Abstract
Recent advances in microscopic techniques have shed light on the roles of specific subcellular structures in the regulation of gene expression. One such structure is the stress granule (SG), which is engaged in stress-triggered translational arrest by sequestering pre-initiation complexes of translation. Recent studies revealed the spatial, compositional, and functional linkage of the SG to the processing body (P-body), another cytoplasmic structure that has been implicated in mRNA degradation and siRNA- or miRNA-mediated gene silencing. In this study, we report that PCBP2, a facilitator of IRES (Internal Ribosomal Entry Site)-mediated translation, is a novel constituent of the SG and P-body. Immunofluorescence studies revealed that while PCBP2 is diffusely distributed throughout the nucleoplasm and the cytoplasm, the protein is enriched in a subset of P-bodies under normal conditions. Upon exposure to heat and arsenic stress, PCBP2 became predominantly accumulated at the SG, but was still present in Dcp1a-positive P-bodies. Live-cell imaging revealed the dynamic association of PCBP2-enriched P-bodies and the SG, and FRAP experiments demonstrated that PCBP2 actively moves in and out of the SG and P-body. Taken together, these results suggest that PCBP2 shuttles between the cytoplasm and the two structures under stress. We propose that PCBP2 may be involved in stress-induced remodeling of mRNP complexes and that it may also play a role in the rapid transition of certain silenced mRNAs into a translationally active state. Additionally, given the property of PCBP2 as a nuclear–cytoplasmic shuttling protein, PCBP2 may play a role in directly targeting nascent mRNPs to specific P-bodies for storage.
Keywords: FRAP, PCBP2, processing body, stress granule
INTRODUCTION
In recent years, a number of subcellular bodies have been identified due to advancements in microscopic techniques (Misteli 2001). Many of these structures were discovered to be involved in gene expression at distinct levels. The processing body, or P-body, is one of the most prominent examples. P bodies were originally identified as cytoplasmic foci that are enriched in RNA-degrading enzymes. Subsequent studies revealed that the P-body serves as a site of mRNA degradation (Sheth and Parker 2003; Cougot et al. 2004) and is also involved in siRNA- and miRNA-mediated repression of gene expression (for review, see Jakymiw et al. 2007). While the P-body is present in cells at rest, some subcellular structures appear specifically under stressful conditions. The stress granule (SG) is one such structure, a cytoplasmic aggregate rapidly formed upon environmental stress (Kedersha et al. 1999). The SG is considered to be a site of translational repression, in which nontranslated transcripts accumulate (Kedersha and Anderson 2002). Recently, close association between the SG and the P-body was demonstrated; it was revealed that the SG and P-body are spatially, compositionally, and functionally related. Based on this observation, a model in which the SG serves as a site of triage, sorting, and mRNP remodeling for silenced mRNA, after which certain transcripts are routed to the neighboring P-bodies for degradation, was proposed recently (Kedersha et al. 2005).
In the present study, we report that PCBP2 (poly-C binding protein 2) is a novel constituent of the SG and P-body. Originally, in an effort to discover SG components through a yeast two-hybrid screen against TIA1, a major component of the SG and a facilitator of SG assembly, we identified PCBP2 (alternately known as hnRNP-E2 and αCP2) as a candidate TIA1-interacting protein. (Interaction between TIA-1 and PCBP2 was verified as shown in Supplemental Figure 1. Further studies are under way and will be reported elsewhere.) PCBP2 was initially identified as a component of the α-complex of the human α-globin mRNA, which greatly increases mRNA stability (Kiledjian et al. 1995). Currently, PCBP2 is best characterized as a facilitator of internal ribosomal entry site (IRES)-mediated translation of both viral and cellular transcripts (Parsley et al. 1997; Evans et al. 2003; Bedard et al. 2004), but it is also implicated in other important biological processes, including transcriptional regulation and translational silencing (Makeyev and Liebhaber 2002). PCBP2 was previously described as a nuclear–cytoplasmic shuttling protein, which allows the protein to fulfill such diverse functions (Chkheidze and Liebhaber 2003). Using immunofluorescence methods, we discovered that while PCBP2 is diffusely distributed in the nucleoplasm and the cytoplasm, it resides in a subset of Dcp1a- and eIF4E-T-positive P-bodies in cells at rest. Upon environmental stress, cytoplasmic PCBP2 became dominantly localized to the SG but was still present in P-bodies. Fluorescence recovery after photobleaching (FRAP) studies revealed that PCBP2 moves in and out of both the SG and P-body. The effect of siRNA-mediated depletion of PCBP2 on P-body and SG integrity, and the sequence requirements for the recruitment to the two structures, was also examined.
