Abstract
The process of mRNA localization typically utilizes cis-targeting elements and trans-recognition factors to direct the compartmental organization of translationally suppressed mRNAs. mRNA localization to the endoplasmic reticulum (ER), in contrast, occurs via a co-translational, signal sequence/signal recognition particle (SRP)-dependent mechanism. We have utilized cell fractionation/cDNA microarray analysis, shRNA-mediated suppression of SRP expression, and mRNA reporter construct studies to define the role of the SRP pathway in ER-directed mRNA localization. Cell fractionation studies of mRNA partitioning between the cytosol and ER demonstrated the expected enrichment of cytosolic/nucleoplasmic protein-encoding mRNAs and secretory/integral membrane protein-encoding mRNAs in the cytosol and ER fractions, respectively, and identified a subpopulation of cytosolic/nucleoplasmic protein-encoding mRNAs in the membrane-bound mRNA pool. The latter finding suggests a signal sequence-independent pathway of ER-directed mRNA localization. Extending from these findings, mRNA partitioning was examined in stable SRP54 shRNA knockdown HeLa cell lines. shRNA-directed reductions in SRP did not globally alter mRNA partitioning patterns, although defects in membrane protein processing were observed, further suggesting the existence of multiple pathways for mRNA localization to the ER. ER localization of GRP94-encoding mRNA was observed when translation was disabled by mutation of the start codon/insertion of a 5′UTR stem–loop structure or upon deletion of the encoded signal sequence. Combined, these data indicate that the mRNA localization to the ER can be conferred independent of the signal sequence/SRP pathway and suggest that mRNA localization to the ER may utilize cis-encoded targeting information.
Keywords: mRNA localization, endoplasmic reticulum, signal sequence
INTRODUCTION
mRNA localization is an evolutionarily conserved phenomenon that serves essential roles in cell polarity determination and cell fate specification (St Johnston 2005; Du et al. 2007). Most prominently, mRNA localization provides a mechanism to compartmentalize protein synthesis and thereby instruct the formation of protein gradients within cells (Palacios and St Johnston 2001; St Johnston 2005; Du et al. 2007). The machinery functioning in mRNA localization is both diverse and complex but shares a common paradigm of cis-localization elements (zipcodes), trans-recognition factors, and, frequently, molecular motors (Czaplinski and Singer 2006). The necessity for localized translation of particular mRNAs, in particular those encoding morphogens, necessitates that localized mRNAs are translationally silenced during the trafficking processes that direct them to their proper location, and implies that final localization is coupled to translational de-repression.
In addition to the established roles of mRNA localization in early development, germline and somatic cells direct a genomewide sorting of mRNAs between the cytosol and endoplasmic reticulum (ER) compartments of the cell. This process, a ubiquitous and essential step in the genesis of secretory and integral membrane proteins, differs from previously established mRNA localization mechanisms in both magnitude and mechanism. With respect to magnitude, ER-directed mRNA localization operates on a substantial fraction (30%–40%) of the genome. And whereas established mRNA localization processes traffic translationally silenced ribonucleoprotein particles (RNPs) to their cellular destinations, ER-directed mRNA localization is thought to require both translation and an encoded signal sequence. In this view, signal sequence recognition via the signal recognition particle (SRP) directs mRNA/ribosome/nascent polypeptide complexes to the ER membrane, thereby partitioning secretory/integral membrane protein-encoding mRNAs to the ER, and enables co-translational protein translocation (Lingappa and Blobel 1980; Walter and Johnson 1994; Blobel 2000). The localization of secretory and integral membrane protein-encoding mRNAs to the ER is therefore an indirect event; localization is conferred by the binding interactions of the ribosome and translation product, rather than the mRNA, with components of the ER (Lingappa and Blobel 1980; Walter and Johnson 1994; Blobel 2000). Evidence in support of this mechanism is substantial. Using both homologous and heterologous in vitro systems, consisting of mammalian rough microsomes, plant or mammal-derived translation-competent cell extracts, and either native or in vitro transcribed mRNAs, it has been demonstrated that translation and a functional signal sequence are both necessary and sufficient to traffic mRNAs to the ER membrane (Walter and Blobel 1981; Gilmore et al. 1982; Meyer et al. 1982).
Analyses of mRNA partitioning between the cytosol and ER membrane compartments of mammalian cells, fly, and yeast have identified cohorts of cytosolic/nucleoplasmic protein-encoding mRNAs that are highly enriched in the ER, suggesting that ER-directed mRNA localization can occur by a mechanism(s) other than the SRP pathway (Mueckler and Pitot 1981; Kopczynski et al. 1998; Diehn et al. 2000; Nicchitta et al. 2005). In support of this proposal, it has been demonstrated that mRNAs retain their association with the ER in the absence of ribosome engagement and thus participate in ribosome-independent interactions with components of the ER (Stephens et al. 2005; Lerner and Nicchitta 2006). The possibility that mRNAs may undergo direct targeting to the ER is also suggested by data obtained in Saccharomyces cerevisae, where genetic inactivation of the SRP pathway revealed compensatory mechanisms for continued functioning of the secretory pathway (Hann and Walter 1991; Ogg et al. 1992; Mutka and Walter 2001). Relatively little is known, however, about how mRNAs encoding secretory and integral membrane proteins are localized in the intact cell and the role(s) played by the SRP pathway in directing mRNA localization to the ER. These questions are the subject of this report.
RESULTS AND DISCUSSION
In current models, mRNA localization to the ER requires translation, a functional signal sequence, and occurs in context of an mRNA/ribosome/nascent chain/signal recognition particle (SRP) complex (Lingappa and Blobel 1980; Walter and Johnson 1994; Blobel 2000). As noted above, it is established that a signal sequence is a necessary and sufficient signal for ER-restricted mRNA translation (Lingappa and Blobel 1980; Walter and Johnson 1994; Blobel 2000). To determine if the SRP pathway represents the sole pathway for mRNA localization to the ER, we first examined the population identities of ER-localized mRNAs. Extending from previous studies comparing the population identities of mRNAs from cytosol and ER-enriched subcellular fractions, we analyzed by cDNA microarray the mRNA population of cytosolic and outer nuclear envelope-bound polyribosomes (Fig. 1A). The rationale for comparisons between cytosolic and the outer nuclear envelope-bound polyribosomes was as follows: nuclei, because of their unique size and density, can be rapidly purified to near homogeneity from cell homogenates, thereby minimizing the contribution of cytosolic polyribosomes to the membrane-bound polyribosome fraction. Second, the ER and outer nuclear envelope are physically and functionally continuous and so by using nuclei as starting material, the contribution of membranous organelles other than ER is minimized.
