan et al. 2006). UNR is a conserved, ubiquitous protein that is recruited to the 3′ UTR of msl-2 by SXL, but its mechanism of action remains obscure.
Most of the current knowledge about UNR derives from mammalian systems. Human UNR (hUNR) is involved in c-fos mRNA destabilization and the translational repression of pabp mRNA (Chang et al. 2004; Patel et al. 2005). In both cases, UNR interacts with PABP within complexes that bind to distinct regions in the target transcripts. Mammalian UNR also regulates translation driven by the internal ribosome entry sites (IRESs) of a number of viral and cellular transcripts, including rhinovirus, poliovirus, c-myc, PITSLRE protein kinase, the pro-apoptotic factor Apaf-1, and UNR itself (Hunt et al. 1999; Boussadia et al. 2003; Evans et al. 2003; Mitchell et al. 2003; Brown and Jackson 2004; Dormoy-Raclet et al. 2005; Tinton et al. 2005; Schepens et al. 2007). At least in the case of Apaf-1, hUNR acts as an RNA chaperone, changing the conformation of the IRES to make it accessible to the activator PTB and, ultimately, the ribosome (Mitchell et al. 2003). RNA binding by hUNR is mediated by its five cold-shock domains (CSDs), an ancient β-barrel fold containing RNP1 and RNP2 motifs (Brown and Jackson 2004). Drosophila UNR (dUNR) contains an additional Q-rich amino terminus that is absent in its mammalian counterpart (Abaza et al. 2006; Duncan et al. 2006).
In order to gain insight into the mechanism of msl-2 translational repression, we have dissected the functional domains of dUNR. We show that dUNR interacts with SXL and msl-2 mRNA via a dedicated CSD, CSD1. Mutational analysis indicates that three residues exposed outside of the β-barrel drive these interactions. However, tethering experiments show that CSD1 is not sufficient for translational repression, indicating that additional elements are involved in translational control. The translational effector domain of dUNR is contained within the first 397 amino acids of the protein, and requires the cooperation of residues from the amino-terminal Q-rich stretch with the two first CSDs. Our results suggest that the lack of the Q-rich domain in mammalian UNR, together with differences in the CSDs, contribute to the poor translational repression of msl-2 observed in mammalian extracts.
RESULTS
dUNR interacts with SXL and msl-2 mRNA via CSD1
Translational repression of msl-2 requires the formation of a SXL:dUNR complex on the 3′ UTR of the transcript (Abaza et al. 2006; Duncan et al. 2006). Both proteins bind to sites located in close proximity within a region referred to as the EF RNA fragment. The interactions of dUNR with SXL and the EF fragment are interdependent. Although dUNR can weakly interact with SXL in the absence of RNA, this interaction is greatly stabilized in the presence of msl-2 mRNA. In addition, no interaction of dUNR with the mRNA is detected in the absence of SXL (Abaza et al. 2006). Thus, interactions between dUNR and SXL can be monitored by the formation of a tripartite complex on EF RNA, which can be visualized as a super-shift of the SXL:EF RNA complex in gel-mobility shift assays (EMSA) (Abaza et al. 2006).
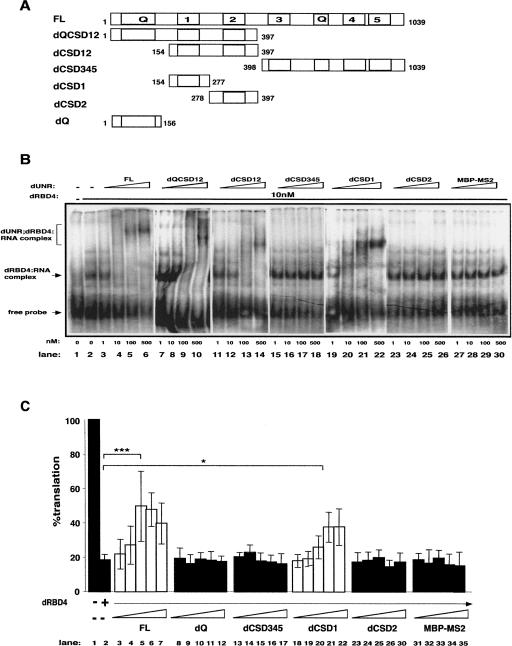
To determine the domain(s) of dUNR that interacts with SXL and msl-2 mRNA, a series of dUNR deletion derivatives was generated (Fig. 1A), their quality assessed by PAGE followed by Coomasie staining (Supplemental Fig. 1A), and their binding to labeled EF RNA tested by EMSA (Fig. 1B). The assays were performed in the presence of a SXL fragment that is fully functional in translational repression (dRBD4) (Grskovic et al. 2003). A complex was observed when nanomolar concentrations of dRBD4 were added to the RNA (Fig. 1B, lanes 1,2). This complex was super-shifted by titration of full-length dUNR (Fig. 1B, FL, lanes 3–6), but not by the unrelated protein MBP-MS2 (Fig. 1B, lanes 27–30). Addition of dUNR fragments covering the C-terminal two thirds of the protein did not change the mobility of the dRBD4:RNA complex (Fig. 1B, lanes 15–18,23–26). However, titration of fragments containing dCSD1, either alone or in combination with other domains, induced super-shifts of the dRBD4:EF complex (Fig. 1B, lanes 7–14,19–22). dCSD1 alone retained the specificity of the full-length protein, because it did not bind to EF RNA in the absence of dRBD4 (Supplemental Fig. 2A). In addition, dCSD1 was unable to bind to EF RNA mutant derivatives lacking SXL (EFmut) or dUNR (mut2456) binding sites (Supplemental Fig. 2B,C). These results indicate that CSD1 mediates the interaction of dUNR with SXL and msl-2 mRNA.
