Abstract
The macrolide erythromycin binds to the large subunit of the prokaryotic ribosome near the peptidyltransferase center (PTC) and inhibits elongation of new peptide chains beyond a few amino acids. Nucleotides A2058 and A2059 (E. coli numbering) in 23S rRNA play a crucial role in the binding of erythromycin, and mutation of nucleotide A2058 confers erythromycin resistance in both Gram-positive and Gram-negative bacteria. There are high levels of sequence and structural similarity in the PTC of prokaryotic and eukaryotic ribosomes. However, eukaryotic ribosomes are resistant to erythromycin and the presence of a G at the position equivalent to E. coli nucleotide A2058 is believed to be the reason. To test this hypothesis, we introduced a G to A mutation at this position of the yeast Saccharomyces cerevisiae 25S rRNA and analyzed sensitivity toward erythromycin. Neither growth studies nor erythromycin binding assays on mutated yeast ribosomes indicated any erythromycin sensitivity in mutated yeast strains. These results suggest that the identity of nucleotide 2058 is not the only determinant responsible for the difference in erythromycin sensitivity between yeast and prokaryotes.
Keywords: ribosome, macrolide, peptidyltransferase, peptide exit tunnel
INTRODUCTION
Macrolides are 14–16 membered lactone ring compounds that bind near the peptidyltransferase center (PTC) of the eubacterial large ribosomal subunit (Schlunzen et al. 2001; Hansen et al. 2002; Tu et al. 2005). Once bound, they inhibit protein synthesis by blocking elongation of the nascent peptide beyond a few amino acids (Tenson et al. 2003). Nucleotides A2058 and A2059 (Escherichia coli numbering) of 23S rRNA are key determinants for erythromycin binding (Vester and Douthwaite 2001; Poehlsgaard and Douthwaite 2005). A2058 is conserved in 99.4% of sequenced eubacteria (Pfister et al. 2005). Those rare bacteria with a G instead are resistant to macrolides such as erythromycin. Furthermore, mutation of A2058 (Sigmund et al. 1984; Vester and Douthwaite 2001) or its modification by a mono- or dimethylase (Weisblum 1995; Pernodet et al. 1996) confers high level macrolide resistance in sensitive species, presumably by sterically disrupting erythromycin binding (Schlunzen et al. 2001). Recent crystal structures of eubacterial and archaeal ribosomes bound by macrolides have provided detailed information about the site of binding (Schlunzen et al. 2001; Auerbach et al. 2002; Hansen et al. 2002; Tu et al. 2005). Nucleotides A2058 and A2059 form a hydrophobic crevice near the PTC-proximal end of the tunnel that is involved in macrolide binding; the desosamine moiety of erythromycin and related macrolides forms a hydrogen bond with A2058.
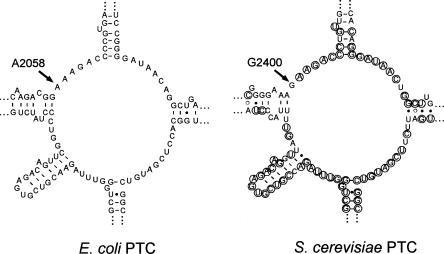
The structure of the PTC and its associated regions are similar in eubacteria, archaea, and eukaryotes. Even though the rRNA sequences in this region are not completely identical, key nucleotides that are proposed to play a role in peptide bond formation are conserved (Poehlsgaard and Douthwaite 2005; Polacek and Mankin 2005). One difference in both archaea and eukaryotes compared with eubacteria is the presence of a G at the position equivalent to E. coli A2058. These organisms are also resistant to macrolide inhibition of protein synthesis. Crystal studies showed that ribosomes from the archaeon Haloarcula marismortui bind macrolides better when the G is mutated to an A (Tu et al. 2005). Thus, it has been proposed that the G accounts for erythromycin resistance of eukaryotic ribosomes (Bottger et al. 2001; Tu et al. 2005). However, this assumption has not been investigated experimentally. To test whether G2058 is sufficient to explain macrolide resistance in eukaryotes, we introduced an A at this position in the 25S rRNA of the yeast Saccharomyces cerevisiae (Fig. 1). We found that this base transition does not confer resistance to erythromycin, suggesting that factors in addition to the base at residue 2058 must contribute to the macrolide resistance of S. cerevisiae.
FIGURE 1.