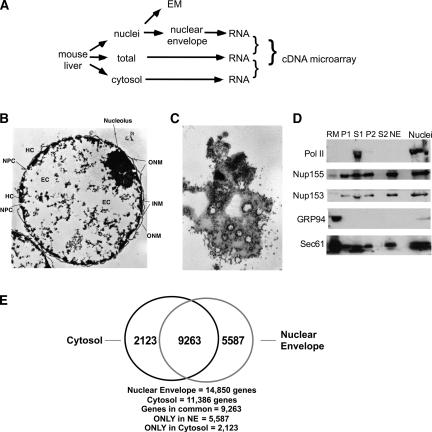
FIGURE 1.
Purification of mouse liver-derived nuclear envelope fraction - a source of membrane-bound RNA for genomic analyses. (A) Schematic of fractionation. (B) Electron micrograph of a cross section of purified nuclei. Scale bar = 0.6 μM. HC, heterochromatin; NPC, nuclear pore complex; EC, euchromatin; ONM, outer nuclear membrane; INM, inner nuclear membrane. (C) Grazing section of nuclei surface, in which abundant polyribosomes (arrows) can be seen, many of which lie in close apposition or in direct association with nuclear pore complexes (asterisks). Scale bar = 0.6 μM. (D) Immunoblot analysis of nuclear envelope isolation fractions. Nuclei were fractionated to yield a nuclear envelope fraction (lanes P1 and NE) and a supernatant fraction containing nucleoplasm (lane S1). The NE fraction was processed to yield an outer (lane P2) and inner (lane S2) nuclear envelope fraction; the former was used for RNA isolation. RM, canine pancreas rough microsomes; lane serves as a positive immunoblot control for ER resident proteins. Fractions were resolved by SDS-PAGE, transferred to PVDF membrane, and the distributions of RNA polymerase II (Pol II), Nup155, Nup153, Sec61p, and GRP94 were determined by immunoblot. (E) Venn diagram representation of mRNA population overlap between cytosolic and membrane-bound mRNA pools. Inclusion in the Venn statistic was determined at more than twofold over background.
Nuclei were isolated by isopycnic density gradient fractionation on iodixanol gradients. As seen in the electron micrographs depicted in Figure 1, B and C, the nuclei isolation procedure yields structurally well-preserved nuclei. In high magnification grazing sections of the outer nuclear envelope, abundant bound polyribosomes were identified in close apposition or physically associated with nuclear pore complexes (Fig. 1C). A high-resolution micrograph illustrating this structural organization is included as Supplemental Figure 1. The nuclear envelope fraction was subsequently prepared from the nuclei fraction using established methods (Matunis 2006). The identity of this fraction as a nuclear envelope was further validated by immunoblot analysis for the ER luminal protein GRP94, the resident ER membrane protein Sec61p, the nucleoporins Nup155 and Nup153, and the nucleoplasmic protein RNA polymerase II. As shown in Figure 1D, rough ER membranes (RM) are enriched in GRP94 and Sec61p, display very low Nup155 levels, and lack RNA polymerase II (pol II). Extraction of the purified nuclei in low-salt buffer supplemented with DNase and heparin yielded a nuclear envelope fraction (P1), enriched in nucleoporins but lacking pol II, and a supernatant fraction highly enriched in pol II (S1). Sec61p, Nup155, and Nup153 were subsequently recovered in the outer nuclear envelope fraction following detergent extraction (Matunis 2006). The cytosol fraction was generated from the liver homogenate by sequential ultracentrifugation. A postnuclear supernatant was initially cleared of all membranous organelles (150,000g, 1 h), and the polyribosome fraction was subsequently isolated from the membrane-free supernatant by ultracentrifugation through a sucrose cushion.
Triplicate cDNA pools were prepared from cytosol and nuclear envelope polyribosome total RNA fractions from three independent mouse liver fractionation experiments, and the mRNA population compositions were compared by cDNA microarray analysis. Sample correlation values exceeded 0.95 for both pools, indicating that sample preparation was highly reproducible. The EASE gene ontology (GO) program was used to compare the compositions of the nuclear envelope (NE)-associated and cytosolic polyribosome fractions (Table 1). As depicted in Table 1, and consistent with its established function in secretory/membrane protein biogenesis, the ER/outer nuclear envelope fraction was highly enriched in mRNAs encoding integral membrane and secreted proteins (EASE values > 3e−12). As expected, the cytosol fraction was highly enriched in mRNAs encoding cytosolic proteins (EASE value > 8e−8). Consistent with previous studies on mRNA partitioning between cytosol and membrane compartments, we observed a partial, but substantial, overlap of cytosolic mRNAs in the nuclear envelope pool (Kopczynski et al. 1998; Diehn et al. 2000, 2006; de Jong et al. 2006). Of the 11,386 genes identified in the cytosol fractions, 9263 of these messages were also detected at significant (more than twofold over background) levels in the nuclear envelope fraction. As reported previously, the cytosol:ER partitioning ratios for the overlap pool varied widely and included mRNAs encoding cytosolic proteins that were highly enriched on the membrane-bound ribosomes (Kopczynski et al. 1998; Diehn et al. 2000, 2006; de Jong et al. 2006; data not shown).
TABLE 1.