FIGURE 1.
dUNR interacts with SXL and msl-2 mRNA via CSD1. (A) Schematic diagram of dUNR and the deletion derivatives used in this study. dUNR contains five cold-shock domains (1–5) and two Glutamine-rich regions (Q). The positions of the first and last amino acid residues of each construct are indicated. (B) Binding of dUNR and its derivatives to msl-2 mRNA. The 3′UTR EF fragment of msl-2, containing the relevant dUNR and SXL-binding sites, was used in EMSA. Recombinant dUNR fragments were expressed as MBP fusions and were added in increasing amounts (indicated at the bottom) in the absence or presence of 10 nM dRBD4. MBP-MS2 was included as a control. Full-length dUNR (FL) was expressed as a His-tagged fusion. The positions of the different complexes are indicated. (C) Excess recombinant dUNR or dCSD1 derepress translation. BLEF mRNA, containing the minimal msl-2 sequences required for translational repression fused to the Firefly luciferase open reading frame, was used as substrate (see Fig. 3A). The minimal sequences consist of 69 nt in the 5′UTR containing SXL-binding site B, and 46 nt in the 3′UTR containing sites E and F (Gebauer et al. 2003). BLEF mRNA was incubated in typical translation reactions in the absence or presence of 15-fold molar excess of dRBD4, and increasing amounts (0.5, 1, 3, 10, and 30 molar excess over dRBD4) of full-length dUNR or its deletion derivatives. MBP-MS2 was carried as a negative control. Firefly luciferase values were corrected for cotranslated Renilla expression. The activity obtained in the absence of recombinant protein was taken as 100%. The confidence between critical values is indicated (FL, P = 0.00015; dCSD1, P = 0.014).
To evaluate the functional significance of the various dUNR fragments in translation inhibition, we first tested the effect of adding them in excess to an in vitro translation reaction programmed with BLEF mRNA. The BLEF transcript contains the ORF of Firefly luciferase fused to the minimal sequences of msl-2 mRNA required for translational repression (see Fig. 1 legend; Fig. 3A, see below). Translation of BLEF was efficiently repressed by dRBD4 (Fig. 1C, cf. lanes 1 and 2). Addition of increasing amounts of full-length dUNR derepressed translation in a dose-dependent manner (lanes 3–7). The same effect was observed when dCSD1 or dUNR fragments containing dCSD1 were added to the reaction (Fig. 1C, lanes 18–22; data not shown). However, addition of dUNR deletion derivatives lacking dCSD1 did not alter translational repression (Fig. 1C, lanes 8–17,23–30), as observed with MBP-MS2 (Fig. 1C, lanes 31–35). Although these data do not rule out a function for other dUNR fragments in translational repression, they highlight the functional relevance of CSD1 and suggest that CSD1 alone can titrate the interaction of dUNR with SXL within the translation repressor complex.
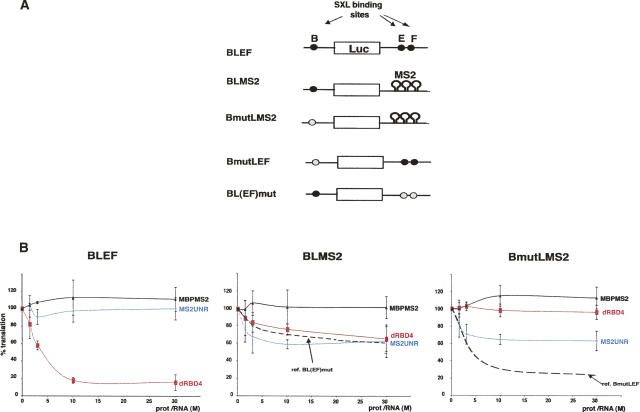
FIGURE 3.