Two-dimensional structures of the rRNA in the peptidyltransferase center of E. coli and S. cerevisiae (http://www.rna.ccbb.utexas.edu/). Circled bases in the S. cerevisiae structure indicate nucleotides also present in E. coli.
RESULTS AND DISCUSSION
To test if an A at G2400 of cytoplasmic large S. cerevisiae rRNA (the residue equivalent to E. coli 2058) confers erythromycin sensitivity, we needed to introduce this mutation into the rRNA genes. The presence of 150–200 copies of rRNA genes in the S. cerevisiae genome makes it impossible to construct such a mutation in wild-type yeast. However, all copies have been deleted from the chromosome in mutant strain NOY770 (Wai et al. 2000), which harbors rRNA genes only on a plasmid (pRDN-hyg1). We introduced the G2400 → A mutation into the yeast rRNA gene carried on another plasmid (pNOY288), transformed the resulting plasmid (p288G2A) into strain NOY770, and eliminated the pRDN-hyg1 plasmid with the wild-type rRNA gene by plating on 5-fluororotic acid (FOA) medium (Boeke et al. 1987).
The resulting strain should contain the desired G2400 → A mutation, but it remained a possibility that the mutation in the rRNA gene on plasmid p288G2A had reverted during the plasmid shuffle process by gene conversion or recombination using the resident pRDN-hyg1 plasmid as source for the wild-type sequence. To ensure that the ribosomes in the mutant strain have the G2400A change, we isolated total RNA from the mutant and the wild-type strains and used it as template for reverse transcriptase-mediated cDNA synthesis followed by PCR. As expected, the PCR products from the G2400G mutant and wild-type cells comigrated in an agarose gel. No product was visible when the reverse transcriptase step was omitted, confirming that the PCR products were copied from the rRNA. The sequence of the PCR products indicated that the G → A-containing 25S rRNA was present in the mutant strain, while rRNA from the wild-type strain carried a G in the same position (data not shown). No other mutations were identified. These results confirm that the ribosomes in the mutant strain contain a G → A mutation at position 2400.