RESULTS AND DISCUSSION
Subcellular localization of PCBP2 in cells at rest
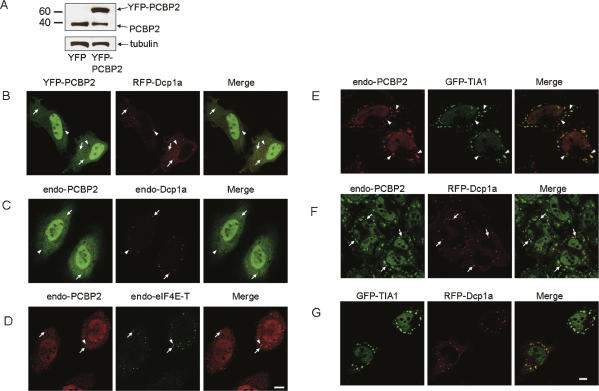
After the identification of PCBP2 as a candidate TIA1-binding protein, full-length PCBP2 was amplified from a human fetal brain cDNA library and a plasmid encoding YFP-tagged PCBP2 was constructed (termed YFP-PCBP2). Expression of the fusion protein in HeLa cells was verified by Western blotting, as shown in Figure 1A. The plasmid encoding YFP-PCBP2 was cotransfected into HeLa cells with a plasmid encoding RFP-tagged Dcp1a, a well-characterized marker for the P-body. Fluorescence microscopy revealed that while PCBP2 was distributed throughout the nucleoplasm and the cytoplasm diffusely, YFP-PCBP2 was present in several Dcp1a-positive P-bodies (Fig. 1B). To verify that PCBP2 is indeed a constituent of the P-body, colocalization studies between endogenous PCBP2 and either RFP-Dcp1a, endogenous Dcp1a, or endogenous eIF4E-T, another prominent component of the P-body (Andrei et al. 2005), were performed. As shown in Figure 1, B and C, and Supplemental Figure 2A, endogenous PCBP2 localized in a number of P-bodies enriched in RFP-tagged Dcp1a (Supplemental Fig. 2A) and endogenous Dcp1a (Fig. 1C) or eIF4E-T (Fig. 1D). We concluded that PCBP2, previously described as a nuclear–cytoplasmic shuttling protein, is a novel component of P-bodies. Interestingly, PCBP2 did not reside in all Dcp1a- and eIF4E-T-positive P-bodies, suggesting the existence of a mechanism that targets PCBP2 only to specific P-bodies.
FIGURE 1.
PCBP2 localizes to both P-bodies and stress granules. (A) Total cell extracts of HeLa cells transfected with a plasmid encoding either EYFP or EYFP-tagged PCBP2 were probed with anti-PCBP2 antibody. The membrane was stripped and reprobed with anti-tubulin antibody. (B) HeLa cells cotransfected with plasmids encoding EYFP-tagged PCBP2 and RFP-tagged Dcp1a were fixed and subjected to microscopic observation. (C) HeLa cells were doubly immunostained using anti-PCBP2 antibody and anti-Dcp1a antibody. (D) HeLa cells were doubly immunostained using anti-PCBP2 antibody and anti-eIF4E-T antibody. (E) HeLa cells transiently transfected with a plasmid encoding GFP-TIA1 were treated with arsenite (0.5 mM, 30 min) and immunostained with anti-PCBP2 antibody. (F) HeLa cells transiently transfected with a plasmid encoding RFP-Dcp1a were treated with arsenite and immunostained with anti-PCBP2 antibody. (G) HeLa cells cotransfected with plasmids encoding GFP-TIA1 and RFP-Dcp1a were treated with arsenite, fixed, and subjected to microscopic observation. (Arrows) Colocalization; (arrowheads) lack of colocalization. Bar, 10 μm.