Subcellular fractions are highly enriched in their expected cohort of genes
In extending previous studies on the mRNA population identity of membrane-bound ribosomes to the outer nuclear envelope membrane, these data further establish that the presence of an encoded signal sequence is very highly correlated with ER localization (EASE values > 3e−12) (Table 1) and that the absence of a signal sequence does not formally prohibit membrane localization (Kopczynski et al. 1998; Diehn et al. 2000, 2006; de Jong et al. 2006). We cannot exclude the interpretation that the presence of cytosolic protein-encoding mRNAs in the outer nuclear envelope fraction reflects, in part, mRNAs in transit through nuclear pore complexes at the point of isolation, although it is not clear why distinct subpopulations of mRNAs, and not others, would be consistently recovered as a nuclear pore-associated fraction. Regardless, prior studies have documented that subsets of cytosolic protein-encoding mRNAs are highly enriched on membrane-bound ribosomes, and so it appears that multiple processes contribute to the phenomenon of ER/outer nuclear envelope-directed mRNA partitioning (Diehn et al. 2000, 2006; Lerner et al. 2003).
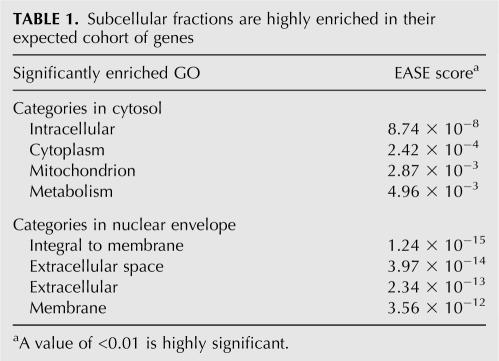
As an alternative approach to investigating the regulation of mRNA partitioning to the ER, we examined mRNA partitioning patterns in HeLa cell lines stably expressing shRNAs targeted to the essential 54-kDa subunit of SRP (Bernstein et al. 1989; Ren et al. 2004). These cell lines have been previously characterized; surprisingly, they display no apparent growth defects (Ren et al. 2004). The lack of a growth defect contrasts with genetic studies in yeast, where the loss of SRP/SRP receptor function has a profound growth defect, and may reflect the phenotype of the knockdown versus knockout (Hann et al. 1989; Ogg et al. 1992). We confirmed that the SRP54 mRNA levels in the stable knockdown cell line are markedly reduced relative to control HeLa cells (Fig. 2A) and also confirmed that cell surface expression of the integral membrane protein DR4 is markedly reduced in these cells (Fig. 2B), whereas cell surface expression of another receptor, FGFR, is not affected (data not shown). Thus, and as previously reported, shRNA-directed reduction in SRP54 levels affects the translocation of some, but not all, signal-bearing proteins (Ren et al. 2004).
FIGURE 2.
Subcellular mRNA distribution in SRP54 knock-down HeLa cell lines. (A) Total RNA from control HeLa (H) or SRP54 (54) stable knock-down cells was analyzed by Northern blot. (B) Cell surface expression of DR4 was determined by flow cytometry using no primary antibody (control) or DR4 antibody. (C) Cells were fractionated by sequential detergent extraction into cytosol (lane C) or membrane-bound (lane M) fractions. RNA isolated from total cells (lane T) or cell fractions was analyzed by Northern blot using the probes listed in Materials and Methods.
To determine the consequences of reduced SRP54 expression on mRNA partitioning, subcellular mRNA distribution patterns were determined in SRP54 knockdown and control cell lines using a well-established sequential detergent extraction technique (Adam et al. 1990; Wilson et al. 1995; Lerner et al. 2003; Stephens et al. 2005, 2007). We examined the distribution of seven mRNAs; four encoding signal sequence bearing proteins (GRP94, calreticulin, BiP, and TRAPδ) and three encoding cytosolic proteins previously reported to undergo noncanonical enrichment on membrane-bound ribosomes (Hsp90, L26, and LDH) (Lerner et al. 2003). For these RNA classes, the message distribution was identical in both control and SRP54 knockdown cells, suggesting that, even in the case of signal sequence-encoding mRNAs, ER localization may occur through multiple pathways (Fig. 2C). However, and because the shRNA approach yielded only a partial SRP54 knockdown, alternative experimental approaches to the question of the mechanism of ER-directed mRNA localization were developed.
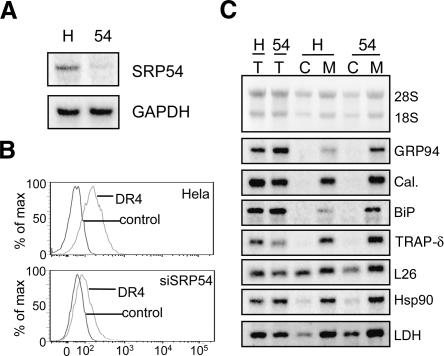
To gain insight into the mechanism(s) of mRNA to the ER, we asked whether mRNA association with the ER persists when mRNA/ribosome/nascent chain complexes are physically dissociated; current models predict that mRNAs are tethered to the ER solely via their functional association with membrane-bound ribosomes (Lingappa and Blobel 1980; Blobel 2000; Alberts et al. 2002). If mRNA association with the ER is mediated solely via translation-driven functional association with membrane-bound ribosomes, subunit dissociation/ribosome disassembly would be predicted to elicit mRNA release (Fig. 3A). In contrast, mRNAs that undergo ribosome-independent binding interactions with the ER would be retained upon ribosomal subunit disassembly. To test these predictions, rough microsomes (RM) were prepared from J558 murine plasmacytoma cells and the relative sensitivities of rRNA and mRNA to EDTA extraction determined. In these experiments, RM were treated with 50 mM EDTA (sufficient to achieve complete dissociation of ribosomes into 40 and 60S subunits) and mRNA release was assayed following separation of the membrane and supernatant fractions by ultracentrifugation. EDTA treatment of the purified RM elicited a marked release of ribosomal subunits (18S, 28S rRNAs) from the RM (Fig. 3B, cf. lanes 1,2 and 3,4). However, mRNAs encoding the resident ER chaperones GRP94, BiP, and calreticulin were wholly retained on the rough microsomes (Fig. 3B, cf. lanes 1,2 and 3,4). In contrast, treatment of the RM with EDTA in the presence of sodium carbonate buffer, pH11, to extract peripheral membrane proteins (Fujiki et al. 1982), or detergent, to solubilize the membranes, yielded the release of RNAs into the supernatant fraction (Fig. 3B, lanes 5–8). These data are consistent with much earlier studies reporting on the association of mRNAs with the ER (Milcarek and Penman 1974; Lande et al. 1975; Cardelli et al. 1976; Lonn 1977) and suggest that mRNAs encoding ER resident proteins have localization information that contributes to their ER partitioning.