To detect if the G → A mutation affects ribosome assembly, we isolated ribosomes from mutant and wild-type cells and analyzed the distribution between subunits, 80S, and polysomes by sucrose gradient sedimentation. No differences between the polysome profile of the mutant and wild-type strains could be observed (Fig. 2).
FIGURE 2.
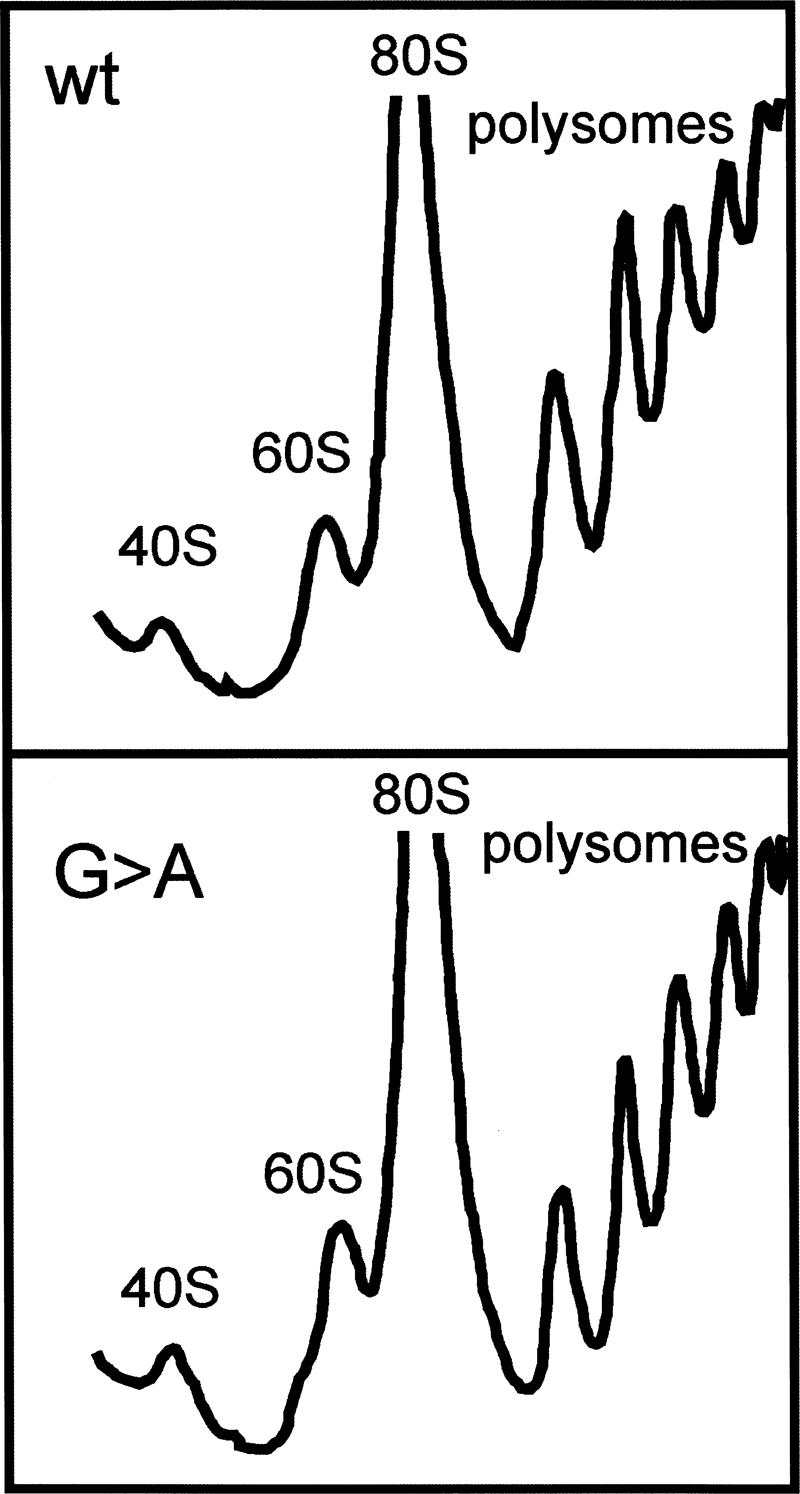
Sucrose gradients of lysates from S. cerevisiae containing a G (YLL979) or an A (YLLG2A) at position 2400 of 25S rRNA.
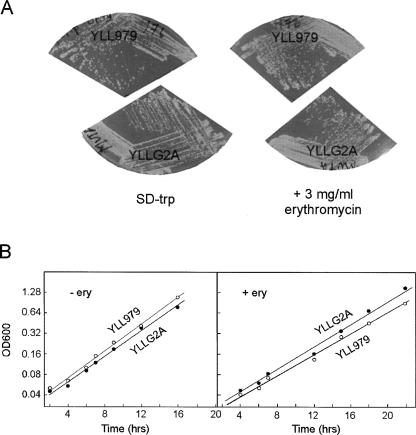
To test if the G → A mutation makes S. cerevisiae more sensitive to erythromycin, we streaked the mutated strain and the wild-type strain on both SD-Trp (Fig. 3A) and YPD (data not shown) supplemented with erythromycin at concentrations up to 3 mg/mL. Irrespective of the media, neither the mutant strain nor the wild-type strain was inhibited by the presence of the antibiotic. Furthermore, growth curves in YPD supplemented with 2 mg/mL of erythromycin showed no difference between the wild-type and mutant strains (Fig. 3B). Thus, the G2400A ribosomes are no more sensitive to erythromycin than are wild-type ribosomes. The resistance to erythromycin cannot be due to failure of the antibiotic to enter the cell, since mitochondrial protein synthesis can be inhibited by erythromycin (Lamb et al. 1968; Linnane et al. 1968).
FIGURE 3.
Growth of S. cerevisiae containing a G (YLL979) or an A (YLLG2A) at position 2400 of 25S rRNA. (A) Colonies on SD-Trp plates with or without 3 mg/mL erythromycin. (B) Growth curves in YPD with or without 2 mg/mL erythromycin.