Subcellular localization of PCBP2 in cells under stress
Next, to examine whether PCBP2 could be recruited to the SG, HeLa cells transiently transfected with a plasmid encoding GFP-tagged TIA-1, a major component of the SG, were treated with arsenite and immunostained with anti-PCBP2 antibody. As shown in Figure 1E, cytoplasmic PCBP2 was predominantly recruited to SGs, but its distribution did not completely overlap with that of TIA-1. This result was confirmed by double staining of arsenite-treated HeLa cells using anti-TIA1 antibody and anti-PCBP2 antibody (Supplemental Fig. 2B). The recruitment of PCBP2 to the SG was also observed in cells subjected to heat stress (30 min at 44°C), ER stress (dithiothreitol [DTT] treatment, 1 mM, 1 h), suggesting that PCBP2 is a general constituent of the SG (data not shown). To examine the presence of PCBP2 in the P-body under stressful conditions, HeLa cells transiently expressing RFP-tagged Dcp1a were treated with arsenite and immunostained with PCBP2 antibody. As shown in Figure 1F, a number of Dcp1a-positive P-bodies clearly colocalized with PCBP2. Since Dcp1a hardly colocalized with TIA-1 (Fig. 1G), we concluded that PCBP2 is dually localized to the SG and P-body under stress.
Mapping of the sequence elements required for the recruitment to the P-body and SG
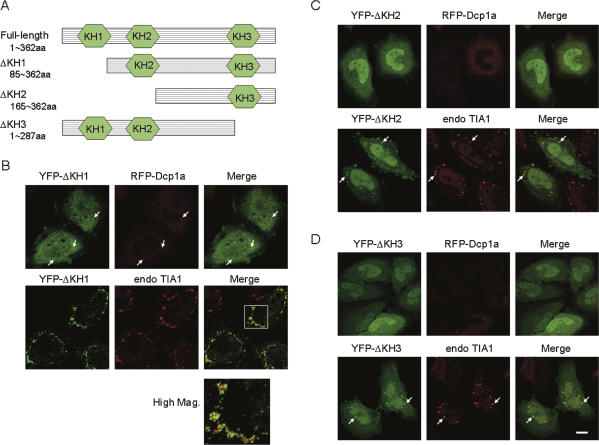
To examine the sequence elements responsible for the targeting to the P-body and SG, plasmids encoding truncation mutants of PCBP2 were constructed (Fig. 2A), and their localization was studied. Deletion of the first KH domain (KH1) had little effect on P-body localization (Fig. 2B, upper row). However, when cells were exposed to arsenic stress, the recruitment to SG was abrogated (Fig. 2B, lower row); while the mutant protein (termed YFP-ΔKH1) was recruited to the vicinity of the SG, it failed to be incorporated into the SG and formed a number of smaller granules on the surface of each SG (Fig. 2B, High Mag). Intriguingly, nuclear localization of YFP-ΔKH1 in arsenite-treated cells was significantly diminished (Fig. 2B). This was also observed in heat-stressed and DTT-treated cells (data not shown), suggesting that KH1 plays a role in stress-triggered nucleo-cytoplasmic export. On the other hand, deletion of the second and third KH domains (KH2 and KH3) did not affect the accumulation at the SG, but did abolish the localization to the P-body (Fig. 2C,D). These results indicate that sequence factors located in KH1 and KH2/3 mediate the recruitment of PCBP2 into distinct RNA granules, SG, and P-body, respectively. More extensive deletion analyses will clearly define the element(s) required for P-body and SG localization.
FIGURE 2.
Mapping of sequence elements required for P-body and SG localization. (A) Structures of plasmids encoding truncation mutants of PCBP2. (B–D) HeLa cells were transfected with a plasmid encoding the indicated YFP-tagged truncation mutant alone (lower row) or together with a plasmid encoding RFP-Dcp1a (upper row). The cells were fixed without (upper row) or with (lower row) arsenite treatment and subjected to fluorescence microscopy. (Arrows) Localization of the indicated mutants either in the P-body (upper row) or SG (lower row). Bar, 10 μm.
