Abstract
During an early step in the evolution of life, RNA served both as genome and as catalyst, according to the RNA world hypothesis. For self-replication, the RNA organisms must have contained an RNA that catalyzes RNA polymerization. As a first step toward recapitulating an RNA world in the laboratory, a polymerase ribozyme was generated previously by in vitro evolution and design. However, the efficiency of this ribozyme is about 100-fold too low for self-replication because of a low affinity of the ribozyme to its primer/template substrate. To improve the substrate interactions by colocalizing ribozyme and substrate on micelles, we attached hydrophobic anchors to both RNAs. We show here that the hydrophobic anchors led to aggregates with the expected size of the corresponding micelles. The micelle formation increased the polymerization yield of full-length products by 3- to 20-fold, depending on substrates and reaction conditions. With the best-characterized substrate, the improvement in polymerization efficiency was primarily due to reduced sequence-specific stalling on partially extended substrates. We discuss how, during the origin of life, micellar ribozyme aggregates could have acted as precursors to membrane-encapsulated life forms.
Keywords: ribozyme, polymerization, RNA world, micelle, self-assembly
INTRODUCTION
According to the RNA world hypothesis, RNA both stored genomic information and catalyzed self-replication at an early stage in the evolution of life. After the discovery of a catalytic RNA (ribozyme) in 1982 (Kruger et al. 1982), a number of experiments were aimed toward creating an RNA world in the laboratory (Bartel and Szostak 1993; Johnston et al. 2001; for reviews see Szostak et al. 2001; Müller 2006). The central molecule in such a system would be a ribozyme that replicates RNA polymers by polymerizing chemically activated units in a template-dependent fashion (James and Ellington 1999). A polymerase ribozyme was developed in 2001 that is able to recognize a primer/template duplex in trans and extend the primer 3′ terminus using nucleoside triphosphates (Johnston et al. 2001). The ribozyme creates 3′–5′ phosphodiester bonds and adds nucleotides complementary to the single-stranded template, with a fidelity of approximately 97%. A bias in the nucleoside triphosphates can increase the fidelity to 99%, which may render the fidelity of polymerization sufficient for self-replication (Johnston et al. 2001; Müller 2006).
However, polymerization efficiency of current ribozymes is too low for self-replication: No primer extension longer than 20 nucleotides (nt) has been observed for a polymerase ribozyme (Zaher and Unrau 2007), whereas the lengths of all known polymerase ribozymes are in the range of 190 nt (Johnston et al. 2001; Lawrence and Bartel 2005; Zaher and Unrau 2007). The limiting factor for polymerization efficiency is a low substrate binding affinity, with a KM in the millimolar range (Lawrence and Bartel 2003). This weak binding causes a mostly distributive reaction mechanism that slows down polymerization so much that, during the limited lifetime of the polymerase (with a half-life of about a day), longer polymerizations could not be achieved (Lawrence and Bartel 2003). In addition, the effective rates of nucleotide addition are sequence dependent, differing by factors of more than 100-fold, which causes the accumulation of specific polymerization intermediates (Lawrence and Bartel 2003; Müller and Bartel 2003). All of the 10 existing polymerase ribozymes (Johnston et al. 2001; Lawrence and Bartel 2005; Zaher and Unrau 2007) appear to suffer from the same problem.
In this study, we tried to improve the polymerization efficiency with a different physical principle. Instead of trying to improve the sequence of the ribozyme, we colocalized ribozymes with their substrates on micelles, with the aim of creating high local ribozyme concentrations and thereby avoiding the necessity for a high substrate affinity. Micelle formation was achieved by attaching hydrophobic anchors to both ribozyme and substrate. Analysis of the polymerization efficiency shows that micellar colocalization can help reduce stalling, causing only a modest increase in catalytic efficiency but a substantial increase of full-length products, by up to 25-fold.
RESULTS
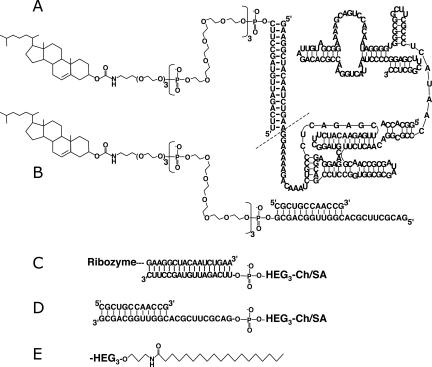
The aims of this study were to engineer variants of the polymerase ribozyme and its substrate that can self-assemble into micelles and to test whether this micellization improves polymerization efficiency. The hydrophobic anchors, either a cholesteryl residue or a stearyl residue, were connected to ribozyme and substrate via oligoethylene glycol linkers and short RNA oligonucleotides that can base pair to either ribozyme or primer (Fig. 1; Table 1).
FIGURE 1.
Attachment of hydrophobic anchors to ribozyme and substrate. (A) Ribozyme with 5′-cholesteryl anchor, connected via a 5′-duplex and three hexaethylene glycol linkers (HEG3). The dashed line denotes the 5′-terminus of the published ribozyme (Johnston et al. 2001). (B) Substrate with 5′-cholesteryl anchor, linked via HEG3. The 5′-orientation is relative to the primer. The substrate consists of a 12mer RNA primer and a 23mer RNA template, coding for the addition of 11 nt. (C) Ribozyme, linked to an anchor at its 3′ terminus, replacing the 3′-terminal stem–loop with a segment that pairs to an oligo linked to a cholesterol anchor (Ch) or a stearate anchor (SA). (D) Substrate 3′-anchor. (E) Stearate anchors replaced cholesteryl anchors. The abbreviations for all constructs are listed (Table 1). Two additional substrate sequences with the same length are described in Materials and Methods.
TABLE 1.
Abbreviations for ribozyme and substrate constructs
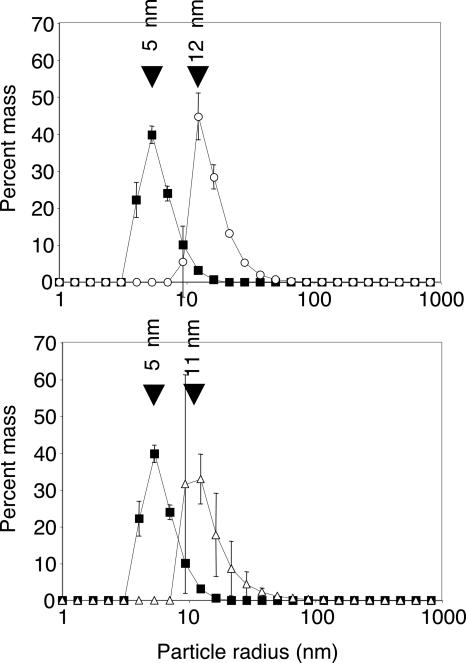
To test the ribozyme constructs for self-assembly into micelles, we dissolved them under the same conditions as used for polymerization reactions and measured the resulting particle sizes by dynamic light scattering (DLS) (Fig. 2). Ribozymes without cholesterol anchor gave particle sizes with a radius of ∼5 nm. This corresponded well with the value expected for the ribozyme monomer (total 206 nt) because the same value has been measured for the P4/P6 domain of the Group I intron, which is 160 nt long (Deras et al. 2000). In contrast, polymerase ribozymes equipped with 5′-terminal cholesterol anchors gave particles with a radius of ∼12 nm, while stearate anchors led to a particle radius of ∼11 nm. These values corresponded well with the size of the expected micellar aggregates: The radius of the hydrophobic core (1–2 nm for both anchors; Tanford 1972; Mazer et al. 1979), the length of the flexible linker (0–5 nm when collapsed or stretched), and the diameter of the ribozyme (∼10 nm, as measured) add up to a radius of 11–17 nm, consistent with the measured values and supporting the interpretation that micelles were formed. Hydrophobic anchors alone did not form detectable particles, probably because the corresponding micelles are too small to give detectable signals at these concentrations (data not shown).