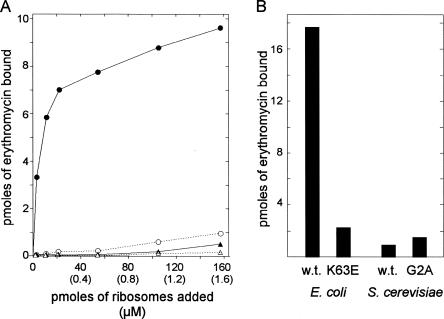
To further characterize the G → A mutant, we compared binding of erythromycin to yeast ribosomes from the G2400 wild-type and the A2400 mutant strains. For comparison, we also measured binding to E. coli ribosomes from strains that are sensitive (EryS) or resistant (EryR) to the antibiotic. Ribosomes isolated from exponentially growing cells were incubated with various concentrations of 14C-labeled erythromycin. Bound radioactivity was measured after passing the incubated mixture through 0.45-μm nitrocellulose filters (Teraoka 1970). Figure 4A shows titration curves of increasing amounts of ribosomes (up to 160 pmol, 1.6 × 10−6 M) added to a fixed amount of erythromycin (9 pmol, 9 × 10−8 M). As expected, essentially all erythromycin bound to wild-type E. coli ribosomes at the highest ribosome concentration, while ribosomes from the E. coli L4 K63E erythromycin-resistant mutant ribosomes bound only about 10% of the erythromycin at the highest ribosome concentration (∼18-fold molar excess of ribosomes). Under the same conditions, both wild-type and G → A yeast ribosomes bound the drug even less than the E. coli EryR ribosomes. We also measured the binding under conditions of erythromycin excess (Fig. 4B). Forty nanomoles of erythromycin (4 × 10−5 M) was mixed with 5.7 × 10−7 M E. coli ribosomes or 4 × 10−7 M yeast ribosomes, i.e., ∼100-fold excess of erythromycin over ribosomes. Consistent with the experiment done under conditions of ribosome excess, we found that ribosomes from wild-type E. coli bound at least eightfold more erythromycin than did ribosomes from the other sources. Again, both wild-type and G → A ribosomes from S. cerevisiae bound even less than did the ribosomes from the erythromycin-resistant E. coli strain (Fig. 4B). Even with the marginal difference between yeast wild-type and G → A ribosomes in Figure 4B, these binding assays show that the G-to-A mutation at 2400 (2058 in E. coli) does not increase the binding of erythromycin to S. cerevisiae ribosomes (Fig. 4A,B) to a degree that is meaningful to the biological ribosome function.
FIGURE 4.
Erythromycin binding to ribosomes from S. cerevisiae containing a G (YLL979) or an A (YLLG2A) at position 2058 of 25S rRNA. For comparison, erythromycin binding to wild-type and EryR ribosomes of E. coli was also determined. (A) A fixed amount of 14C-erythromycin (9 pmol; concentration = 9 × 10−8 M) was mixed with 0–160 pmol (1.6 × 10−6 M) ribosomes from the indicated sources. Ribosome concentrations were calculated from A260 measurement and assuming that A260 = 1 in a 10-mm cuvette corresponds to 23 pmol ribosomes per milliliter for E. coli (Haselkorn et al. 1963) and 16 pmol per milliliter for yeast (Domínguez et al. 1999). Closed triangles indicate wild-type (YLL979) yeast; open triangles, YLLG2A yeast; closed circles, wild-type E. coli; and open circles, EryR E. coli. (B) Forty nanomoles of 14C-erythromycin (4 × 10−5 M) was mixed with 5.7 × 10−7 M E. coli ribosomes or 4 × 10−7 M yeast ribosomes. In all cases, erythromycin binding was measured by passing the mixture through nitrocellulose filters.
Together, the experiments reported here show that a G at the position equivalent to A2058 in E. coli neither renders S. cerevisiae sensitive to erythromycin nor measurably increases the affinity of the ribosomes for the antibiotic. Thus yeast, and presumably other eukaryotes, differs from the archaeon H. marismortui in that G2058 is not the only key determinant of macrolide resistance. It is likely that, in spite of the sequence and structural similarity among prokaryotic and eukaryotic PTCs, slight nuances have a profound influence on antibiotic binding. For example, the presence of nonconserved bases near the PTC (Fig. 1) and additional rRNA sequences elsewhere may influence the structural dimensions of the hydrophobic pocket formed by A2058/A2059. The influence of ribosomal proteins L4 and L22 also cannot be ruled out, since mutations in these proteins affect erythromycin sensitivity and binding in bacteria (Chittum and Champney 1994; Gabashvili et al. 2001; Zaman et al. 2007).