FIGURE 3.
mRNAs retain physical interactions with the endoplasmic reticulum following the disassembly of membrane-bound ribosomes. (A) Schematic depicting possible fates of membrane-associated mRNAs following extraction of RM with EDTA. (B) Purified rough microsomes (RM) were treated with EDTA, sodium carbonate, pH 11, and/or a NP40/DOC detergent admixture (det.) as indicated. RM were centrifuged through a 0.5 M sucrose pad, and bound (pellet, lanes P) and released (supernatant, lanes S) fractions were collected. Total RNA extracted from each fraction was analyzed by Northern blot (upper panels). The recovery of the integral membrane protein fraction was assayed by immunoblot analysis of the ER integral membrane protein TRAPα (bottom panel).
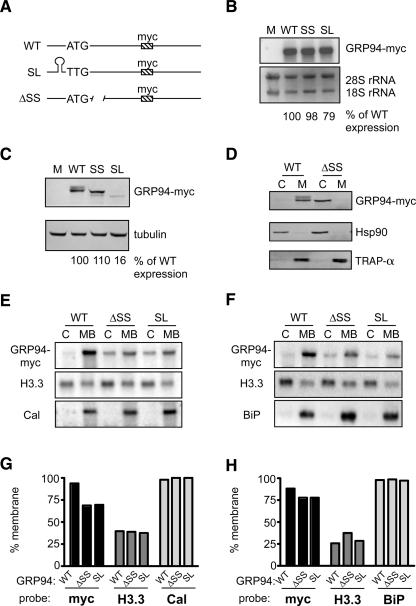
Using a representative ER enriched transcript, GRP94 mRNA, we examined the role of the SRP pathway in mRNA localization to the ER. Because the GRP94 ORF encodes a canonical signal sequence, its partitioning to the ER would be expected to be both translation- and signal sequence-dependent. As depicted in Figure 4A, all reporter constructs contain the entire ORF and UTRs of GRP94, as well as a myc epitope tag to allow direct detection of the mRNA and mRNA translation products. Using the detergent extraction fractionation protocol described above, we observed that GRP94-myc mRNA and GRP94-myc protein are both found exclusively in the membrane-bound fraction (Fig. 4; data not shown).
FIGURE 4.
Endoplasmic reticulum localization of GRP94 mRNA occurs in the absence of signal sequence function or translation. (A) Schematic of GRP94-myc reporter constructs. WT, wild-type GRP94-myc; SL, GRP94-myc with addition of a stem–loop and ATG→TTG start codon mutation; ΔSS, GRP94-myc with a deletion of the signal sequence. (B) Total RNA was extracted from Cos-7 cells transfected with constructs as listed and analyzed by Northern blot. Expression was normalized to the 28S rRNA band and wild-type levels set at 100% for quantification. (C) Total cell lysates from Cos-7 cells transfected with constructs as listed were analyzed by immunoblot. Expression was normalized to tubulin and wild-type levels set at 100% for quantification. (D) Cos-7 cells transfected with WT or ΔSS constructs were fractionated into cytosol (lanes C) and membrane-bound (lanes M) fractions. Proteins were analyzed by immunoblot using anti-myc, anti-Hsp90 (cytosolic marker), or anti-TRAPα (ER marker) antibodies. (E,F) Cos-7 (E) or HeLa (F) cells transfected with WT, ΔSS, or SL GRP94-myc constructs were fractionated into cytosol and membrane-bound fractions. RNA isolated from each fraction was analyzed by Northern blot using probes against myc, histone 3.3 (cytosolic mRNA), calreticulin, or BiP (ER-bound mRNAs). (G,H) The relative enrichment of each message in the membrane-bound fraction for Cos-7 cells (G) or HeLa cells (H).
To determine if translation is required for mRNA localization to the ER, the ATG start codon of the GRP94 reporter was mutated to a TTG and a stable stem–loop structure (ΔG = −52kcal/mol) (Mathews et al. 1999; Zuker 2003), similar to that previously shown to inhibit translation (Pelletier and Sonenberg 1985; Dostie and Dreyfuss 2002), was inserted into the 5′UTR (Fig. 4A, GRP94-myc SL). No protein product was detected in cells transfected with the GRP94-myc SL construct, although the wild-type form was readily detected (Fig. 4C), confirming that the GRP94-myc SL construct is translated weakly, or not at all, in tissue culture cells. The faint, faster migrating band present in the SL lane likely represents an internal initiation product. Importantly, the GRP94 primary amino acid sequence indicates that all internal initiation products lack a signal sequence, and therefore internal initiation would not be expected to serve as a mechanism whereby mRNAs could be ER localized.
To determine whether ER-directed localization of GRP94 mRNA requires a signal sequence, a cDNA was constructed where the nucleotides encoding amino acids 4–19, the GRP94 signal sequence, were deleted (Fig. 4A, GRP94-myc ΔSS). Cells transfected with the ΔSS construct were fractionated into cytosol and membrane bound compartments, and the partitioning of the translation products between the cytosol and endoplasmic reticulum was determined. In the absence of a signal sequence, translocation of GRP94 into the ER lumen was disabled and the GRP94-myc ΔSS translation product was recovered exclusively in the cytosol fraction (Fig. 4D). In contrast, the signal sequence encoding form of GRP94-myc was recovered entirely in the membrane fraction. Paired control analyses, where the cytosolic protein Hsp90, or the resident ER membrane protein TRAPα, were assayed, confirmed both the validity of the fractionation and the ascribed locations of the wild-type (WT) and ΔSS forms of the GRP94-myc reporters (Fig. 4D).