Effect of siRNA-mediated depletion of PCBP2 on SG and P-body integrity
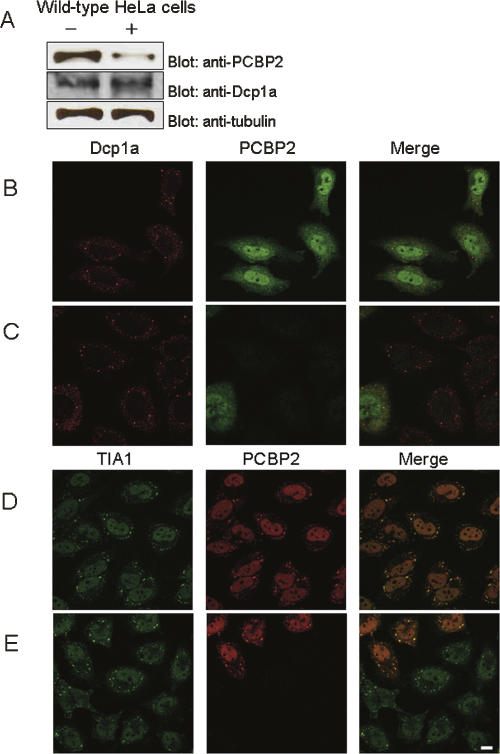
To study the role of PCBP2 in SG and P-body integrity, we attempted to knock down endogenously expressed PCBP2 by siRNA transfection. siRNA transfection was performed on HeLa cells as described in the Materials and Methods. As shown in Figure 3A, the expression level of PCBP2 was dramatically reduced when cells were transfected with the siRNA designed to deplete PCBP2, whereas those of tubulin and Dcp1a were unaffected. Sixty hours after transfection of either control siRNA or PCBP2 siRNA, the cells were immunostained using anti-PCBP2 and anti-Dcp1a antibody (Fig. 3B,C). As shown in Figure 3C, P-bodies enriched in Dcp1a remained intact, indicating that depletion of PCBP2 did not affect the integrity of P-bodies. Next, to examine the effect of PCBP2 knockdown on SG integrity, wild-type HeLa cells were similarly transfected with siRNA and doubly immunostained using anti-PCBP2 and anti-TIA1 antibody (Fig. 3D,E). Again, PCBP2 depletion had little effect on SG assembly upon arsenic stress (Fig. 3E). Taken together, these results indicate that PCBP2 does not play a significant role in maintaining the structural integrity of the P-body and SG.
FIGURE 3.
Effect of siRNA-mediated depletion of PCBP2 on P-body and SG integrity. (A) Verification of siRNA-mediated depletion of PCBP2. Total cell extracts of HeLa cells transfected either with a control siRNA (−) or PCBP2 siRNA (+) were probed with anti-PCBP2 antibody. The membrane was stripped and reprobed with anti-Dcp1a and anti-tubulin antibodies. (B,C) HeLa cells were transfected either with a control siRNA (B) or PCBP2 siRNA (C) and immunostained using anti-PCBP2 antibody. (D,E) Wild-type HeLa cells transfected either with a control siRNA (D) or PCBP2 siRNA (E) were arsenite-treated and doubly immunostained using anti-PCBP2 antibody and anti-TIA1 antibody. Bar, 10 μm.