MATERIALS AND METHODS
S. cerevisiae strain NOY770 (MATα ade2-1 ura3-1 trp1-1 his3-11 leu2-3 can100 rdnΔΔ∷HIS3) carries plasmid pRDN-hyg1. This plasmid contains the entire rRNA transcription unit, including the RNA polymerase I (Pol I) promoter, with the wild-type (G2400) 25S rRNA and the URA3 selectable marker (Wai et al. 2000). Plasmid pRDN-wt (alias pNOY288) also carries the entire transcription unit with a Pol I promoter and a wild-type 25S, and selectable markers TRP 1 and leu2-d (Chernoff et al. 1994). Plasmid p288G2A is a derivative of pNOY288 carrying a G2400A mutation in 25S generated by QuikChange XL Kit (Stratagene). Strains YLL979 and the isogenic YLLG2A mutant were constructed by transforming NOY770 with plasmids pNOY288 or p288G2A, respectively, and eliminating the resident plasmid pRDN-hyg1 by growth on FOA medium (Boeke et al. 1987). E. coli strains were AB301 (A19 Hfr met−) and the N282 mutant of this strain carrying a K63E mutation in the L4 gene (Wittmann et al. 1973; Chittum and Champney 1994).
To test for erythromycin sensitivity, YLL979 and YLLG2A were streaked on YPD or SD-Trp plates supplemented with erythromycin at concentrations ranging from 0–3 mg/mL and incubated for 3–4 d at 30°C. To measure the effect of erythromycin on growth rates, YPD or SD-Trp cultures with or without 2 mg/mL erythromycin were inoculated with 2 μL of saturated overnight culture and incubated with shaking at 30°C. The OD600 was followed for up to 36 h. Growth curves were determined in YPD cultures with or without erythromycin (2 mg/mL).
Yeast ribosomes were prepared essentially as described by Warner and Gorenstein (1978). Cultures were grown at 30°C in YPD to mid-log phase (OD600 ∼ 1.0; ∼2 × 107 cells/mL), harvested on ice, centrifuged, and resuspended in Lysis buffer (50 mM Tris-HCl at pH 7.5, 30 mM MgCl2, 100 mM NaCl, 1 mM PMSF, 1 μg/mL pepstatin A, 1 μg/mL leupeptin; Sigma). Cells were lysed by three passages through a French press (18,000 psi), and the cell debris was removed by centrifugation (30,000g, 4°C). The clarified supernatant was then layered onto an equal volume of 20 mM Tris-HCl (pH 7.5), 500 mM potassium chloride, 5 mM magnesium chloride, and 10% sucrose and centrifuged at 200,000g in a Beckman SW40Ti rotor for 4 h at 4°C. The ribosome pellets were resuspended in Lysis buffer and stored at −80°C. E. coli ribosomes were isolated from cultures grown at 37°C in LB to midlog phase (OD450 = 1.5 − 2.0; ∼3 × 108 cells/mL). Cells were harvested on ice, pelleted, and lysed by two passages through a French press (18,000 psi). Cell debris was removed by centrifugation at 22 K for 30 min, and the clarified supernatant was processed as described for yeast ribosomes.
Erythromycin binding assays were done by the method of Teraoka (1970). Briefly, ribosomes isolated from yeast or E. coli strains were incubated with the indicated concentrations of [N-methyl-14C] erythromycin (New England Nuclear, 55 mCi/mmol, but for the experiment in Fig. 4B diluted with nonradioactive erythromycin to a final specific activity of 11 mCi/mmol) in 100 μL erythromycin binding buffer (50 mM Tris-HCl at pH 7.8, 16 mM Mg acetate, 60 mM KCl). After incubation at 30°C for 15 min, the assay mix was poured through a 0.45 μm nitrocellulose filter (Millipore). Filters were washed three times each with 2 mL wash buffer (25 mM Tris-HCl at pH 7.8, 20 mM Mg-acetate, 60 mM KCl, containing 30% ethanol). The amount of radioactive erythromycin bound to the ribosomes was measured by liquid scintillation counting of the filters (Beckman-Coulter LS 6500). Assays were always done in duplicate.
Total RNA from cells was prepared by a hot phenol procedure (Lindahl et al. 1992). Integrity of the RNA was determined by agarose gel electrophoresis and ethidium bromide staining. Two micrograms of total RNA was treated with RNase-free DNase (Fermentas) and used for cDNA synthesis using BIORAD iScript cDNA Synthesis Kit (170–8890) with random primers. The relevant region of the rRNA sequence was then amplified by PCR using gene specific primers (5′-GGATTAACGAGATTCCCACTGT and 5′-GGTTAGTCGATCCTAAGAGATG). The PCR product was analyzed by cycle sequencing.