FIGURE 2.
Particle sizes resulting from ribozymes without and with hydrophobic anchors, measured by DLS. (Filled squares) |Rz. (Circles) Ch–|Rz. (Triangles) SA–|Rz. The same data set for |Rz is displayed in both graphs for clarity. The percent mass values are averages of triplicate experiments, and error bars denote the standard deviations.
An upper limit for the number of monomers per micelles was estimated from the measured particle sizes. Assuming that the ribozymes with a diameter of 10 nm completely filled the volume of a sphere with a diameter of 22 nm (stearate anchors) or 24 nm (cholesteryl anchors), the number of monomers was 11 or 14, respectively. However, because the micelles may not have been spherical and the particle shape also influences the diffusion constants (Perrin 1934) measured by DLS, these upper limits are only rough estimates.
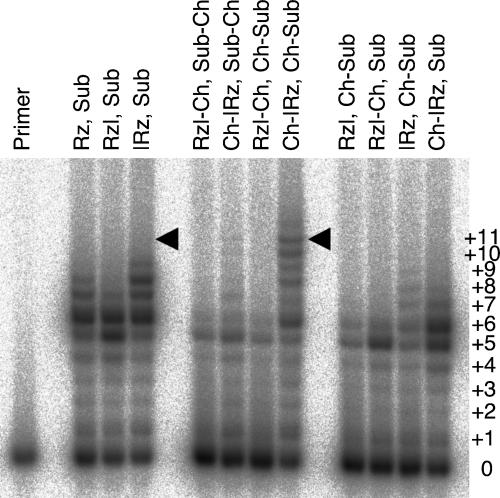
To test the effects of micellar colocalization on polymerization efficiency, we incubated the constructs with and without hydrophobic anchors under polymerization conditions (Fig. 3). Only in one out of four orientations of ribozyme and substrate we observed improved polymerization. When the polymerase and the substrate were anchored on their 5′ terminus (relative to ribozyme sequence and primer sequence, respectively), full-length extension was improved by more than threefold. This improvement required both ribozyme and substrate to carry hydrophobic anchors. If only one of them carried a hydrophobic anchor, no improved polymerization was visible, confirming that micelle formation was necessary to show an increase in full-length products.
FIGURE 3.
Effects of cholesteryl anchors on polymerization. The three left lanes show polymerization products without anchors. The four middle lanes show the products of polymerization with cholesteryl anchors, in all four orientations on the 5′ terminus and 3′ terminus on the ribozyme and the substrate, respectively. The four right lanes show polymerization products with only one cholesteryl anchor on either ribozyme or substrate. The positions of full-length extended products are indicated by arrowheads. The bands are annotated by the number of nucleotides that have been added. Reaction conditions were 184 mM free MgCl2 and 50 mM Tris/HCl, pH 8.5, at 12°C.
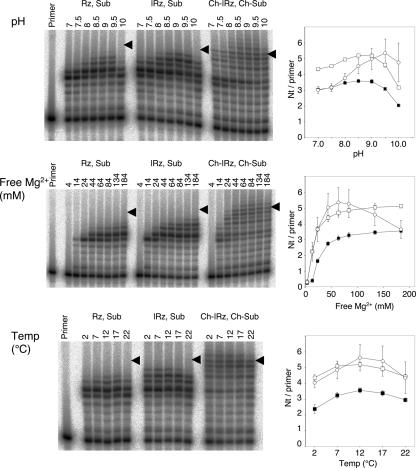
We optimized the reaction temperature, Mg2+ concentration, and pH for polymerization without and with hydrophobic anchors (Fig. 4). As a measure for polymerization efficiency, we quantified the average number of nucleotides added per primer molecule. Three different ribozyme constructs were employed, (1) the published ribozyme without anchor (Johnston et al. 2001); (2) the ribozyme with no hydrophobic anchor but the anchor duplex at its 5′ terminus; and (3) the ribozyme with cholesterol modification linked to its 5′ terminus. While the first two constructs were incubated with unmodified RNA substrates, the third construct was incubated with a substrate that carried a 5′-terminal cholesteryl anchor. Both ribozyme constructs without hydrophobic anchors operated best at 200 mM total magnesium (corresponding to 184 mM free Mg2+; Khan and Martell 1962) and a pH of 8.5, consistent with published values (Johnston et al. 2001). In contrast, the ribozymes with hydrophobic anchors operated most efficiently at a pH of 9.5 and a total Mg2+ concentration of 80 mM (64 mM free Mg2+). The optimal values of these parameters were interdependent. For example, a pH above 9.5 did not allow a free Mg2+ concentration of 184 mM because it caused precipitation of Mg(OH)2 and reduced polymerization efficiency. The best reaction temperature was always 12°C, regardless of the other conditions. Although the error bars describing the reactions of cholesterol-anchored constructs were often overlapping between adjacent conditions (e.g., between pH 9.0 and pH 9.5), the optimum of the average curve was identical to the optimum in each single experiment, confirming the positions of these optima.
FIGURE 4.
Optimization of reaction conditions without and with cholesteryl anchors. The positions of full-length extended products are indicated by arrowheads. (Filled squares) Rz, Sub. (Squares) |Rz, Sub. (Circles) Ch–|Rz, Ch–Sub. The values are the averages of triplicate experiments, with error bars denoting the standard deviation.
The 17-bp anchor duplex at the 5′ terminus of the ribozyme reduced the magnesium requirement for polymerization (Fig. 4). In the absence of the duplex, half-maximal polymerization efficiency was measured at a free Mg2+ concentration of 27 ± 2 mM, while the concentration was reduced to 16 ± 2 mM in the presence of the duplex (the same as 17 ± 5 mM with duplex and hydrophobic anchor).
The optimum of 64 mM free Mg2+, pH 9.5, was specific for the construct with hydrophobic anchor, whereas the optimum of 184 mM free Mg2+ and pH 8.5 was specific for the constructs without hydrophobic anchor. The construct with hydrophobic anchor yielded 100% more full-length product and 27% more nucleotides per primer at its own optimum, relative to the optimum of the reaction without anchors. The construct without hydrophobic anchors showed a weaker preference for its own optimal conditions relative to the optimum of the reaction with anchors, with 6% and 15% preference, respectively (data not shown).