Tethered dUNR represses translation. (A) Schematic diagram of the RNA constructs used in translation assays. BLEF mRNA (see Fig. 1 legend) and derivatives are depicted. Wild-type and mutated SXL-binding sites are shown as black and gray ovals, respectively. Hairpins denote the substitution of the 3′UTR by nine MS2 binding sites. (B) Tethered dUNR represses translation. BLEF, BLMS2, and BmutLMS2 mRNAs were translated in Drosophila embryo extracts in the presence of increasing amounts of either MS2-dUNR (blue lines) or dRBD4 (red lines). MBP-MS2 was used as a negative control (solid black lines). As reference, the translational repression of BL(EF)mut and BmutLEF mRNAs by SXL was tested (dashed black lines). The translation rate was measured as indicated in the legend of Figure 1 and was plotted against the molar ratio of recombinant protein/RNA.
Adding increasing amounts of dRBD4 to a reaction that was derepressed by excess dUNR reinstated translation inhibition, indicating that a given stoichiometry between dUNR and SXL must be preserved for efficient translational repression, and suggesting that SXL and dUNR are the limiting components of the repressor complex assembled on msl-2 3′ UTR (Supplemental Fig. 3).
Residues within CSD1 involved in SXL and msl-2 interactions
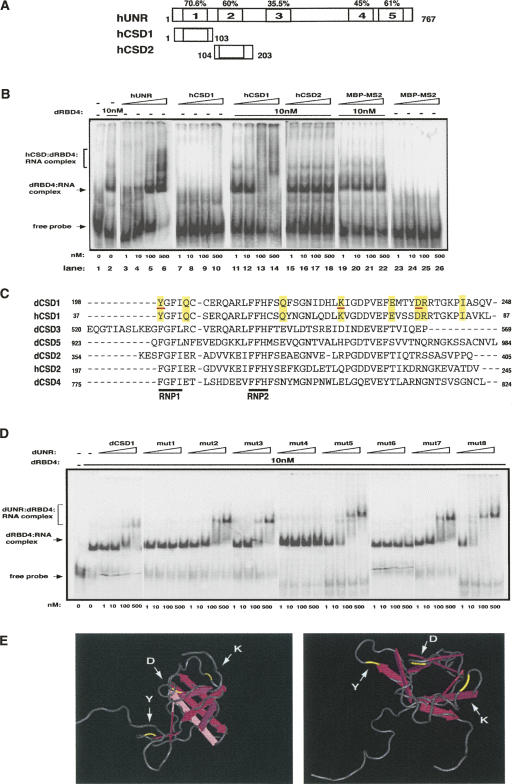
To map more finely the residues within dCSD1 that interact with SXL and msl-2, we took advantage of the mammalian UNR homolog. Human UNR shares an overall identity of 45% with its Drosophila counterpart and lacks Q-rich regions. The conservation rises in the CSDs, being highest for CSD1 (70.6% identity) (Fig. 2A). EMSA assays indicated that, contrary to dUNR, hUNR binds to msl-2 in the absence of dRBD4 (Fig. 2B, lanes 3–6). However, similar to Drosophila, hCSD1 in isolation does not bind to msl-2 but requires dRBD4 (Fig. 2B, cf. lanes 7–10 and 11–14). These effects are specific for hCSD1, because hCSD2 or the unrelated protein MBP-MS2 do not produce a super-shift of the dRBD4:RNA complex (Fig. 2B, lanes 15–22). In addition, although the binding affinity of hCSD1 for the EF RNA seems lower than that of dCSD1, hCSD1 does not bind to EFmut or mut2456 probes, even in the presence of SXL (Supplemental Fig. 2). These data indicate that, similar to Drosophila, hCSD1—but not hCSD2—interacts with SXL. Thus, residues present in CSD1 of UNR from both species, but absent in hCSD2 or the remaining dCSDs should be responsible for the interaction with SXL and msl-2. This group of eight residues is shown in Figure 2C (highlighted in yellow). Individual mutation of each of these residues to alanine and subsequent EMSA analysis of the mutants revealed that three of them were indeed required (Fig. 2D; underlined in red in Fig. 2C). Modeling the position of these residues on the crystal structure of human CSD1 (available at http://www.rcsb.org/pdb/explore.do?structureId=1WFQ) shows that they are likely to be exposed in the outer surface of the β-barrel, at the edges of the cylinder (Fig. 2E, highlighted in yellow).
FIGURE 2.
Residues of dCSD1 involved in SXL and msl-2 interactions. (A) Schematic representation of hUNR and its fragments. The positions of the first and last amino acids of each construct as well as the conservation of each CSD are indicated. (B) The CSD1 of hUNR interacts with SXL. Recombinant proteins were expressed as His-tagged fusions and tested as indicated in the legend of Figure 1. hCSD1 and hCSD2 also contained a MS2-tag. MBP-MS2 was included as a negative control. The positions of the different complexes are indicated. (C) Alignment of CSDs of Drosophila and human UNR. The indicated CSDs were aligned using ClustalW (EBI). The RNP motifs (RNP1 and RNP2) are underlined in black. The amino acids common for dCSD1 and hCSD1, but different for the rest of the CSDs are shadowed in yellow. The amino acids interacting with SXL and msl-2 are underlined in red (see below). (D) Binding of single-point dCSD1 mutants to msl-2. The amino acids shadowed in yellow in C were named 1–8 and were mutated to alanine. Mutant proteins were expressed as MBP fusions and used for EMSA as described in the legend of Figure 1. (E) Stereo-view of hCSD1 showing the position of relevant amino acids. Five anti-parallel β-strands (pink arrows) give an overall β-barrel fold. The SXL and msl-2 interacting residues are depicted in yellow.