The consequences of suppressed translation/loss of signal sequence function on ER directed RNA localization was examined in Cos-7 and HeLa cells transfected with the different GRP94-myc reporter constructs and fractionated by the techniques noted above, to assess partitioning between the cytosol and ER-enriched fractions (Fig. 4E,F). As shown in Figure 4E (Cos-7) and Figure 4F (HeLa), both the GRP94-myc SL and Grp94-myc ΔSS, like the WT mRNA, are highly enriched in membrane-bound fractions. For comparison, calreticulin and BiP messages are found exclusively in ER-bound fractions, and histone H3.3 mRNA is highly enriched in cytosolic fractions. These results serve to validate the fractionation protocol. In HeLa cells, the results are striking—there is little change in the distribution of GRP94-myc mRNA for either mutation (from 89% membrane bound for the wild type to 78% for both ΔSS or SL constructs) (Fig. 4G). In Cos-7 cells, the ΔSS and SL constructs are less efficiently partitioned to the membrane bound fraction, as compared to the wild-type (∼ 60% versus 90% for the wild-type construct); they are nonetheless substantially enriched in the membrane bound fraction (Fig. 4B).
To determine if the observed partitioning patterns were a function of expression levels, experiments were conducted in which cells were transfected with serial dilutions of plasmid DNA. Extensive comparisons of the partitioning patterns confirmed that the differences between HeLa and Cos7 cells in the relative efficiency of cytosol:ER partitioning were not due to variations in reporter expression level between the two cell lines and were not, for example, artifacts of over-expression (data not shown). These data indicate that mRNAs may themselves encode localization information specific for the ER compartment. Thus, although the signal sequence/SRP pathway is sufficient to support ER-directed RNA localization, the absence of a signal sequence and/or suppression of mRNA translation only modestly affects ER-directed mRNA localization. On the basis of these data, we propose that signal-sequence encoding mRNAs can undergo direct, translation-independent localization to the ER. At present, we do not know the relative contributions of translation-dependent versus translation-independent pathways to the global partitioning of mRNAs to the ER. A full understanding of the global regulation of mRNA partitioning to the ER will require the identification of the presumed cis-localization signals which direct this class of mRNAs to the ER as well as the trans-acting recognition factors which decode this localization information.
On the basis of these data, we propose that mRNAs contain localization determinants that govern their partitioning to the ER membrane (Nicchitta et al. 2005). In this view, cis-localization elements, and their cognate trans-recognition factors, provide the primary control of mRNA partitioning between the cytosol and ER compartments; the SRP pathway would then function to direct the translation product into the exocytic pathway. The separation of these two processes, mRNA localization and protein translocation, suggests a degree of plasticity in the organization of the subcellular compartmentalization of protein synthesis not anticipated in current models. Importantly, such a model explains the now-established phenomenon of “noncanonical” mRNA partitioning to the ER and is consistent with established modes of mRNA localization in the cell (Palacios and St Johnston 2001). Of particular interest, a recent global analysis of mRNA localization in the Drosophila embryo identified an unexpected and rich diversity of mRNA localization patterns, leading these authors to postulate fundamental roles for mRNA localization in the genesis of the complex compartmentalization processes that are characteristic of metazoans (Lecuyer et al. 2007). In this context, it will be critical to further investigate the mechanisms and the functionality of such localization pathways. Possible functions of such targeting pathways include the compartmentalization of metabolic pathways to the 2D surface of the ER membrane, localization of protein synthesis to discrete subdomains of the ER with either different physiological functions (Choi et al. 2000) or which initiate the formation of organized secretory units that contribute to both structural and functional polarity (Herpers and Rabouille 2004; Levine and Rabouille 2005).
MATERIALS AND METHODS
Cell culture
HeLa human epithelial cells and Cos-7 monkey kidney cells (ATCC) were cultured in DMEM supplemented with 10% fetal calf serum. J558 murine plasmacytoma cells (ATCC) were cultured in DMEM supplemented with 10% calf serum. HeLa cells stably expressing a short hairpin RNA against SRP54 (Ren et al. 2004) were obtained from Q. Deveraux, through MTA agreement with Novartis Research Foundation (San Diego, CA). All cells were maintained at 37°C and 5% CO2 and were subcultured at 2–3-d intervals. Tissue culture reagents were from Invitrogen or Mediatech.
Plasmid construction and transfections
Molecular biology reagents were from Promega, New England Biolabs, and Invitrogen, unless specified otherwise. Oligonucleotides were obtained from IDT. Transfections were conducted using Lipofectamine 2000 (Invitrogen), according to the manufacturer's directions with the exception that plasmid DNA/lipofectamine additions were performed in the absence of serum and media was replaced after 4 h of incubation. Typically, cells were split at a 1:2 ratio at this time.
GRP94 was amplified by RT-PCR of total RNA purified from HeLa cells. cDNA was amplified from total RNA using Superscript RNAse H- reverse transcriptase (Invitrogen) and oligo dT primers. PCR amplification of GRP94 was done using the following oligos: 5′-CGCAGATCTGTGGGCGGACCGCGCGGC-3′ and 5′- CGCAGATCTATTTGGGATCTTTTTTATTTTTATACA-3′. The PCR product was cloned using a PCR-SMART cloning kit (Lucagen). A myc tag was inserted into the unique NdeI site of GRP94 using the following oligos: 5′-TATGAACAAAAACTCATCTCAGAAGAGGATCTGGCA-3′ and 5′-TATGCCAGATCCTCTTCTGAGATGAGTTTTTGTTCA-3′. GRP94-myc was isolated from the PCR-SMART vector using BglII and inserted into the BamHI site of pcDNA6/myc-HIS C (Invitrogen).