Live-cell imaging of PCBP2 under stress; FRAP experiments
To study the dynamics of PCBP2 in living cells under stress, HeLa cells doubly transfected with plasmids encoding YFP-PCBP2 and RFP-Dcp1a were treated with arsenite and subjected to live-cell imaging using confocal microscopy. As shown in Figure 4A, YFP-tagged PCBP2 exhibited dynamic movements together with Dcp1a. As reported by Kedersha et al. (2005), some of the Dcp1a- and PCBP2-positive P-bodies were attached to the surface of the PCBP2-enriched SG, while other P-bodies remained unbound. Occasional fusion and fission between the SG and P-body was observed. Interestingly, PCBP2 localized in the P-body never became depleted upon association with the SG, suggesting that PCBP2 does not dissociate from P-bodies upon fusion with SGs. Next, FRAP analyses on SG-localized PCBP2 were performed. A target SG that was not in proximity to any of the Dcp1a-positive P-bodies was defined as a region of interest (ROI) and was photobleached by scanning at the maximum laser power for 60 iterations. As shown in Figure 4B (and Supplemental Fig. 3A), the fluorescence of YFP-PCBP2 rapidly recovered to ∼80% of the pre-bleach level in 20 sec, demonstrating that PCBP2 is very actively exchanged between the SG and the cytoplasm. FRAP analyses were also performed on YFP-PCBP2 accumulated at the P-body. In consideration of the motility of the P-body, a 2-μm square area containing one P-body was defined as a ROI. The ROI was photobleached and the fluorescence recovery was measured. Since YFP-PCBP2 localized to the P-body was more resistant to photobleaching than the SG, the number of iterations was set to 100. As shown in Figure 4C (and Supplemental Fig. 3B), YFP-PCBP2 in the P-body also exhibited rapid fluorescence recovery, comparable to that of SG-localized PCBP2. These results suggest that PCBP2 very rapidly moves in and out of both the SG and the P-body, and given the close association between the two structures, the protein may shuttle between the SG and the P-body. It was previously proposed by Kedersha et al. (2005) that mRNA released from polysomes during stress is recruited to SGs for triage, sorting, and mRNP remodeling, after which certain transcripts are selectively exported to associated P-bodies for degradation. PCBP2 may play a role in remodeling mRNP in the SG and transferring certain mRNA species from the SG to the P-body for degradation.
FIGURE 4.
Dynamic movement of PCBP2 under stress. (A) HeLa cells cotransfected with plasmids encoding YFP-PCBP2 and RFP-Dcp1a were arsenite-treated (0.5 mM, 30 min) and subjected to microscopic observation using confocal microscopy. (Yellow arrows) P-body, (white arrowheads) SG. Bar, 10 μm. (B) Recovery kinetics of YFP-PCBP2 accumulated at the SG. A single SG without a juxtaposed P-body was photobleached, and the fluorescence recovery was monitored as described in the Materials and Methods. (C) Recovery kinetics of YFP-PCBP2 accumulated at the P-body. A P-body that was not in proximity to any of the PCBP2-positive P-bodies was chosen as a target, and the ROI was set as a 2 μm square area containing the target P-body at the center. The ROI was bleached and the fluorescence recovery was monitored as described in the Materials and Methods.
CONCLUSIONS
In the present study, we studied the subcelullar localization of PCBP2 and found that PCBP2 is a novel constituent of the P-body and SG. In previous reports, PCBP2 was shown to reside exclusively in the nucleus (Chkheidze and Liebhaber 2003) or in the cytoplasm (Gamarnik and Andino 1997). However, our studies using immunofluorescence and expression of YFP- and HA-tagged (data not shown) fusion proteins consistently detected PCBP2 in both compartments, with pronounced localization in the nucleus. Nevertheless, a considerable portion of PCBP2 was cytoplasmic and was enriched in the P-body under normal conditions. Intriguingly, PCBP2 was not present in all of the Dcp1a- or eIF4E-T-enriched P-bodies. Such selective distribution may represent the functional diversity of P-bodies, as proposed by Pillai et al. (2005). Further, given the fact that PCBP2 is a member of the hnRNP (heterogeneous ribonucleoprotein) and shuttles between the nucleus and the cytoplasm, the protein may play a role in directly targeting nascent mRNPs to particular P-bodies for storage. Upon exposure to stress, cytoplasmic PCBP2 became concentrated in the P-body and SG. siRNA-mediated depletion of PCBP2 did not affect either P-body or SG integrity, suggesting that the expression of PCBP2 is not a prerequisite for the assembly of these structures. In addition, overexpression of PCBP2 did not lead to prominent fusion of the P-body with the SG (Supplemental Fig. 4), indicating that PCBP2, unlike TTP or BRF, does not promote the interaction of the P-body and SG. Thus, PCBP2 is not likely to play a role in the structural basis of the P-body and SG. Nevertheless, live-cell imaging revealed that PCBP2-positive P-bodies actively associate with the SG, and subsequent FRAP experiments demonstrated that PCBP2 dynamically moves in and out of the P-body and SG, strongly suggesting that PCBP2 shuttles between the two structures. Even though these results do not directly indicate the involvement of PCBP2 in mRNA degradation, they do suggest a novel role for PCBP2 in the stress-induced rearrangement of mRNP at the interface between the P-body and SG. The presence of PCBP2 in the P-body in cells at rest may help explain the protein's translational silencing activity (Ostareck-Lederer et al. 1998; Perrotti et al. 2002). Meanwhile, involvement of PCBP2 in IRES-mediated translation initiation and its mRNA-stabilizing function (Wang et al. 1999) point to the possibility that PCBP2 plays a part in rapidly shifting certain untranslated mRNAs into the translationally active state.