The translational repressor domain of dUNR
To identify the domain(s) of dUNR involved in translational repression, we performed functional tethering analysis. dUNR was fused to the 13.7 kDa bacteriophage MS2 coat protein, which binds to a specific stem–loop structure, the MS2 site, with high affinity. The MS2 site was placed in several copies at the 3′ UTR of msl-2 reporters (Fig. 3A). These reporters were based on the BLEF construct (Gebauer et al. 2003) and contained or lacked the SXL binding site B at the 5′ UTR (BLMS2 and BmutLMS2, respectively). Tethering dUNR to the 3′ UTR of msl-2 reporters in this manner allows the analysis of its effects on translation independently of binding to RNA or SXL. As references, we used RNAs lacking MS2-binding sites, but containing SXL-binding sites either at the 5′ UTR, the 3′ UTR, or both [BL(EF)mut, BmutL(EF), and BLEF, respectively] (Fig. 3A). The ability of the MS2-dUNR fusion protein to repress the translation of the reporters was tested in vitro (Fig. 3B). Tethered dUNR repressed the translation of the MS2 reporter mRNAs by 40% (Fig. 3B, middle and right panels, blue lines). This repression was specific because MS2-dUNR did not repress the translation of BLEF (Fig. 3B, left panel, blue line), and because the translation of MS2 reporter mRNAs was not repressed by a different MS2 fusion protein (MBP-MS2, observe the black lines in the middle and right panels). In addition, dRBD4 did not repress the translation of the MS2 reporter lacking SXL-binding sites (Fig. 3B, right panel, red line). dRBD4 repressed translation to a similar level as with the reference BL(EF)mut mRNA when SXL-binding sites were provided in the 5′ UTR, indicating that the MS2 sites placed at the 3′ UTR do not interfere with repression by the 5′ UTR (Fig. 3B, middle panel, cf. red line and discontinuous black line). Furthermore, when dRBD4 and dUNR were added together to a reporter containing binding sites for both, an additive effect was observed (Supplemental Fig. 4). Although repression by tethered dUNR was less efficient than repression by SXL (Fig. 3B, right panel, cf. blue line and dashed black line), it was consistent and specific. These results indicate that tethered dUNR can repress translation in the absence of SXL.
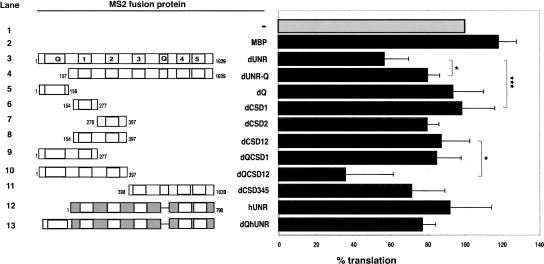
To determine which domains of dUNR are required for translational repression of msl-2, we fused fragments of dUNR to the MS2 coat protein and analyzed their ability to inhibit the translation of BmutLMS2 mRNA (Fig. 4). Interestingly, tethered dCSD1 did not inhibit translation (Fig 4, lane 6), indicating that the translational effector domain can be distinguished from the SXL interaction domain. Moderate translational repression was observed with tethered dCSD345 (Fig 4, lane 11). Deletion of the Q-rich N terminus partially abrogated translational repression (Fig 4, cf. lanes 3 and 4). However, the Q-rich domain alone was unable to repress translation (Fig 4, lane 5). These data suggest that the Q-rich domain contributes to translational repression in combination with other domains. Indeed, robust translation inhibition was only observed when a fragment containing the Q-rich domain and CSDs 1 and 2 was tethered, while these domains in isolation showed at best a weak activity (Fig 4, cf. lane 10 and lanes 5–9). In addition, tethered hUNR did not repress translation, and addition of the Drosophila Q-rich domain did not increase repression significantly (Fig 4, lanes 12,13), suggesting that residues within CSDs 1 and 2 that are specific to the Drosophila protein are involved in repression. Altogether, the data show that the amino-terminal 397 amino acids of dUNR strongly contribute to translational repression.
FIGURE 4.
The translational repressor domain of dUNR is embedded within its amino-terminal 397 amino acids. The translation inhibition of BmLMS2 mRNA by increasing amounts of tethered MS2-UNR derivatives was measured. The result obtained at a 30-fold molar excess of protein to RNA is shown. The confidence between critical values are depicted (lanes 3,4, P = 0.027; lanes 3,6, P = 0.001; lanes 8,10, P = 0.035). Similar results were obtained when BLMS2 mRNA was used (data not shown). A schematic representation of the different constructs is shown at the left. Proteins were expressed as His-tagged MS2 fusions.