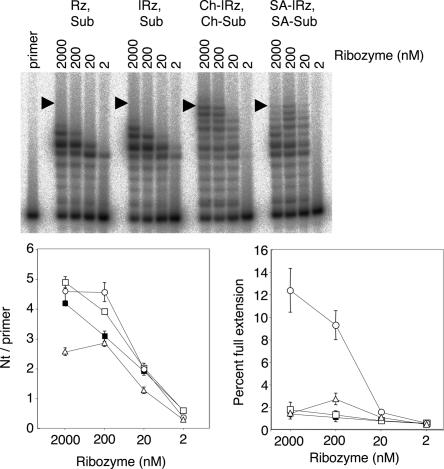
The pattern of polymerization intermediates showed that the micelle-based reaction was less sequence dependent than the reaction without hydrophobic anchors (Fig. 5). The latter led to the accumulation of intermediates after 5 and 6 nt had been added; this stalling has been analyzed before (Lawrence and Bartel 2003; Müller and Bartel 2003). In contrast, the polymerization with hydrophobic anchors did not show an accumulation of these intermediates (see Fig. 8, below). To quantitate this phenomenon, we expressed the polymerization efficiency in two ways, first by quantifying how many nucleotides were added on average per primer molecule, and second by quantifying the fraction of primers that were extended to full length of the template. The attachment of cholesterol anchors only modestly increased the number of nucleotides added per primer (from 4.9 nt/primer ± 0.1 to 5.6 nt/primer ± 0.2) but increased full-length extension by at least 10-fold (from ≤1.6% ± 0.2% to 17.1% ± 1.0%). However, the increase in the number of nucleotides per primer was not the main cause for the increase in the fraction of full-length extended primer because an increase in full-length extension sometimes coincided with a decrease in the nucleotides added per primer (e.g., Fig. 5, lanes 1,3,5,6).
FIGURE 5.
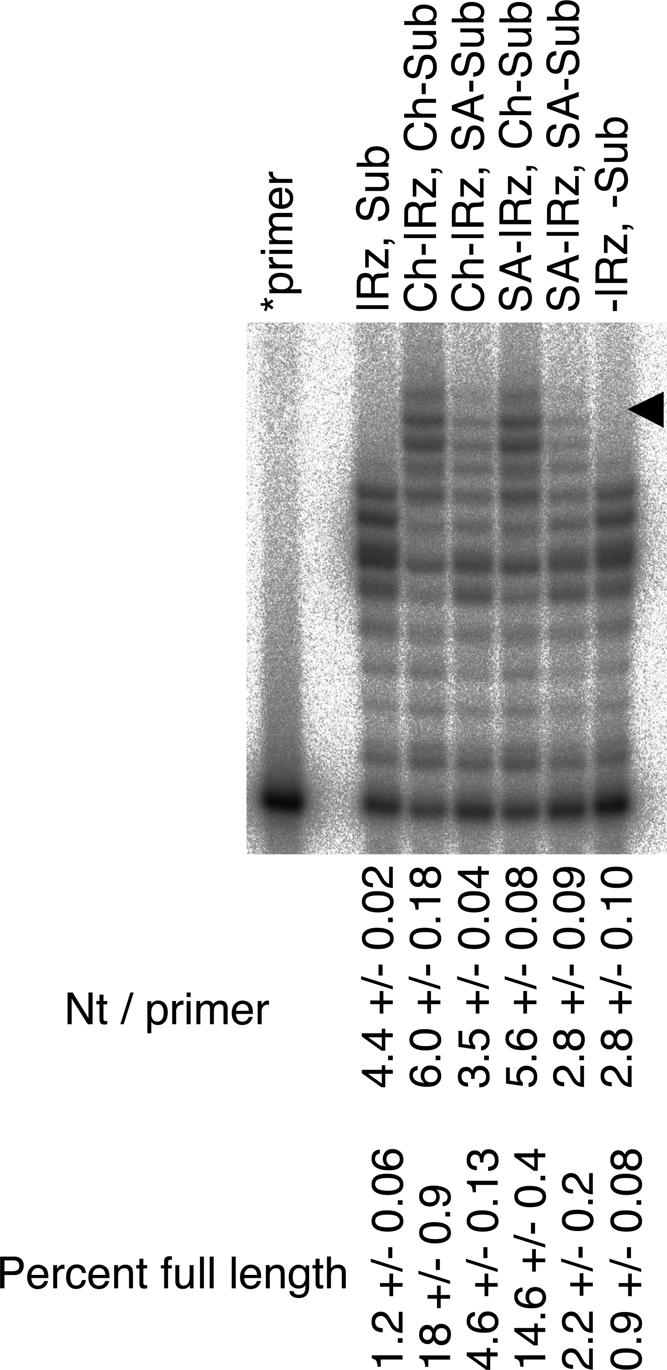
Effects of different anchor components on polymerization efficiency. The positions of full-length extended products are indicated by arrowhead. Reaction conditions were 64 free mM MgCl2, pH 9.5, and 12°C. The values are the averages of triplicate experiments (+/− standard deviation).
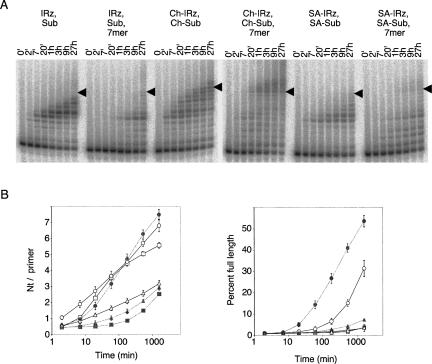
FIGURE 8.
Polymerization kinetics of the reactions without and with hydrophobic anchors. The positions of full-length extended products are indicated by arrowheads. (Squares) |Rz, Sub. (Gray squares) |Rz, Sub, 7mer. (Circles) Ch–|Rz, Ch–Sub. (Gray circles) Ch–|Rz, Ch–Sub, 7mer. (Triangles) SA–|Rz, SA–Sub. (Gray triangles) SA–|Rz, SA–Sub, 7mer. The reaction conditions were 184 mM free MgCl2, pH 8.5, 12°C without hydrophobic anchors and 64 mM free MgCl2, pH 9.5, 12°C with hydrophobic anchor. The values are the averages of triplicate experiments, with error bars denoting the standard deviation.
The length of the flexible linkers was not at a critical threshold concerning polymerization efficiency. When comparing the described reaction (with 21 ethylene glycol units per linker) to a reaction with shorter linkers (with 15 ethylene glycol units per linker), we found no significant differences (data not shown).
The effects of the cholesterol anchors could be general for hydrophobic groups, but they could also be specific to cholesterol or could be mediated by the ethylene glycol linkers. We addressed this question by substituting the cholesteryl units with stearyl units. The polymerization assay (Fig. 5) showed an increased full-length extension for stearyl anchors, similar to but not as pronounced as in the reaction using cholesteryl anchors. This could be explained by a smaller hydrophobic effect of stearyl groups relative to cholesteryl groups, resulting in lower micelle formation (see Discussion). Combinations of cholesterol anchors and stearyl anchors led to similar results. The ethylene glycol linkers in the absence of a hydrophobic residue did not improve full-length extension (Fig. 5). These results showed that hydrophobicity with consequent aggregation, and not other characteristics of the anchor, mediated the improved full-length polymerization.
At lower ribozyme concentrations, the polymerization efficiency of the non-anchored ribozyme decreases because substrate binding is limiting (Lawrence and Bartel 2003). However, it is possible that the micelle-based reaction could sustain good polymerization efficiency at low ribozyme concentrations if the micelles were stable. To test this, we reduced the concentrations of ribozyme in the polymerization reactions and compared the polymerization efficiencies without and with hydrophobic anchors (Fig. 6). The results showed a comparable drop in polymerization efficiency for all reactions upon lowering of ribozyme concentration, indicating that the micelles were not stable at low concentrations. For both reactions with hydrophobic anchors, this drop was shifted to lower concentrations, suggesting a slight improvement in substrate affinity with hydrophobic anchors. The reaction with stearate anchors was optimal with 200 nM ribozyme at 100 nM template. However, at all ribozyme concentrations the reaction with stearate anchors had lower efficiencies than the reaction with cholesterol anchors, and this did not change at a template concentration of 10 nM (data not shown). Because we were interested in the highest yield of full-length products, we focused our attention on the reaction with cholesterol anchors at 2 μM ribozyme concentration.