MATERIALS AND METHODS
Reagents and antibodies
Restriction enzymes were purchased from Takara Shuzo. Sodium arsenite was purchased from Sigma, and DTT from Wako Pure Chemicals. siRNAs were obtained from Ambion (Cat. Number 4635 for negative control and 143,968 for PCBP2 knockdown). The following primary antibodies were used: polyclonal goat anti-TIA1, polyclonal goat anti-eIF4E-T (Santa Cruz Biotechnology), mouse monoclonal anti-tubulin (Sigma), rabbit polyclonal anti-GFP (MBL), rat monoclonal anti-HA antibody (Roche Diagnostics), rabbit polyclonal anti-Dcp1a antibody (a kind gift from Dr. Jens-Lykke Andersen, University of Colorado), and mouse monoclonal anti-PCBP2 (Abnova). Secondary antibodies used were: Cy2- and Cy3-conjugated anti-mouse antibodies (Chemicon), Cy2-conjugated anti-goat antibody (Jackson ImmunoResearch), and AlexaFluor546-conjugated anti-goat antibody (Molecular Probes) for immunofluorescence; and HRP-conjugated anti-mouse, anti-rat, or anti-rabbit antibody (Santa Cruz Biotechnology) for Western blotting.
Plasmid construction
Full-length TIA-1 (NM022173), Dcp1a (NM018403), and PCBP2 (NM031989) cDNAs were PCR-amplified from a human fetal brain cDNA library (Clontech) and cloned into vectors EGFP-C3, dsRed-monomer C1, and EYFP-C1 (Clontech), resulting in plasmids that encode GFP-TIA1, RFP-Dcp1a, and YFP-PCBP2, respectively. PCBP2 cDNA was also cloned into vectors pCMV-HA and pGADT7, and TIA-1 cDNA into pGBKT7 (Clontech). Deletion mutants of PCBP2 were constructed by PCR amplification of the indicated regions and cloned into EYFP-C1 vector.
Yeast two-hybrid methods
The bait vector that encodes full-length human TIA-1 fused to the GAL4 DNA-binding domain was transformed into Saccharomyces cerevisae strain AH109. The screening procedures were carried out according to the manufacturer's instructions (Clontech). Briefly, the transformants bearing the bait vector were sequentially transformed with the MATCHMAKER library containing human fetal brain cDNA fused to the GAL4 activation domain using the lithium–acetate method. The doubly transformed yeasts were spread on plates containing selective medium lacking histidine, leucine, and tryptophan, and were incubated for 5 d at 30°C. Subsequently, the transformants were replica-plated on selective medium plates, streaked repeatedly to eliminate false-positive clones, and plasmids were isolated and transformed into Escherichia coli strain DH5α. The plasmid DNA was isolated from E. coli and subjected to sequencing analysis. For survival assay, expression vectors encoding the SV40 large T antigen fused to the GAL4-activating domain (pGADT7-SV40TAg), p53, and lamin C fused to the GAL4-binding domain (pGBKT7-p53 and pGBKT7-lamC, respectively) provided from the manufacturer were used as controls where indicated.