GRP94-myc-ΔSS was created by using a Quik-Change site-directed mutagenesis kit (Stratagene) to add an ApaI site at nt 56 of GRP94-myc in the PCR-SMART vector. GRP94-myc (+ApaI) was cut out of the PCR-SMART vector using BglII and inserted into the BamHI site of pcDNA6/myc-HIS C. The signal sequence was deleted by digesting with ApaI and re-ligating the resulting product to create GRP94-myc-ΔSS. The resulting construct encodes a protein with the following initial sequence: MRARAD (wild type=MRALWVLGLCCVLLTFGSVRAD). GRP94-myc SL was created by using a Quik-Change site-directed mutagenesis kit (Stratagene) to change the ATG start codon of GRP94-myc in the PCR-SMART vector to a TTG. GRP94-myc ΔATG Δ3′UTR was amplified by PCR using the 5′ start oligo listed above and 5′-CGCAGATCTTTACAATTCATCTTTTTCAGCTGTAG-3′. This fragment was digested with BglII and inserted into the BamHI site of pcDNA6/myc-HIS C. A stem–loop was inserted into the BamHI site in the 5′UTR of GRP94 using the following oligo: 5′-GATCCGGCCGGATCCGGCCGGATCCGGCCGGATCCGGCCG-3′. The 3′UTR of GRP94 was then inserted into this construct by ligating a PstI fragment from full-length GRP94-myc into a PstI-digested plasmid.
Nuclei isolation and electron microscopy
Nuclei were isolated from pooled, freshly isolated mouse livers by iodixanol (OptiPrep) gradient isolation of isotonic liver homogenates, as described in the manufacturer's application guide (Axis-Shield). Briefly, fresh mouse livers were homogenized in a buffer containing 10 mM HEPES/KOH, pH 7.4, 250 mM sucrose, 1 mM DTT, 1 mM EDTA, 0.5 mM PMSF, 0.1 μM leupeptin, 25 mM KCl, and 5 mM MgCl2. The homogenate was filtered through four layers of cheesecloth and centrifuged at 1000g for 10 min to yield a pellet that was resuspended and recentrifuged. The pellet was then made to 25% iodixanol, underlayed with equal volumes of 30% and 35% iodixanol in homogenization buffer, and centrifuged for 10,000g for 20 min. The nuclei were recovered at the 30%–35% interface, resuspended and washed in homogenization buffer. Nuclei prepared in this manner are virtually free from cytosolic contamination, as determined by enzymatic assay of the cytosolic enzyme lactate dehydrogenase (data not shown).
Nuclear envelopes were prepared from freshly isolated mouse liver nuclei as described in Matunis (2006), with the exception that RNase was omitted during the nuclease digestion step. In brief, freshly isolated nuclei were resuspended in a hypotonic buffer (0.1 mM MgCl2, 1 mM DTT, 0.1 mM PMSF), subsequently diluted into buffered 10% sucrose, and chromatin-hydrolyzed by addition of RNase-free DNase I. The nuclear envelope fraction was isolated by centrifugation, and the outer nuclear envelope fraction was obtained by the detergent extraction protocols detailed in Matunis (2006). Thin section transmission electron microscopy of isolated mouse liver nuclei were performed as described previously (Lerner et al. 2003). Briefly, nuclei were fixed in solution with glutaraldehyde, and following post-fixation staining, samples were embedded and processed for electron microscopic analysis.
RNA isolation, amplification, and generation of cDNAs, probe preparation, and microarray hybridization
Total RNA from murine liver, purified cytosolic polyribosomes, purified nuclei, and purified nuclear envelope fractions were isolated using TRIzol (Invitrogen). Total RNA quality was assayed using an Agilent bioanalyzer (Silicon Genetics), as performed in the Duke Microarray Core Facility (DMCF). Amplified mRNA was analyzed on oligonucleotide DNA microarrays using the Operon Murine Genome Oligo Set Version 3.0 (Operon). Total RNA from each sample and a reference RNA (Universal Murine Reference RNA, Stratagene) was used in probe preparation using direct labeling. Detailed protocols are available on the DMCF Web site (http://microarray.genome.duke.edu/spotted-arrays/protocols).
Data processing and statistical analysis
Genespring v7.2 software (Agilent Technologies, Silicon Genetics) was used to perform data analysis. The array files are freely accessible at http://data.cgt.duke.edu/CN.php. Intensity-dependent (Lowess) normalization was done on the entire data set. Based on triplicates of each condition, a threshold of twofold increase or decrease in expression relative to the total RNA and a two-way ANOVA with a P cutoff of 0.05 was done. The cytosol and nuclear envelope-derived mRNA pools were characterized using EASE analysis (Hosack et al. 2003). EASE calculates overrepresentation statistics relative to the total gene number and allows paired comparisons of GO categories via use of EASE scores, a one-tailed Fisher exact probability statistical measure. EASE-based GO analyses of the mRNA fractions were performed to define the most prominent categories/members of the cytosol and nuclear envelope-associated mRNA fractions, with overall presence or absence of any given mRNA, in any fraction, defined through a cutoff twofold expression over background as a minimum signal.
Sequential detergent extraction and sucrose density gradients
These protocols have been described previously (Lerner et al. 2003; Stephens et al. 2005, 2007; Lerner and Nicchitta 2006).
RNA–endoplasmic reticulum interaction assay
Rough microsomes (RM) were purified from J558 cells as described previously (Lerner et al. 2003; Stephens et al. 2005, 2007). RM from ∼ 1.8 × 108 cells were recovered by centrifugation at 40,000 rpm in a Beckman TLA 100.3 for 20 min and resuspended in 1.5 mL of SHKM buffer (0.25 M sucrose, 50 mM HEPES, pH 6.8, 150 mM KOAc, 5 mM MgOAc2) using a Dounce homogenizer. As indicated, EDTA was added to a final concentration of 50 mM and incubated for 20 min on ice. The indicated samples were then incubated in 100 mM sodium carbonate, pH 11, or 1% NP40 and 0.5% sodium deoxycholate for 20 min on ice. RM were collected by centrifugation at 44,000 rpm in a TLA100 for 10 min over a 0.5 M sucrose cushion. RNA or proteins were recovered from pellet and supernatant fractions and analyzed by Northern blotting or immunoblotting.