FIGURE 6.
Effects of ribozyme concentration on the polymerization reaction without and with hydrophobic anchors. The positions of full-length extended products are indicated by arrowheads. (Filled squares) Rz, Sub. (Squares) |Rz, Sub. (Circles) Ch–|Rz, Ch–Sub. (Triangles) SA–|Rz, SA–Sub. The reaction conditions were 184 mM free MgCl2, pH 8.5, 12°C without hydrophobic anchors and 64 mM free MgCl2, pH 9.5, 12°C with hydrophobic anchors. The values are the averages of triplicate experiments, with error bars denoting the standard deviation.
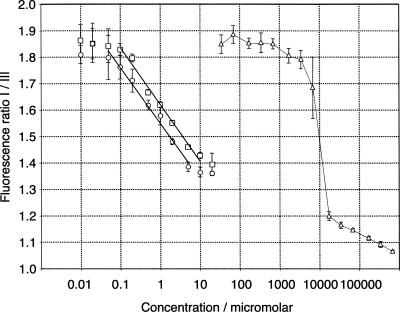
To test whether the micelle stability may control this concentration dependence of polymerization on the micelles, we measured the concentration dependence of micelle aggregation by pyrene fluorescence (Kalyanasundaram and Thomas 1977). At the buffer conditions used in the polymerization reaction, cholesterol-modified RNA anchors were titrated in the absence and presence of the polymerase ribozyme (Fig. 7). The intensity ratio of the first and the third vibronic band of pyrene was used to detect hydrophobic aggregates. We found that aggregation was detectable above a concentration of 100 nM of the cholesterol-modified RNA and increases up to at least 10 μM. This gradual aggregation over a more than 100-fold concentration range stands in contrast to the sharp phase transition during the aggregation of sodium dodecyl sulfate (SDS) micelles, at a defined critical micelle concentration (cmc) of 8 mM (Honda et al. 2004). For the cholesterol-modified RNA, the presence of the polymerase ribozyme increases the aggregation concentration by only 2.2-fold (Fig. 7, linear fits). These results show that micelles that are formed with cholesterol anchors under polymerization conditions with 2 μM ribozyme are not stable toward dilution. Therefore, micellar stability appears limiting for all described ribozyme polymerizations with cholesterol anchors.
FIGURE 7.
Aggregation of cholesterol-modified RNAs, as measured by pyrene fluorescence. The vertical axis denotes the ratio of the peak intensities from the first and the third vibronic peaks of pyrene. (Open circles) Cholesterol-modified RNA anchor. (Open squares) Equimolar concentrations of cholesterol-modified anchor and ribozyme. (Open triangles) sodium dodecyl sulfate. Error bars denote standard deviations of triplicate experiments. The straight lines are linear curve fits to the dynamic region of the data for the cholesterol-modified RNAs.
To analyze the accumulation kinetics of polymerization intermediates and to see whether the reaction time used in the previous experiments (15 h) gave representative results, we recorded polymerization kinetics for three different ribozyme constructs (Fig. 8). The accumulation kinetics of polymerization intermediates led to three observations: First, as observed in Figure 3, the reactions without anchors accumulated the polymerization intermediates +5 and +6, while the reaction with hydrophobic anchors prevented the accumulation of these intermediates. Second, the pattern observed at 9 h resembled that observed at 27 h. Therefore, the results of the previous experiments were not contingent on the 15-h time point used in those experiments.
The third observation regarded the effects of adding the 7mer oligonucleotide 5′-GGCACCA to the reaction. This 7mer is a part of the polymerase ribozyme (Fig. 1) and serves to restore a stem that was lost when constructing the polymerase ribozyme from its parent, a ligase ribozyme (Ekland and Bartel 1995; Johnston et al. 2001). Because the 7mer was not essential for the reaction, it was left out of the previous experiments in this study. However, in the context of the 5′-duplex (|Rz), the cholesterol anchor (Ch–|Rz), and the stearate anchor (SA–|Rz), the 7mer had strong effects on the reaction yield. While the previously published ribozyme is stimulated by the 7mer (Johnston et al. 2001), the presence of the 5′-duplex rendered the 7mer inhibitory on polymerization (Fig. 8A, first and second time courses). However, in the presence of 5′-duplex and cholesterol anchor, the 7mer improved polymerization efficiency to a level higher than all other reactions, adding on average more than 7 nt per primer and reaching more than 50% full-length product (Fig. 8A, third and fourth time courses). In the presence of the 5′-duplex and the stearate anchor, the 7mer caused a small but significant increase in both the added nucleotides per primer and the full-length product (fifth and sixth time courses), indicating that the 7mer might generally improve the efficiency of the micelle-based reaction.
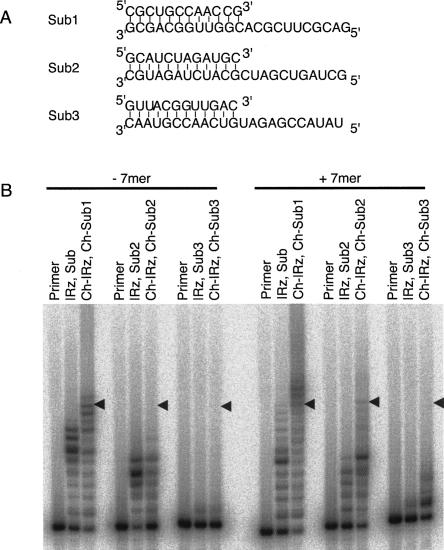
To test whether the effects of the hydrophobic anchor were specific for the substrate sequence used, we tested the effect of cholesterol anchors on the polymerization with three different substrate sequences (Fig. 9). The results showed that, in the absence of the 7mer, the hydrophobic anchors can have positive (Fig. 9A, substrate 1), negative (Fig. 9A, substrate 2), or no effects (Fig. 9A, substrate 3). In the presence of the 7mer, the cholesterol anchor always improved polymerization for all three templates three- to fourfold, when measuring added nucleotides per primer. Full-length products could not accurately be quantified for substrates 2 and 3, but substrate 1 yielded 50% of the primers extended to the full length in the presence of the 7mer. This was an increase of more than 20-fold over the ribozyme without micellar attachment.
FIGURE 9.
Effect of cholesteryl anchors and the 7mer GGCACCA on polymerizations with different substrate sequences. (A) Sequences of the three used substrates, with Sub1 being equal to the substrate used in the previous experiments. The substrate sequences are designed to have low nucleotide bias and low tendencies to form secondary structures. (B) Polymerization patterns with the use of the indicated substrate sequences. The positions of full-length extended products are indicated by arrowheads. All reaction conditions were 64 mM free MgCl2, pH 9.5, and 12°C. The experiments were performed in triplicate, and representative results are shown.