Cell culture and transfection
HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM, purchased from Nissui) supplemented with 10% FCS (Gibco–Invitrogen). Transient transfections of plasmids were performed using LipofectAMINE-PLUS reagent (Invitrogen) following the manufacturer's instructions. For siRNA transfection, 6 × 104 cells per well were seeded in a 24-well plate, cultured for 24 h, and transfected with 50 pmol of siRNA using Lipofectamine 2000 (Invitrogen). Following siRNA transfection, cells were trypsinized and replated on 35-mm dishes either with or without glass coverslips and cultured for 60 h.
Immunoprecipitation and western blotting
Immunoprecipitation was performed using μMACs tag isolation kit (Miltenyi Biotec) according to the manufacturer's instructions. Briefly, HeLa cells grown on 10-cm dishes were transfected with the indicated plasmids, lysed with 1 mL of lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton-X 100), and immunoprecipitated using magnetic beads coupled to anti-HA or anti-GFP antibodies. For Western blotting, samples were resolved on a 10% SDS-PAGE gel (Wako Pure Chemicals), transferred to nitrocellulose membrane using the semi-dry blotting method, and incubated with 5% skim milk in TBST for 1 h at room temperature. Primary antibody was added at the dilution of 1:1000 and incubated overnight at 4°C. After extensive washing with TBST, the secondary antibody conjugated to horseradish peroxidase (HRP) was added at the dilution of 1:2000 and incubated for 1 h at room temperature. Finally, proteins were detected by chemiluminescence (Amersham). Where indicated, the membrane was stripped and reprobed with anti-tubulin or anti-Dcp1a antibody.
Immunofluorescence
Cells were grown either on glass coverslips or on 35-mm glass-bottomed dishes (Iwaki). After being rinsed with PBS, cells were fixed with 3% paraformaldehyde (PFA) in PBS for 20 min at room temperature. Following permeabilization with 0.2% Triton-X 100, cells were incubated in 5% skim milk (alternatively, 2.5% FCS) in PBS for 30 min to eliminate a nonspecific reaction, and the primary antibody was added at a 1:300 dilution and stored overnight at 4°C. After an extensive wash, secondary antibody was added at a 1:400 dilution and incubated for 1 h at room temperature. Finally, cells were washed three times with PBS and subjected to microscopic observation.
Microscopic observation
Samples were viewed with a 63× oil-immersion PlanApo lens on an Axiovert135 fluorescence microscope (Carl Zeiss). Images were acquired with a confocal laser-scanning microscope (LSM510, Carl Zeiss). For live-cell imaging of HeLa cells transiently expressing RFP-Dcp1a and YFP-PCBP2, images were collected every 10 sec (with 2× averaging) with the laser power set at 1% of the 25-mW argon ion laser (488-nm line) and 30% of the 1-mW helium–neon laser (543-nm line).
FRAP experiments
HeLa cells seeded on 35-mm glass-bottomed dishes (Iwaki) were cotransfected with plasmids encoding YFP-PCBP2 and RFP-Dcp1a and cultured overnight. After treatment with arsenite, the transfectants were subjected to FRAP experiments utilizing a confocal microscope (LSM510) using the 488-nm line (25 mW, argon ion laser) for bleaching and visualizing YFP and the 543-nm line (1 mW, helium–neon laser) for visualizing RFP, with a pinhole adjustment resulting in a 2-μm optical slice. For FRAP experiments of SG-localized YFP-PCBP2, a single SG that was not in close proximity to any of the RFP-Dcp1a-positive P-bodies was defined as a region of interest (ROI). Afterward, the target SG was photobleached by scanning at the maximum laser power for 60 iterations, and fluorescence recovery was measured every 4 sec for 100 sec (pixel time 1.6 μsec) with the laser power set at 1% of the argon ion laser and 30% of the helium–neon laser. For FRAP experiments of P-body-localized YFP-PCBP2, a single P-body that contained both YFP-PCBP2 and RFP-Dcp1a was chosen. A 2-μm square area that enclosed the target P-body was defined as the ROI and was photobleached by scanning for 100 iterations, and the fluorescence recovery was measured every 4 sec. The above procedure was repeated for 10 different cells. For quantitative analyses, background intensity was subtracted, and intensities of selected SGs or P-bodies were measured over time and normalized using intensities of a ROI in the nucleus of a transfected but unbleached cell.