DISCUSSION
Inhibition of msl-2 expression is essential for development of female Drosophila flies, since expression of MSL2 causes the assembly of the DCC on both X chromosomes and lethality (Kelley and Kuroda 1995). A complex of SXL and dUNR binds to the 3′ UTR of msl-2 mRNA and plays an essential role in repression (Abaza et al. 2006; Duncan et al. 2006). We have analyzed the functional domains of dUNR and show that interaction with SXL and msl-2 is not sufficient for translational repression. Residues within the amino-terminal fragment containing the Q-rich domain and CSDs 1 and 2 are essential for translation inhibition.
The CSD is a domain highly conserved in evolution used to bind single stranded nucleic acids (Ermolenko and Makhatadze 2002). In addition, the CSD can support protein–protein interactions. Indeed, the CSD1 of Drosophila UNR sustains both binding to msl-2 mRNA and SXL (Figs. 1, 2). Three specific residues within dCSD1 are responsible for these interactions: a tyrosine (Y) that is part of the RNP1 motif, and a lysine (K), and aspartic acid (D), which lay outside the RNP motifs (Fig. 2C). Although the assay we have used does not allow distinction between mRNA and protein binding, the location of these amino acids suggests that Y likely mediates msl-2 binding, while K and D may be involved in SXL interaction. Our data do not formally rule out that other domains of dUNR contribute separately to bind either SXL or msl-2. However, this possibility is unlikely because the efficiency of binding of dCSD1 alone is identical to that of the full-length protein (Fig. 1B). The use of a dedicated CSD for RNA binding contrasts with the known properties of mammalian UNR. All five CSDs of hUNR are required to bind to the rhinovirus IRES (Brown and Jackson 2004). The fact that hUNR can bind to msl-2 mRNA in isolation while dUNR cannot, indeed suggests that the two proteins have different modes of RNA binding.
In order to map the translational effector domain of UNR we performed tethering analysis. Translational repression by tethered dUNR was less efficient than that observed for SXL in its natural context (Fig. 3B), suggesting that SXL function in 3′ UTR-mediated repression is not limited to the recruitment of dUNR. Alternatively, the lesser efficiency of dUNR in repression could be due to aberrant conformation of the recombinant protein or to geometry constraints imposed on the tethered complex. In support for the latter, even though SXL is critical for msl-2 translational repression, it does not function when tethered to the 3′ UTR (Grskovic et al. 2003).
Tethering assays show that dCSD1 is not sufficient for translational repression, indicating that elements in addition to SXL and msl-2 binding are required for inhibition (Fig. 4). These could include the interaction with other corepressors or with components of the translational apparatus. Similar to dCSD1, tethered hUNR could not support translational repression, implying that the translational effector domain is lacking from hUNR. An obvious domain absent in hUNR but present in its Drosophila counterpart is the N-terminal Q-rich domain. This domain contains 52 glutamines interrupted mainly by histidines, resulting in a highly polar stretch suitable for interactions. Certainly, Q-rich domains are present in proteins with diverse roles in gene expression and serve as protein–protein interaction and multimerisation modules (Pascal and Tjian 1991; Emili et al. 1994; Stott et al. 1995; Strom et al. 1996). To test whether the Q-rich domain could confer translational repression, we deleted it from dUNR and fused it to hUNR. dUNR lacking the Q-rich domain repressed translation less efficiently than the intact protein (Fig. 4), indicating that the Q-rich domain was necessary for optimal repression. However, the Q-rich domain did not confer a significant translational repression activity to hUNR, suggesting that residues within the CSDs specific to the Drosophila protein were also relevant. Importantly, the fragment containing the Q-rich domain fused to dCSDs 1 and 2 showed a strong translational repression activity, indicating that the translational effector domain of dUNR is embedded within the first 397 amino acids of the protein. Consistent with these results, analysis of UNR mutant flies indicates that the N-terminal half of UNR exerts robust repression of dosage compensation in females (S. Patalano, Y. Belacortu, N. Paricio, and F. Gebauer, in prep.).
TIA-1, a splicing and translation regulator, contains a Q-rich C-terminal domain that interacts with the protein U1C facilitating the recruitment of the U1 snRNP to the 5′ splice site (Forch et al. 2002). By analogy, the Q-rich domain of dUNR could facilitate the recruitment of corepressors, or components of the translation machinery that are so sequestered, to the 3′ UTR of msl-2. One such component could be PABP. This translation factor has been shown to interact with hUNR in complexes binding to the coding region of c-fos mRNA and the 5′ UTR of pabp mRNA, which are involved in destabilization and translational repression, respectively (Chang et al. 2004; Patel et al. 2005). However, it is not immediately obvious how PABP recruitment to the 3′ UTR of msl-2 would result in repression, because PABP stimulates translation when tethered to the 3′ as it does when it binds to the poly(A) tail (Gray et al. 2000). Furthermore, substantial translational repression by the UNR:SXL complex occurs on nonadenylated msl-2 mRNA (Gebauer et al. 1999; I. Abaza and F. Gebauer, unpubl.). Thus, even though PABP could play a role, additional factors are involved in translational repression by dUNR.