DISCUSSION
The polymerase ribozyme developed by Johnston et al. (2001) is limited in its polymerization efficiency, apparently because of weak substrate interactions (Lawrence and Bartel 2003), which are thought to be even weaker in specific sequence contexts, causing polymerization to stall. We show here that hydrophobic anchors can force ribozymes onto micelles and that the colocalization of ribozyme and substrate on these micelles can improve ribozyme polymerization.
The results showed that the presence of a 5′-terminal duplex decreases the required Mg2+ concentration and can improve polymerization efficiency. This is reminiscent of the recent finding by Zaher and Unrau (2007) that a 4-nt sequence inserted at the same position increases polymerization efficiency.
The micellar colocalization of ribozyme and substrate helps resolve stalls during polymerization, which in turn increases the full-length primer extension, for at least one substrate (Fig. 9). This effect is equivalent to reduced sequence dependence; however it is unclear how this is caused by micellar colocalization. We favor a combination of two explanations: First, the colocalization makes all substrates experience high local ribozyme concentrations, regardless of their sequence. The local ribozyme concentrations should increase from 2 μM in the nonanchored reaction to 0.5–3 mM in the anchored reaction, assuming that 2–14 ribozymes are limited to a sphere with a radius of 12 nm (Fig. 2). Whereas at micromolar ribozyme concentrations without anchors, the sequence-specific substrate binding affinity would dominate the polymerization kinetics and lead to stalling, the polymerization efficiency on the crowded surface of micelles would be limited by other factors that are less sequence dependent (see following paragraphs). A second cause for the decreased sequence dependence could be orientation effects. On the micelle, ribozyme and substrate can be aligned in the correct orientation for catalysis, which would reduce nonproductive interactions and increase productive interactions. This explanation is supported by our finding that the orientation of ribozyme and substrate on the micelle is important for polymerization (Fig. 3).
Why did the polymerization reaction not improve more, similar to a 1000-fold increase in local ribozyme concentration? Several factors might have prevented such a high increase in polymerization efficiency.
First, the high local concentration of RNAs colocalized on the micelles might favor inhibitory RNA–RNA interactions. This interpretation is supported by the observation that ribozyme concentrations above 15 μM are inhibitory for ribozyme polymerization (Lawrence and Bartel 2003).
Second, the micelles are not fully formed under the conditions used, with 2 μM cholesteryl RNA conjugates (Fig. 7). Our pyrene fluorescence data suggest a lower aggregation limit of 100 nM for cholesteryl RNAs, which is 10-fold higher than the concentration at which unmodified cholesterol starts to aggregate (Renshaw et al. 1983). In addition, the concentration at which micelles are fully formed is at least 10 μM, and thereby at least 10-fold higher than the value measured for cholesterol–oligoethylene glycol conjugates (Beugin et al. 1998). This difference is probably caused by the attachment of RNA to the cholesterol, which should entropically disfavor micelle formation. This hypothesis is supported by our finding that an increase in RNA size, by attaching the polymerase ribozyme to the RNA anchor, raises the aggregation concentrations about 2.2-fold (Fig. 7). In our experiments, the assembly of cholesteryl–RNA micelles is taking place over a more than 100-fold concentration range (Fig. 7). Such a stepwise aggregation behavior appears to be typical for cholesterol derivatives (Matsuoka et al. 2006).
The stearate anchor is expected to force a smaller fraction of RNAs into micelles because the cmc of stearate micelles is much higher, around 200 μM (Evans 1956). However, the true cmc of the stearate/ribozyme conjugates might be very different because it can be decreased by interactions of the ethylene glycol chains (Weil et al. 1958) that links stearate anchors with ribozymes or by interactions between the bound RNAs, and it can be increased because the large size of the ribozymes sterically prevents their micelles from reaching the optimum aggregation number of around 100 (Tanford 1972). These results and the weaker effects of stearate anchors on polymerization anchors support the interpretation that the stearate conjugates form less stable micelles than the cholesterol conjugates.
The 7mer oligonucleotide GGCACCA enhances polymerization efficiency if no 5′-terminal duplex is present (Johnston et al. 2001). This 7mer pairs to a sequence on the ribozyme, adjacent to the catalytic site, to form a helix termed P2. Two explanations for the stimulatory effect on polymerization are that (1) the helix improves the substrate-binding site on the ribozyme and (2) the 7mer shields the single-stranded sequence from inhibitory interactions with other sequences. However, the effects of the 7mer on the micelle-anchored reaction present a more complicated picture: In the presence of the 5′-duplex, the 7mer inhibits polymerization if no hydrophobic anchor is present, while it improves polymerization with a hydrophobic anchor (Fig. 8). This could be explained by a competition between the 5′-terminal duplex and the 7mer for the same position near the substrate-binding site. In the absence of cholesterol anchors, this inhibitory effect would be dominating, while in the presence of cholesterol anchors, the double strand could be pulled away from the site of competition by the hydrophobic anchors at the 5′-terminal duplex. Alternatively, the 7mer could be more important in the crowded conditions on the micelles because of the potential shielding effect of the 7mer against unspecific interactions.
Implications for the origin of life
For a molecular system to qualify as living, self-replication is not sufficient. A second essential requirement for life is the ability to undergo Darwinian evolution. A population of polymerases replicating each other free in solution could not evolve into improved polymerases. Any improved polymerase that emerges would replicate other molecules in the vicinity more efficiently but would have no advantage itself and thus would not increase in relative abundance. A physical link between the improved enzyme and the molecules that encoded it would enable this preferential replication to occur and thereby enable Darwinian evolution. In contemporary biology, this physical link comes in the form of cellular encapsulations, but in early life forms it might have existed in the form of stable micelles, base-pairing, adsorption to suspended particles, or colonies adsorbed to mineral surfaces (Orgel 2004).
A number of characteristics make micelles attractive as evolutionary precursors of membrane encapsulations. First, micelles are easier to replicate than vesicles. The only experimentally validated mechanism that could have divided vesicles in the early origin of life is based on physical shearing forces, but is of limited prebiotic plausibility (Hanczyc et al. 2003). In contrast, micelles should be much easier to divide because two mechanisms limit the maximum size of ribozyme-coated micelles, one being the chain length of the amphiphile (Tanford 1972), the other one being steric constraints of the ribozymes on the surface (see Fig. 2 legend). The stochastic inheritance of ribozymes from mother micelle to daughter micelles could ensure that enough daughter micelles obtain all the different RNAs that are necessary for self-replication. For example, under the assumption that the autocatalytic set of RNAs consists of two RNA species (a polymerase and a second RNA that encodes the polymerase), and the micelle contains a maximum of 14 RNAs with 7 RNAs being passed to each daughter micelle, the likelihood of both daughter micelles obtaining both polymerase and substrate would be more than 99%. If three or four RNA species are linked on the micelles, 67% or 19% of the daughter micelles would contain the whole set of RNAs, respectively.
A second characteristic that could favor micelles over membrane encapsulations in an early RNA world is that micelles tolerate much higher magnesium concentrations than vesicles, while magnesium is important for ribozyme catalysis. Vesicles have a tendency to aggregate and fuse even at magnesium concentrations below 2 mM and monovalent ion concentrations below 200 mM (Monnard et al. 2002). Only with alcohol modifications of the headgroup have functional vesicles been found to withstand magnesium concentrations of up to 4 mM (Chen et al. 2005). However, ribozyme catalysis is usually most efficient at higher magnesium concentrations. Although ribozymes can function in the absence of magnesium at high monovalent ion concentrations (Murray et al. 1998; Nakano et al. 2001), these conditions do not support stable vesicles either (Monnard et al. 2002). Therefore, stable vesicles can coexist with functional ribozymes only in a narrow range of conditions, with trade-offs for both vesicles and ribozymes (Chen et al. 2005). In contrast, we show here that ribozyme-coated micelles do not fuse and can improve ribozyme catalysis at free magnesium concentrations up to at least 184 mM.