SUPPLEMENTAL DATA
Supplemental material can be found online at http://bio.c.u-tokyo.ac.jp/labs/murata/literature/index.html.
ACKNOWLEDGMENTS
We thank Dr. Jens-Lykke Andersen (University of Colorado) for kindly providing anti-Dcp1a antibody, Ms. Junko Mochida for technically supporting our experiments leading to this research, and all the past and present laboratory members for continuous encouragement.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.780708.
REFERENCES
- Andrei, M.A., Ingelfinger, D., Heintzmann, R., Achsel, T., Rivera-Pomar, R., Luhrmann, R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard, K.M., Walter, B.L., Semler, B.L. Multimerization of poly(rC) binding protein 2 is required for translation initiation mediated by a viral IRES. RNA. 2004;10:1266–1276. doi: 10.1261/rna.7070304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chkheidze, A.N., Liebhaber, S.A. A novel set of nuclear localization signals determine dstributions of the αCP RNA-binding proteins. Mol. Cell. Biol. 2003;23:8405–8415. doi: 10.1128/MCB.23.23.8405-8415.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot, N., Babajko, N., Seraphin, B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, J.R., Mitchell, S.A., Spriggs, K.A., Ostrowski, J., Bomsztyk, K., Ostarek, D., Willis, A.E. Members of the poly (rC) binding protein family stimulate the activity of the c-myc internal ribosome entry segment in vitro and in vivo. Oncogene. 2003;22:8012–8020. doi: 10.1038/sj.onc.1206645. [DOI] [PubMed] [Google Scholar]
- Gamarnik, A.V., Andino, R. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- Jakymiw, A., Pauley, K.M., Li, S., Ikeda, K., Lian, S., Eystathioy, T., Satoh, M., Fritzler, M.J., Chan, E.K.L. The role of GW/P-bodies in RNA processing and silencing. J. Cell Sci. 2007;120:1317–1323. doi: 10.1242/jcs.03429. [DOI] [PubMed] [Google Scholar]
- Kedersha, N., Anderson, P. Stress granules: Sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- Kedersha, N.L., Gupta, M., Li, W., Miller, I., Anderson, P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J. Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N., Stoecklin, G., Ayodele, M., Yacono, P., Lykke-Andersen, J., Fritzler, M.J., Scheuner, D., Kaufman, R.J., Golan, D.E., Anderson, P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian, M., Wang, X., Liebhaber, S.A. Identification of two KH domain proteins in the α-globin mRNP stability complex. EMBO J. 1995;14:4357–4364. doi: 10.1002/j.1460-2075.1995.tb00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev, A.V., Liebhaber, S.A. The poly(C)-binding proteins: A multiplicity of functions and a search for mechanisms. RNA. 2002;8:265–278. doi: 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T. Protein dynamics: Implications for nuclear architecture and gene expression. Science. 2001;291:843–847. doi: 10.1126/science.291.5505.843. [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer, A., Ostareck, D.H., Hentze, M.W. Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem. Sci. 1998;23:409–411. doi: 10.1016/s0968-0004(98)01301-2. [DOI] [PubMed] [Google Scholar]
- Parsley, T.B., Towner, J.S., Blyn, L.B., Ehrenfeld, E., Semler, B.L. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- Perrotti, D., Cesi, V., Trotta, R., Guerzoni, C., Santilli, G., Campbell, K., Iervolino, A., Condorelli, F., Gambacorti-Passerini, C., Caligiuri, M.A., et al. BCR-ABL suppresses C/EBPα expression through inhibitory action of hnRNP E2. Nat. Genet. 2002;30:48–58. doi: 10.1038/ng791. [DOI] [PubMed] [Google Scholar]
- Pillai, R.S., Bhattacharyya, S.N., Artus, C.G., Zoller, T., Cougot, N., Basyuk, E., Bertrand, E., Filipowicz, W. Inhibition of translational initiation by Let-7 microRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Sheth, U., Parker, R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Day, D., Trifillis, P., Kiledjian, M. An mRNA stability complex functions with the poly (A)-binding protein to stabilize mRNA in vitro. Mol. Cell. Biol. 1999;19:4552–4560. doi: 10.1128/mcb.19.7.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]