In summary, our data delimit the functional domains of dUNR in msl-2 translational repression. Finding out which factors interact with the translational effector domain will help us to gain insight into the molecular mechanism of translation inhibition by this essential protein.
MATERIALS AND METHODS
Plasmids
BLEF and BL(EF)mut constructs have been previously described (Gebauer et al. 2003). BmutLEF was obtained by exchanging the 5′ UTR of BLEF with a similar segment generated by hybridization of complementary oligonucleotides containing a substitution of site B (T16) by (CT)8. BLMS2 and BmutLMS2 were generated by replacing the 3′ UTRs of BLEF and BmutLEF, respectively, with nine copies of the MS2 binding site obtained from plasmid pLucMS2(9) (Collier et al. 2005).
dUNR deletion derivatives were obtained by PCR amplification of relevant fragments from plasmid pET15b-dUNR (FL, Abaza et al. 2006) and cloned into pMALc. pET21d-hUNR was kindly provided by Anne Willis. Full-length MS2-tagged dUNR and hUNR were generated by PCR amplification of MS2 from pMALc-MS2 (a kind gift from Josep Vilardell, CGR, Barcelona, Spain) and subsequent cloning into the NdeI site of pET15b-dUNR or the NcoI site of pET21d-hUNR, respectively. MS2-UNR deletion derivatives were obtained by PCR amplification of UNR fragments that were cloned into the AgeI and XhoI sites of pET30a-MS2-GFP after releasing the GFP insert (Bertrand et al. 1998). To obtain the dQhUNR construct, the MS2-dQ fragment was PCR amplified from pET30a-MS2-dQ and cloned into the NcoI site of pET21d-hUNR. Constructs containing point mutations in dCSD1 (mut1–mut8) were obtained by site-directed mutagenesis from pMALc-dCSD1.
Protein expression and purification
dRBD4 was expressed in Escherichia coli as an N-terminal GST-tagged fusion and purified as described previously (Grskovic et al. 2003).
Full-length dUNR, hUNR, and MS2 fusion proteins (used in Figs. 2B, 4) were expressed as N-terminal His-tagged fusions and purified following the pET system user's manual (Novagen). Most MS2-fusion proteins were largely insoluble. Thus, except for hCSD1, MS2-fusion proteins were purified on Ni2+ columns under denaturing conditions (6 M Urea) and slowly renatured on the column by step washes of decreasing Urea concentrations. In addition, MS2 fusion dCSD2, dCSD12, dQCDS12, and dCSD345 proteins were further purified on S-agarose beads following the recommendations of the vendor (Novagen). MBP-dUNR deletion derivatives (used in Figs. 1, 2D) were purified following the pMALc system user's manual (New England Biolabs). Appropriate purification of all proteins was assessed by PAGE, followed by Coomasie staining (Supplemental Fig. 1). All proteins were dialyzed against buffer D (20 mM HEPES at pH 8.0, 20% glycerol, 1 mM DTT, 0.2 mM EDTA, 0.01% NP-40).
In vitro translation
In vitro translation reactions in Drosophila embryo extracts were performed as described (Abaza et al. 2006). Renilla mRNA was cotranslated as an internal control. The translation efficiency was determined by measuring the luciferase activity using the dual luciferase assay system (Promega), and Firefly luciferase values were corrected for Renilla expression. For translation competition assays, Drosophila embryo extracts were supplemented with increasing amounts of recombinant full-length dUNR or derivatives prior to assembling the translation reactions.
Gel mobility-shift assays
One femptomole of 32P-labeled msl-2 3′UTR RNA (positions 909–954) containing the relevant SXL and UNR binding sites either in wild-type (EF) (Grskovic et al. 2003) or mutated versions (EFmut and mut2456, respectively) was incubated with increasing amounts of dUNR, hUNR, or their derivatives, and processed as described previously (Valcarcel et al. 1993). Where indicated, 10 nM dRBD4 was included.
SUPPLEMENTAL DATA
Supplemental material can be found at http://pasteur.crg.es/images/FIGURES/Functionaldomains_SupplementaryFigures.pdf.
ACKNOWLEDGMENTS
We are grateful to Drs. Nicola Gray, Anne Willis, Raúl Mendez, and Josep Vilardell for generously providing pLuc-MS2(9), pET21d-hUNR, pET31a-MS2-GFP, and pMALc-MS2 plasmids, respectively. We also thank Juan Valcárcel for carefully reading of this manuscript. I.A. was supported by a FI-IQUC fellowship from the Generalitat of Catalunya. This work was supported by grants BMC203-04108 and BFU2006-01874 from the Spanish Ministry of Education and Science (MEC), and grant 2005SGR00669 from the Department of Universities, Information and Sciences of the Generalitat of Catalunya (DURSI).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.802908.