Northern and immunoblot analysis
Northern blots were conducted by standard protocols and have been described previously (Stephens et al. 2007). Probes were labeled with [α-32P]dCTP (MP Biomedical) using a random hexanucleotide primer kit (Roche) and either a KpnI fragment from canine GRP94 or a PstI/EcoRI fragment of BiP. Alternatively, probes were labeled with [γ-32P]ATP (MP Biomedical) using T4 polynucleotide kinase (NEB) and the oligonucleotide sequences listed below. Phosphorimaging was performed with a Typhoon 9400 (GE Healthcare), and data analysis was performed using ImageQuant TL software (GE Healthcare).
Immunoblots were conducted by standard protocols using antibodies against myc (ab9106, Abcam), tubulin (Developmental Studies Hybridoma Bank), TRAPα or Hsp90 as previously described (Lerner et al. 2003). Quantification was performed with the ImageQuant TL software package (GE Healthcare).
Northern blot oligonucleotide probes
Flow cytometry
Surface expression of DR4 was determined by flow cytometry using a BD LSR II flow cytometer (BD Biosciences). Cells were harvested, washed with PBS, and incubated with 20 μg/mL mouse monoclonal antibody to DR4 (DJR1, Abcam) in growth media for 45 min on ice. Cells were then washed with PBS and incubated with 1 μg/mL biotin conjugated goat anti-mouse secondary antibody (Invitrogen) for 30 min on ice. Cells were then washed with PBS and incubated with 1:100 streptavidin-PE (BioRad) for 30 min on ice. Cells were washed with PBS and analyzed by flow cytometry.
SUPPLEMENTAL DATA
Supplemental material, including a high-resolution micrograph of Figure 1C, depicting polyribosomes in association with the outer nuclear envelope and the nuclear pore complex, can be found at http://www.cellbio.duke.edu/faculty/research/Nicchitta.html (listed as Pyhtila et al. Nuclear Envelope Micrograph).
ACKNOWLEDGMENTS
We thank Angela Jockheck-Clark and Deanna Crossman for help with flow cytometry and Sam Stephens and Rebecca Dodd for critical comments. This work was supported by grants from the National Institutes of Health (GM072243 to B.P; GM077382 to C.V.N.; AR14317 to M.C.R.).
Footnotes
Abbreviations: ER, endoplasmic reticulum; SRP, signal recognition particle; RM, rough microsome; UTR, untranslated region.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.721108.
REFERENCES
- Adam, S.A., Marr, R.S., Gerace, L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K., Walter, P. Molecular Biology of the Cell. Garland Science; New York: 2002. [Google Scholar]
- Bernstein, H.D., Poritz, M.A., Strub, K., Hoben, P.J., Brenner, S., Walter, P. Model for signal sequence recognition from amino acid sequence of 54 K subunit of signal recognition particle. Nature. 1989;340:482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- Blobel, G. Protein targeting (Nobel lecture) ChemBioChem. 2000;1:86–102. doi: 10.1002/1439-7633(20000818)1:2<86::AID-CBIC86>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Cardelli, J., Long, B., Pitot, H.C. Direct association of messenger RNA labeled in the presence of fluoro-orotate with membranes of the endoplasmic reticulum in rat liver. J. Cell Biol. 1976;70:47–58. doi: 10.1083/jcb.70.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, S.B., Wang, C., Muench, D.G., Ozawa, K., Franceschi, V.R., Wu, Y., Okita, T.W. Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature. 2000;407:765–767. doi: 10.1038/35037633. [DOI] [PubMed] [Google Scholar]
- Czaplinski, K., Singer, R.H. Pathways for mRNA localization in the cytoplasm. Trends Biochem. Sci. 2006;31:687–693. doi: 10.1016/j.tibs.2006.10.007. [DOI] [PubMed] [Google Scholar]
- de Jong, M., van Breukelen, B., Wittink, F.R., Menke, F.L., Weisbeek, P.J., Van den Ackerveken, G. Membrane-associated transcripts in Arabidopsis: Their isolation and characterization by DNA microarray analysis and bioinformatics. Plant J. 2006;46:708–721. doi: 10.1111/j.1365-313X.2006.02724.x. [DOI] [PubMed] [Google Scholar]
- Diehn, M., Bhattacharya, R., Botstein, D., Brown, P.O. Genome-scale identification of membrane-associated human mRNAs. PLoS Genet. 2006;2:e11. doi: 10.1371/journal.pgen.0010087.g001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn, M., Eisen, M.B., Botstein, D., Brown, P.O. Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nat. Genet. 2000;25:58–62. doi: 10.1038/75603. [DOI] [PubMed] [Google Scholar]
- Dostie, J., Dreyfuss, G. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr. Biol. 2002;12:1060–1067. doi: 10.1016/s0960-9822(02)00902-8. [DOI] [PubMed] [Google Scholar]
- Du, T.G., Schmid, M., Jansen, R.P. Why cells move messages: The biological functions of mRNA localization. Semin. Cell. Dev. Biol. 2007;18:171–177. doi: 10.1016/j.semcdb.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Fujiki, Y., Hubbard, A.L., Fowler, S., Lazarow, P.B. Isolation of intracellular membranes by means of sodium carbonate treatment: Application to endoplasmic reticulum. J. Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore, R., Blobel, G., Walter, P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J. Cell Biol. 1982;95:463–469. doi: 10.1083/jcb.95.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann, B.C., Walter, P. The signal recognition particle in S. cerevisiae . Cell. 1991;67:131–144. doi: 10.1016/0092-8674(91)90577-l. [DOI] [PubMed] [Google Scholar]