A third characteristic of micelles that would make them good precursors of vesicles is that micelles can act as microreactors in an aqueous environment. Some reactions that are inhibited in aqueous solution proceed efficiently in micellar microenvironments, the hydrophobic core, or the micellar surface (for review, see Tascioglu 1996). One example that might be of prebiotic relevance is the catalysis of aldol reactions by proline, which proceeds most efficiently in organic solvents (Hajos and Parrish 1974). This reaction does not proceed well in water but gives high yields in the presence of surfactants such as SDS (Cordova et al. 2002; Peng et al. 2003), with excellent enantioselectivity if the pyrrolidine moiety of proline is covalently linked to the surfactant (Mase et al. 2006). The micellar microenvironment has also been observed to help ribozyme catalysis, specifically the self-cleavage of a 4-nt bulge in the anticodon stem of tRNATyr precursor from Arabidopsis thaliana (Riepe et al. 1999). However, the mechanism of this micellar reaction enhancement appears to be different from the colocalization and orientation effects seen in the current study. For micellar catalysis of pre-tRNATyr self-cleavage, amphiphile micelles are added to the pre-tRNATyr solution, and the hydrophobic core of the micelles appears to be directly involved in catalysis, through changing the local polarity at the catalytic site. Therefore, the hydrophobic core of micellar RNA aggregates can serve the same roles as the generally hydrophobic core of globular proteins (Dill 1990), as structural organizing center (this study) and as hydrophobic pockets that directly assist in catalysis (Riepe et al. 1999).
Fourth, micelles would also fit the role as evolutionary precursors of vesicles because the evolutionary pathway from micelles to vesicles does not appear to be problematic. Small changes in pH, solvent polarity (Hargreaves and Deamer 1978), chemistry of the hydrophobic tail (Walter et al. 2000; Kleinschmidt and Tamm 2002), or amphiphilic additives (Gantz et al. 1999) can determine whether amphiphiles assemble in micelles or double-layered membrane vesicles. Additionally, the micelle–vesicle transition can be aided by catalysis from a variety of minerals (Hanczyc et al. 2003). This means that an early, micellar evolutionary stage might be chemically well equipped to transition into a cellular stage.
A few problems face the model of micelles as evolutionary units. First, micelles do not protect their attached ribozymes against molecular parasites. Molecular parasites are RNAs with excellent templating ability but without useful catalytic abilities. Such molecules can quickly overtake a replicating system (Mills et al. 1967; Breaker and Joyce 1994) and prevent replication of the catalysts. This problem may be insignificant for early stages of micellar life when the concentrations of free RNA are low, reducing the encounter frequency between micelles and parasitic nucleic acids. However, a membrane encapsulation would be required as soon as higher population densities are reached. Second, the kinetic stabilities of micellar assemblies can be a problem for self-replicating systems. If the half-life of the micelle is shorter than the time required for self-replication, the genotype–phenotype link that rewards “fit” genomic molecules with replication will be lost. The life times of the most stable micelles reach only a few minutes (Tao and Uhrich 2006), which is below the hours of time that the current polymerase ribozyme requires to extend 50% of all primer molecules by only 1/20 of the ribozyme length. However, when vesicles rather than micelles are considered, cholesterol can have interchange kinetics between vesicles of several hours (Wattenberg and Silbert 1983; Yeagle and Young 1986), indicating that sufficiently slow kinetics may be achievable. Additional forces that stabilize the micelles are either additives that stabilize the hydrophobic core, such as alkyl alcohols (Patist et al. 1998), or interactions between the ribozymes on the surface of micelles, such as short complementary sequences between ribozymes. A low rate of micellar instability could also act in favor of early life, by facilitating the “budding-off” of daughter micelles, as well as by facilitating the exchange of genetic material between micellar units, potentially leading to more fit combinations of ribozymes. A small-molecule metabolism, which would require an encapsulation, is not essential at this early stage of evolution: When a first self-replicating unit of ribozymes originated from prebiotic processes (or a non-RNA organism), all molecules necessary for RNA formation must be present in the solution. This makes it unnecessary to synthesize these molecules through a metabolism. In fact, it appears unlikely that a macromolecule-catalyzed metabolism existed at this initial stage because it would assume the simultaneous appearance of a polymerase ribozyme and a set of metabolic ribozymes. As efficiently self-replicating ribozyme systems reduce available precursors, a membrane encapsulation would become more important so that metabolically competent ribozyme systems can harvest the fruits of their own catalysis.
MATERIALS AND METHODS
Ribozymes were synthesized by in vitro transcription from PCR products, using T7 polymerase as described (Johnston et al. 2001). Transcription products were purified by 7 M urea 10% polyacrylamide gel electrophoresis. Sequence modifications were introduced into the ribozyme sequence by amplification of the PCR template with modified PCR primers (purchased from IDT). The primer sequences were 5′-TTCTAATACGACTCACTATAGGAAAAAGACAAATCTGCCCTCAGA-3′ and 5′-TTCAGATTGTAGCCTTCGGGG-3′ for the 5′-anchored ribozyme and 5′-TTCTAATACGACTCACTATAGAAGGCtACAAtCtGAAGGAAAAAGACAAATCTGCCCTCAGA-3′ and 5′-GGAGCCGAAGCTCCGGGGATTATGACC-3′ for the 3′-anchored ribozyme.
RNA oligonucleotides were purchased from Dharmacon, including RNAs with cholesterol anchors. RNA sequences for substrates 1, 2, and 3 are as described in Figure 8. The cholesterol anchors include three ethylene glycol linkers and are linked to the RNA via further 18 ethylene glycol linkers, if not indicated otherwise. The ethylene glycol linkers are connected via phosphodiester groups every six ethylene glycol units, due to their stepwise phosphoamidite synthesis.
Stearate conjugates were synthesized from amino-modified RNAs (including 20 ethylene glycol linkers as described for the cholesterol-modified RNAs) via NHS-activated stearic acid. The stearic acid activation was modified from a published protocol (Sehgal and Vijay 1994). A solution of 56 mM EDC (1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride), 32 mM NHS (N-hydroxysuccinimide), and 20 mM stearic acid in anhydrous DMF was heated for 90 min at 70°C. After cooling to room temperature, 1 volume of this mixture was added to 19 volumes of a solution of 10 μM amino-modified RNA in 100 mM MES (2-(4-Morpholino)ethanesulfonic acid)/KOH (pH 6.5). After incubation for 12 h at 22°C, the RNA was precipitated and washed with anhydrous THF, dissolved in water, and purified by reverse-phase HPLC on a C18 column, using an acetonitrile gradient (8%–80%) in 100 mM TEAA (triethyl ammonium acetate) at pH 7.0. The products were vacuum-dried and dissolved in water. Yields were 30%–70%. The products ran as single band on 1% SDS 7 M urea 20% polyacrylamide gel electrophoresis after radioactive labeling of the 3′ terminus using 5′-[32P]pCp and T4 RNA ligase.