REFERENCES
- Abaza, I., Coll, O., Patalano, S., Gebauer, F. Drosophila UNR is required for translational repression of male-specific lethal 2 mRNA during regulation of X-chromosome dosage compensation. Genes & Dev. 2006;20:380–389. doi: 10.1101/gad.371906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashaw, G.J., Baker, B.S. The regulation of the Drosophila msl-2 gene reveals a function for Sex-lethal in translational control. Cell. 1997;89:789–798. doi: 10.1016/s0092-8674(00)80262-7. [DOI] [PubMed] [Google Scholar]
- Beckmann, K., Grskovic, M., Gebauer, F., Hentze, M.W. A dual inhibitory mechanism restricts msl-2 mRNA translation for dosage compensation in Drosophila . Cell. 2005;122:529–540. doi: 10.1016/j.cell.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Bertrand, E., Chartrand, P., Schaefer, M., Shenoy, S.M., Singer, R.H., Long, R.M. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- Boussadia, O., Niepmann, M., Creancier, L., Prats, A.C., Dautry, F., Jacquemin-Sablon, H. UNR is required in vivo for efficient initiation of translation from the internal ribosome entry sites of both rhinovirus and poliovirus. J. Virol. 2003;77:3353–3359. doi: 10.1128/JVI.77.6.3353-3359.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, E.C., Jackson, R.J. All five cold-shock domains of UNR (upstream of N-ras) are required for stimulation of human rhinovirus RNA translation. J. Gen. Virol. 2004;85:2279–2287. doi: 10.1099/vir.0.80045-0. [DOI] [PubMed] [Google Scholar]
- Chang, T.C., Yamashita, A., Chen, C.Y., Yamashita, Y., Zhu, W., Durdan, S., Kahvejian, A., Sonenberg, N., Shyu, A.B. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Genes & Dev. 2004;18:2010–2023. doi: 10.1101/gad.1219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier, B., Gorgoni, B., Loveridge, C., Cooke, H.J., Gray, N.K. The DAZL family proteins are PABP-binding proteins that regulate translation in germ cells. EMBO J. 2005;24:2656–2666. doi: 10.1038/sj.emboj.7600738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormoy-Raclet, V., Markovits, J., Jacquemin-Sablon, A., Jacquemin-Sablon, H. Regulation of Unr expression by 5′- and 3′-untranslated regions of its mRNA through modulation of stability and IRES mediated translation. RNA Biol. 2005;2:e27–e35. [PubMed] [Google Scholar]
- Duncan, K., Grskovic, M., Strein, C., Beckmann, K., Niggeweg, R., Abaza, I., Gebauer, F., Wilm, M., Hentze, M.W. Sex-lethal imparts a sex-specific function to UNR by recruiting it to the msl-2 mRNA 3′ UTR: Translational repression for dosage compensation. Genes & Dev. 2006;20:368–379. doi: 10.1101/gad.371406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili, A., Greenblatt, J., Ingles, C.J. Species-specific interaction of the glutamine-rich activation domains of Sp1 with the TATA box-binding protein. Mol. Cell. Biol. 1994;14:1582–1593. doi: 10.1128/mcb.14.3.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolenko, D.N., Makhatadze, G.I. Bacterial cold-shock proteins. Cell. Mol. Life Sci. 2002;59:1902–1913. doi: 10.1007/PL00012513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, J.R., Mitchell, S.A., Spriggs, K.A., Ostrowski, J., Bomsztyk, K., Ostarek, D., Willis, A.E. Members of the poly (rC) binding protein family stimulate the activity of the c-myc internal ribosome entry segment in vitro and in vivo. Oncogene. 2003;22:8012–8020. doi: 10.1038/sj.onc.1206645. [DOI] [PubMed] [Google Scholar]
- Forch, P., Puig, O., Martinez, C., Seraphin, B., Valcarcel, J. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J. 2002;21:6882–6892. doi: 10.1093/emboj/cdf668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer, F., Merendino, L., Hentze, M.W., Valcarcel, J. The Drosophila splicing regulator sex-lethal directly inhibits translation of male-specific-lethal 2 mRNA. RNA. 1998;4:142–150. [PMC free article] [PubMed] [Google Scholar]
- Gebauer, F., Corona, D.F., Preiss, T., Becker, P.B., Hentze, M.W. Translational control of dosage compensation in Drosophila by Sex-lethal: Cooperative silencing via the 5′ and 3′ UTRs of msl-2 mRNA is independent of the poly(A) tail. EMBO J. 1999;18:6146–6154. doi: 10.1093/emboj/18.21.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer, F., Gskovic, M., Hentze, M.W. Drosophila Sex-lethal inhibits the stable association if the 40S ribosomal subunit with msl-2 mRNA. Mol. Cell. 2003;11:1397–1404. doi: 10.1016/s1097-2765(03)00176-x. [DOI] [PubMed] [Google Scholar]