- Hann, B.C., Poritz, M.A., Walter, P. Saccharomyces cerevisiae and Schizosaccharomyces pombe contain a homologue to the 54-kDa subunit of the signal recognition particle that in S. cerevisiae is essential for growth. J. Cell Biol. 1989;109:3223–3230. doi: 10.1083/jcb.109.6.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herpers, B., Rabouille, C. mRNA localization and ER-based protein sorting mechanisms dictate the use of transitional endoplasmic reticulum-Golgi units involved in Gurken transport in Drosophila oocytes. Mol. Biol. Cell. 2004;15:5306–5317. doi: 10.1091/mbc.E04-05-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosack, D.A., Dennis G., Jr, Sherman, B.T., Lane, H.C., Lempicki, R.A. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopczynski, C.C., Noordermeer, J.N., Serano, T.L., Chen, W.Y., Pendleton, J.D., Lewis, S., Goodman, C.S., Rubin, G.M. A high-throughput screen to identify secreted and transmembrane proteins involved in Drosophila embryogenesis. Proc. Natl. Acad. Sci. 1998;95:9973–9978. doi: 10.1073/pnas.95.17.9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande, M.A., Adesnik, M., Sumida, M., Tashiro, Y., Sabatini, D.D. Direct association of messenger RNA with microsomal membranes in human diploid fibroblasts. J. Cell Biol. 1975;65:513–528. doi: 10.1083/jcb.65.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer, E., Yoshida, H., Parthasarathy, N., Alm, C., Babak, T., Cerovina, T., Hughes, T.R., Tomancak, P., Krause, H.M. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Lerner, R.S., Nicchitta, C.V. mRNA translation is compartmentalized to the endoplasmic reticulum following physiological inhibition of cap-dependent translation. RNA. 2006;12:775–789. doi: 10.1261/rna.2318906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner, R.S., Seiser, R.M., Zheng, T., Lager, P.J., Reedy, M.C., Keene, J.D., Nicchitta, C.V. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA. 2003;9:1123–1137. doi: 10.1261/rna.5610403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, T., Rabouille, C. Endoplasmic reticulum: One continuous network compartmentalized by extrinsic cues. Curr. Opin. Cell Biol. 2005;17:362–368. doi: 10.1016/j.ceb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Lingappa, V.R., Blobel, G. Early events in the biosynthesis of secretory and membrane proteins: The signal hypothesis. Recent Prog. Horm. Res. 1980;36:451–475. doi: 10.1016/b978-0-12-571136-4.50018-8. [DOI] [PubMed] [Google Scholar]
- Lonn, U. Direct association of Balbiani ring 75S RNA with membranes of the endoplasmic reticulum. Nature. 1977;270:630–631. doi: 10.1038/270630a0. [DOI] [PubMed] [Google Scholar]
- Mathews, D.H., Sabina, J., Zuker, M., Turner, D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Matunis, M.J. Isolation and fractionation of rat liver nuclear envelopes and nuclear pore complexes. Methods. 2006;39:277–283. doi: 10.1016/j.ymeth.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Meyer, D.I., Krause, E., Dobberstein, B. Secretory protein translocation across membranes—The role of the “docking protein.”. Nature. 1982;297:647–650. doi: 10.1038/297647a0. [DOI] [PubMed] [Google Scholar]
- Milcarek, C., Penman, S. Membrane-bound polyribosomes in HeLa cells: Association of polyadenylic acid with membranes. J. Mol. Biol. 1974;89:327–338. doi: 10.1016/0022-2836(74)90522-1. [DOI] [PubMed] [Google Scholar]
- Mueckler, M.M., Pitot, H.C. Structure and function of rat liver polysome populations. I. Complexity, frequency distribution, and degree of uniqueness of free and membrane-bound polysomal polyadenylate-containing RNA populations. J. Cell Biol. 1981;90:495–506. doi: 10.1083/jcb.90.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutka, S.C., Walter, P. Multifaceted physiological response allows yeast to adapt to the loss of the signal recognition particle-dependent protein-targeting pathway. Mol. Biol. Cell. 2001;12:577–588. doi: 10.1091/mbc.12.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicchitta, C.V., Lerner, R.S., Stephens, S.B., Dodd, R.D., Pyhtila, B. Pathways for compartmentalizing protein synthesis in eukaryotic cells: the template-partitioning model. Biochem. Cell Biol. 2005;83:687–695. doi: 10.1139/o05-147. [DOI] [PubMed] [Google Scholar]
- Ogg, S.C., Poritz, M.A., Walter, P. Signal recognition particle receptor is important for cell growth and protein secretion in Saccharomyces cerevisiae . Mol. Biol. Cell. 1992;3:895–911. doi: 10.1091/mbc.3.8.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios, I.M., St Johnston, D. Getting the message across: The intracellular localization of mRNAs in higher eukaryotes. Annu. Rev. Cell Dev. Biol. 2001;17:569–614. doi: 10.1146/annurev.cellbio.17.1.569. [DOI] [PubMed] [Google Scholar]
- Pelletier, J., Sonenberg, N. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985;40:515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- Ren, Y.G., Wagner, K.W., Knee, D.A., Aza-Blanc, P., Nasoff, M., Deveraux, Q.L. Differential regulation of the TRAIL death receptors DR4 and DR5 by the signal recognition particle. Mol. Biol. Cell. 2004;15:5064–5074. doi: 10.1091/mbc.E04-03-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston, D. Moving messages: The intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- Stephens, S.B., Dodd, R.D., Brewer, J.W., Lager, P.J., Keene, J.D., Nicchitta, C.V. Stable ribosome binding to the endoplasmic reticulum enables compartment-specific regulation of mRNA translation. Mol. Biol. Cell. 2005;16:5819–5831. doi: 10.1091/mbc.E05-07-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, S., Dodd, R., Lerner, R., Pyhtila, B., Nicchitta, C. Analysis of mRNA partitioning between the cytosol and endoplasmic reticulum compartments of mammalian cells. In: Wilusz J., editor. Methods in molecular biology. Vol. 419. Humana Press; Totowa, NJ: 2007. [DOI] [PubMed] [Google Scholar]
- Walter, P., Blobel, G. Translocation of proteins across the endoplasmic reticulum. II. Signal recognition protein (SRP) mediates the selective binding to microsomal membranes of in vitro–assembled polysomes synthesizing secretory protein. J. Cell Biol. 1981;91:551–556. doi: 10.1083/jcb.91.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, P., Johnson, A.E. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- Wilson, R., Allen, A.J., Oliver, J., Brookman, J.L., High, S., Bulleid, N.J. The translocation, folding, assembly and redox-dependent degradation of secretory and membrane proteins in semi-permeabilized mammalian cells. Biochem. J. 1995;307:679–687. doi: 10.1042/bj3070679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]