Dynamic light scattering experiments were performed on a Dynapro temperature controlled microsampler (Protein Solutions). Ribozyme constructs were pretreated as in polymerization experiments and dissolved in the same buffer (200 mM MgCl2 and 50 mM Tris/HCl at pH 8.5) but without addition of NTPs. In each experiment, scattering data were collected from 200 measurements, each of which lasted 1 sec. The data were converted to percent mass distributions with the software supplied by the manufacturer. The viscosity was approximated with that of 1× PBS, but other plausible entries did not significantly change the results.
Pyrene fluorescence measurements were performed on a fluorescence spectrophotometer from JY Horiba (model FL3-11) with a standard photomultiplier (Hamamatsu R928). Excitation was done at 337 nm with a slit width of 5 nm. Emission was monitored from 350 nm to 450 nm, using a slit width of 1 nm, an increment of 1 nm, and an integration time of 1 sec. All measurents were done at 20°C–22°C. Buffer conditions were 64 mM MgCl2 and 50 mM Tris/HCl (pH 8.5) for cholesterol–RNA samples, and water for SDS samples. All samples contained an additional 1% ethanol and 100 nM pyrene. Reliable measurements were not obtained above a concentration of 20 μM RNA–cholesterol conjugates due to increased turbidity. After subtracting the water spectrum from all measured spectra, the maximum values of vibronic peak I (∼373 nm) were divided by the maximum values of vibronic peak III (∼383 nm). The linear fits to both aggregation curves of cholesteryl-modified RNAs were done by least squares fitting, using the values in the dynamic range of each curve (50 nM–5 μM for the cholesteryl anchor and 100 nM–10 μM for the cholesteryl anchor with ribozyme).
Ribozyme reactions were performed as described (Johnston et al. 2001). All RNAs were dissolved in water at the appropriate concentration, heat denatured (2 min/80°C) and slowly cooled (at 0.1°C/sec) to the reaction temperature. Reactions were started by adding a 2.5× reaction buffer containing magnesium chloride, buffer (Tris/HCl for pH 7.0–8.5; CHES (N-cyclohexyl-2-aminoethanesulfonic acid)/KOH for pH 9.0–10), and NTPs so that the final concentrations were 50 mM buffer and 4 mM of each NTP. Reaction times were 15 h, if not indicated otherwise. The reactions were stopped by the addition of an equal volume of stop buffer (8 M urea, 200 mM Na2EDTA at pH 8.4) and a template-complementary RNA added in 20-fold excess over the template. The mixtures were heat denatured (2 min/80°C) and cooled to room temperature before loading and separating on 7 M urea 1× TBE 20% PAGE. Autoradiographs of the separations were recorded by PhosphorImaging (Fuji BAS-2500) and quantitated using the software Image Gauge v4.2. Shifts higher than the full-length extension by 11 nt were counted as full-length extension. The values for “nucleotides per primer” were obtained by multiplying the fraction of intensity for each band with the number of added nucleotides corresponding to that band. The background signal was around 0.3 for the average number of nucleotides and ∼0.7% for the fraction of full-length extended primers.
ACKNOWLEDGMENTS
We thank Inese Smukste and Matt Hartmann for chemical advice, Mary Rozenman and Mike Lawrence for help with HPLC purifications, Judy Kim for helping us to use her fluorescence spectrophotometer, and Burckhard Seelig, Martin Hancyc, Scott Baskerville, Ed Curtis, and Graham Ruby for helpful discussions. Financial support was provided by a postdoctoral fellowship (MU-1708 1/1) from the German Research Council DFG, a postdoctoral fellowship (GM071283), and a research grant (GM061835) from the NIH.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.494508.
REFERENCES
- Bartel, D.P., Szostak, J.W. Isolation of new ribozymes from a large pool of random sequences. Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- Beugin, S., Edwards, K., Karlsson, G., Ollivon, M., Lesieur, S. New sterically stabilized vesicles based on nonionic surfactant, cholesterol, and poly(ethylene glycol)-cholesterol conjugates. Biophys. J. 1998;74:3198–3210. doi: 10.1016/S0006-3495(98)78026-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker, R.R., Joyce, G.F. Emergence of a replicating species from an in vitro RNA evolution reaction. Proc. Natl. Acad. Sci. 1994;91:6093–6097. doi: 10.1073/pnas.91.13.6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, I.A., Salehi-Ashtiani, K., Szostak, J.W. RNA catalysis in model protocell vesicles. J. Am. Chem. Soc. 2005;127:13213–13219. doi: 10.1021/ja051784p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordova, A., Notz, W., Barbas C.F., 3rd Direct organocatalytic aldol reactions in buffered aqueous media. Chem. Commun. (Camb.) 2002:3024–3025. doi: 10.1039/b207664k. [DOI] [PubMed] [Google Scholar]
- Deras, M.L., Brenowitz, M., Ralston, C.Y., Chance, M.R., Woodson, S.A. Folding mechanism of the Tetrahymena ribozyme P4–P6 domain. Biochemistry. 2000;39:10975–10985. doi: 10.1021/bi0010118. [DOI] [PubMed] [Google Scholar]
- Dill, K.A. Dominant forces in protein folding. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- Ekland, E.H., Bartel, D.P. The secondary structure and sequence optimization of an RNA ligase ribozyme. Nucleic Acids Res. 1995;23:3231–3238. doi: 10.1093/nar/23.16.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, H.C. Alkyl sulphates. Part I. Critical micelle concentration of the sodium salts. J. Chem. Soc. 1956;78:579–586. [Google Scholar]
- Gantz, D.L., Wang, D.Q., Carey, M.C., Small, D.M. Cryoelectron microscopy of a nucleating model bile in vitreous ice: Formation of primordial vesicles. Biophys. J. 1999;76:1436–1451. doi: 10.1016/S0006-3495(99)77304-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajos, Z.G., Parrish, D.R. Asymmetric synthesis of bicyclic intermediates of natural product chemistry. J. Org. Chem. 1974;39:1615–1621. [Google Scholar]
- Hanczyc, M.M., Fujikawa, S.M., Szostak, J.W. Experimental models of primitive cellular compartments: Encapsulation, growth, and division. Science. 2003;302:618–622. doi: 10.1126/science.1089904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves, W.R., Deamer, D.W. Liposomes from ionic, single-chain amphiphiles. Biochemistry. 1978;17:3759–3768. doi: 10.1021/bi00611a014. [DOI] [PubMed] [Google Scholar]
- Honda, C., Kamizono, H., Matsumoto, K., Endo, K. Studies on bovine serum albumin-sodium dodecyl sulfate complexes using pyrene fluorescence probe and 5-doxylstearic acid spin probe. J. Colloid Interface Sci. 2004;278:310–317. doi: 10.1016/j.jcis.2004.06.023. [DOI] [PubMed] [Google Scholar]