- Gray, N.K., Coller, J.M., Dickson, K.S., Wickens, M. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 2000;19:4723–4733. doi: 10.1093/emboj/19.17.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grskovic, M., Hentze, M.W., Gebauer, F. A co-repressor assembly nucleated by Sex-lethal in the 3′ UTR mediates translational control of Drosophila msl-2 mRNA. EMBO J. 2003;22:5571–5581. doi: 10.1093/emboj/cdg539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada, F.N., Park, P.J., Gordadze, P.R., Kuroda, M.I. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster . Genes & Dev. 2005;19:2289–2294. doi: 10.1101/gad.1343705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, S.L., Hsuan, J.J., Totty, N., Jackson, R.J. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes & Dev. 1999;13:437–448. doi: 10.1101/gad.13.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, R., Wang, J., Bell, L., Kuroda, M. Sex-lethal controls dosage copensation in Drosophila by a nonsplicing mechanism. Nature. 1997;387:195–199. doi: 10.1038/387195a0. [DOI] [PubMed] [Google Scholar]
- Kelley, R.L., Kuroda, M. Equality for X chromosomes. Science. 1995;270:1607–1610. doi: 10.1126/science.270.5242.1607. [DOI] [PubMed] [Google Scholar]
- Kuersten, S., Goodwin, E.B. The power of the 3′ UTR: Translational control and development. Nat. Rev. Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- Larsson, J., Meller, V.H. Dosage compensation, the origin and the afterlife of sex chromosomes. Chromosome Res. 2006;14:417–431. doi: 10.1007/s10577-006-1064-3. [DOI] [PubMed] [Google Scholar]
- Mendjan, S., Akhtar, A. The right dose for every sex. Chromosoma. 2007;116:95–106. doi: 10.1007/s00412-006-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, S.A., Spriggs, K.A., Coldwell, M.J., Jackson, R.J., Willis, A.E. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and UNR. Mol. Cell. 2003;11:757–771. doi: 10.1016/s1097-2765(03)00093-5. [DOI] [PubMed] [Google Scholar]
- Pascal, E., Tjian, R. Different activation domains of Sp1 govern formation of multimers and mediate transcriptional synergism. Genes & Dev. 1991;5:1646–1656. doi: 10.1101/gad.5.9.1646. [DOI] [PubMed] [Google Scholar]
- Patel, G.P., Ma, S., Bag, J. The autoregulatory translational control element of poly(A)-binding protein mRNA forms a heteromeric ribonucleoprotein complex. Nucleic Acids Res. 2005;33:7074–7089. doi: 10.1093/nar/gki1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepens, B., Tinton, S.A., Bruynooghe, Y., Parthoens, E., Haegman, M., Beyaert, R., Cornelis, S. A role for hnRNP C1/C2 and UNR in internal initiation of translation during mitosis. EMBO J. 2007;26:158–169. doi: 10.1038/sj.emboj.7601468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stott, K., Blackburn, J.M., Butler, P.J., Perutz, M. Incorporation of glutamine repeats makes protein oligomerize: Implications for neurodegenerative diseases. Proc. Natl. Acad. Sci. 1995;92:6509–6513. doi: 10.1073/pnas.92.14.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub, T., Becker, P.B. Dosage compensation: The beginning and end of generalization. Nat. Rev. Genet. 2007;8:47–57. doi: 10.1038/nrg2013. [DOI] [PubMed] [Google Scholar]
- Straub, T., Gilfillan, G.D., Maier, V.K., Becker, P.B. The Drosophila MSL complex activates the transcription of target genes. Genes & Dev. 2005;19:2284–2288. doi: 10.1101/gad.1343105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom, A.C., Forsberg, M., Lillhager, P., Westin, G. The transcription factors Sp1 and Oct-1 interact physically to regulate human U2 snRNA gene expression. Nucleic Acids Res. 1996;24:1981–1986. doi: 10.1093/nar/24.11.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, B., Wickens, M., Kimble, J. Translational control in development. In: Mathews M.B., Sonenberg N., Hershey J.W.B., editors. Translational control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 507–544. [Google Scholar]
- Tinton, S.A., Schepens, B., Bruynooghe, Y., Beyaert, R., Cornelis, S. Regulation of the cell-cycle-dependent internal ribosome entry site of the PITSLRE protein kinase: Roles of UNR (upstream of N-ras) protein and phosphorylated translation initiation factor eIF-2α. Biochem. J. 2005;385:155–163. doi: 10.1042/BJ20040963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcarcel, J., Singh, R., Zamore, P.D., Green, M.R. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature. 1993;362:171–175. doi: 10.1038/362171a0. [DOI] [PubMed] [Google Scholar]
- Wilhelm, J.E., Smibert, C.A. Mechanisms of translational regulation in Drosophila . Biol. Cell. 2005;97:235–252. doi: 10.1042/BC20040097. [DOI] [PubMed] [Google Scholar]