- James, K.D., Ellington, A.D. The fidelity of template-directed oligonucleotide ligation and the inevitability of polymerase function. Orig. Life Evol. Biosph. 1999;29:375–390. doi: 10.1023/a:1006544611320. [DOI] [PubMed] [Google Scholar]
- Johnston, W.K., Unrau, P.J., Lawrence, M.S., Glasner, M.E., Bartel, D.P. RNA-catalyzed RNA polymerization: Accurate and general RNA-templated primer extension. Science. 2001;292:1319–1325. doi: 10.1126/science.1060786. [DOI] [PubMed] [Google Scholar]
- Kalyanasundaram, K., Thomas, J.K. Environmental effects on vibronic band intensities in pyrene monomer fluorescence and their application in studies of micellar systems. J. Am. Chem. Soc. 1977;99:2039–2044. [Google Scholar]
- Khan, M.M.T., Martell, A.E. Metal chelates of adenosine triphosphate. J. Phys. Chem. 1962;66:10–15. [Google Scholar]
- Kleinschmidt, J.H., Tamm, L.K. Structural transitions in short-chain lipid assemblies studied by (31)P-NMR spectroscopy. Biophys. J. 2002;83:994–1003. doi: 10.1016/S0006-3495(02)75225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger, K., Grabowski, P.J., Zaug, A.J., Sands, J., Gottschling, D.E., Cech, T.R. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena . Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Lawrence, M.S., Bartel, D.P. Processivity of ribozyme-catalyzed RNA polymerization. Biochemistry. 2003;42:8748–8755. doi: 10.1021/bi034228l. [DOI] [PubMed] [Google Scholar]
- Lawrence, M.S., Bartel, D.P. New ligase-derived RNA polymerase ribozymes. RNA. 2005;11:1173–1180. doi: 10.1261/rna.2110905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mase, N., Nakai, Y., Ohara, N., Yoda, H., Takabe, K., Tanaka, F., Barbas C.F., 3rd Organocatalytic direct asymmetric aldol reactions in water. J. Am. Chem. Soc. 2006;128:734–735. doi: 10.1021/ja0573312. [DOI] [PubMed] [Google Scholar]
- Matsuoka, K., Suzuki, M., Honda, C., Endo, K., Moroi, Y. Micellization of conjugated chenodeoxy- and ursodeoxycholates and solubilization of cholesterol into their micelles: Comparison with other four conjugated bile salts species. Chem. Phys. Lipids. 2006;139:1–10. doi: 10.1016/j.chemphyslip.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Mazer, N.A., Carey, M.C., Kwasnick, R.F., Benedek, G.B. Quasielastic light scattering studies of aqueous biliary lipid systems. Size, shape, and thermodynamics of bile salt micelles. Biochemistry. 1979;18:3064–3075. doi: 10.1021/bi00581a024. [DOI] [PubMed] [Google Scholar]
- Mills, D.R., Peterson, R.L., Spiegelman, S. An extracellular Darwinian experiment with a self-duplicating nucleic acid molecule. Proc. Natl. Acad. Sci. 1967;58:217–224. doi: 10.1073/pnas.58.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnard, P.A., Apel, C.L., Kanavarioti, A., Deamer, D.W. Influence of ionic inorganic solutes on self-assembly and polymerization processes related to early forms of life: Implications for a prebiotic aqueous medium. Astrobiology. 2002;2:139–152. doi: 10.1089/15311070260192237. [DOI] [PubMed] [Google Scholar]
- Murray, J.B., Seyhan, A.A., Walter, N.G., Burke, J.M., Scott, W.G. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- Müller, U.F. Re-creating an RNA world. Cell. Mol. Life Sci. 2006;63:1278–1293. doi: 10.1007/s00018-006-6047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, U.F., Bartel, D.P. Substrate 2′-hydroxyl groups required for ribozyme-catalyzed polymerization. Chem. Biol. 2003;10:799–806. doi: 10.1016/s1074-5521(03)00171-6. [DOI] [PubMed] [Google Scholar]
- Nakano, S., Proctor, D.J., Bevilacqua, P.C. Mechanistic characterization of the HDV genomic ribozyme: Assessing the catalytic and structural contributions of divalent metal ions within a multichannel reaction mechanism. Biochemistry. 2001;40:12022–12038. doi: 10.1021/bi011253n. [DOI] [PubMed] [Google Scholar]
- Orgel, L.E. Prebiotic chemistry and the origin of the RNA world. Crit. Rev. Biochem. Mol. Biol. 2004;39:99–123. doi: 10.1080/10409230490460765. [DOI] [PubMed] [Google Scholar]
- Patist, A., Axelberd, T., Shah, D.O. Effect of long chain alcohols on micellar relaxation time and foaming properties of sodium dodecyl sulfate solutions. J. Colloid Interface Sci. 1998;208:259–265. doi: 10.1006/jcis.1998.5830. [DOI] [PubMed] [Google Scholar]
- Peng, Y.-Y., Ding, Q.-P., Li, Z., Wang, P.G., Cheng, J.-P. Proline catalyzed aldol reactions in aqueous micelles: An environmentally friendly reaction system. Tetrahedron Lett. 2003;44:3871–3875. [Google Scholar]
- Perrin, F. Brownian movement of an ellipsoid. I. Dielectric dispersion of an ellipsoidal molecule. J. Physique Radium. 1934;5:497–511. [Google Scholar]
- Renshaw, P.F., Janoff, A.S., Miller, K.W. On the nature of dilute aqueous cholesterol suspensions. J. Lipid Res. 1983;24:47–51. [PubMed] [Google Scholar]
- Riepe, A., Beier, H., Gross, H.J. Enhancement of RNA self-cleavage by micellar catalysis. FEBS Lett. 1999;457:193–199. doi: 10.1016/s0014-5793(99)01038-8. [DOI] [PubMed] [Google Scholar]
- Sehgal, D., Vijay, I.K. A method for the high efficiency of water-soluble carbodiimide-mediated amidation. Anal. Biochem. 1994;218:87–91. doi: 10.1006/abio.1994.1144. [DOI] [PubMed] [Google Scholar]
- Szostak, J.W., Bartel, D.P., Luisi, P.L. Synthesizing life. Nature. 2001;409:387–390. doi: 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- Tanford, T. Micelle shape and size. J. Phys. Chem. 1972;76:3020–3024. [Google Scholar]
- Tao, L., Uhrich, K.E. Novel amphiphilic macromolecules and their in vitro characterization as stabilized micellar drug delivery systems. J. Colloid Interface Sci. 2006;298:102–110. doi: 10.1016/j.jcis.2005.12.018. [DOI] [PubMed] [Google Scholar]
- Tascioglu, S. Micellar solutions as reaction media. Tetrahedron. 1996;52:11113–11152. [Google Scholar]
- Walter, A., Kuehl, G., Barnes, K., VanderWaerdt, G. The vesicle-to-micelle transition of phosphatidylcholine vesicles induced by nonionic detergents: Effects of sodium chloride, sucrose, and urea. Biochim. Biophys. Acta. 2000;1508:20–33. doi: 10.1016/s0304-4157(00)00005-8. [DOI] [PubMed] [Google Scholar]
- Wattenberg, B.W., Silbert, D.F. Sterol partitioning among intracellular membranes. Testing a model for cellular sterol distribution. J. Biol. Chem. 1983;258:2284–2289. [PubMed] [Google Scholar]
- Weil, J.K., Bistline R.G., Jr, Stirton, A.J. The critical micelle concentration of ether alcohol sulfates, (ROC2H4)i–OSO3Na. J. Phys. Chem. 1958;62:1083–1085. [Google Scholar]
- Yeagle, P.L., Young, J.E. Factors contributing to the distribution of cholesterol among phospholipid vesicles. J. Biol. Chem. 1986;261:8175–8181. [PubMed] [Google Scholar]
- Zaher, H.S., Unrau, P.J. Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA. 2007;13:1017–1026. doi: 10.1261/rna.548807. [DOI] [PMC free article] [PubMed] [Google Scholar]