Sucrose gradient analysis was performed essentially as described (Deshmukh et al. 1993).
ACKNOWLEDGMENTS
This work was supported by grant MCB 03499443 from the National Science Foundation. We thank M. Nomura for strains and plasmids, and R. Pollock and R. Karpel for helpful discussions.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.786408.
REFERENCES
- Auerbach, T., Bashan, A., Harms, J., Schluenzen, F., Zarivach, R., Bartels, H., Agmon, I., Kessler, M., Pioletti, M., Franceschi, F., et al. Antibiotics targeting ribosomes: Crystallographic studies. Curr. Drug Targets Infect. Disord. 2002;2:169–186. doi: 10.2174/1568005023342506. [DOI] [PubMed] [Google Scholar]
- Boeke, J.D., Truehart, J., Natsoulis, G., Fink, G.R. 5-fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Bottger, E.C., Springer, B., Prammananan, T., Kidan, Y., Sander, P. Structural basis for selectivity and toxicity of ribosomal antibiotics. EMBO Rep. 2001;2:318–323. doi: 10.1093/embo-reports/kve062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff, Y.O., Vincent, A., Liebman, S.W. Mutations in eukaryotic 18S ribosomal RNA affect translational fidelity and resistance to aminoglycoside antibiotics. EMBO J. 1994;13:906–913. doi: 10.1002/j.1460-2075.1994.tb06334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittum, H.S., Champney, W.S. Ribosomal protein gene sequence changes in erythromycin-resistant mutants of Escherichia coli . J. Bacteriol. 1994;176:6192–6198. doi: 10.1128/jb.176.20.6192-6198.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh, M., Tsay, Y.F., Paulovich, A.G., Woolford J.L., Jr Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol. Cell. Biol. 1993;13:2835–2845. doi: 10.1128/mcb.13.5.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez, J.M., Gómez-Lorenzo, M.G., Martín, J.J. Sordarin inhibits fungal protein synthesis by blocking translocation differently to fusidic acid. J. Biol. Chem. 1999;274:22423–22427. doi: 10.1074/jbc.274.32.22423. [DOI] [PubMed] [Google Scholar]
- Gabashvili, I.S., Gregory, S.T., Valle, M., Grassucci, R., Worbs, M., Wahl, M.C., Dahlberg, A.E., Frank, J. The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22. Mol. Cell. 2001;8:181–188. doi: 10.1016/s1097-2765(01)00293-3. [DOI] [PubMed] [Google Scholar]
- Hansen, J.L., Ippolito, J.A., Ban, N., Nissen, P., Moore, P.B., Steitz, T.A. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell. 2002;10:117–128. doi: 10.1016/s1097-2765(02)00570-1. [DOI] [PubMed] [Google Scholar]
- Haselkorn, R., Fried, V.A., Dahlberg, J.E. Cellfree protein synthesis: The association of viral RNA and E. coli ribosomes. Proc. Natl. Acad. Sci. 1963;49:511–517. doi: 10.1073/pnas.49.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, A.J., Clark-Walker, G.D., Linnane, A.W. The biogenesis of mitochondria. 4. The differentiation of mitochondrial and cytoplasmic protein synthesizing systems in vitro by antibiotics. Biochim. Biophys. Acta. 1968;161:415–427. [PubMed] [Google Scholar]
- Lindahl, L., Archer, R.H., Zengel, J.M. A new rRNA processing mutant of Saccharomyces cerevisiae . Nucleic Acids Res. 1992;20:295–301. doi: 10.1093/nar/20.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnane, A.W., Lamb, A.J., Christodoulou, C., Lukins, H.B. The biogenesis of mitochondria, VI. Biochemical basis of the resistance of Saccharomyces cerevisiae toward antibiotics which specifically inhibit mitochondrial protein synthesis. Proc. Natl. Acad. Sci. 1968;59:1288–1293. doi: 10.1073/pnas.59.4.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernodet, J.L., Fish, S., Blondelet-Rouault, M.H., Cundliffe, E. The macrolide-lincosamide-streptogramin B resistance phenotypes characterized by using a specifically deleted, antibiotic-sensitive strain of Streptomyces lividans . Antimicrob. Agents Chemother. 1996;40:581–585. doi: 10.1128/aac.40.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister, P., Corti, N., Hobbie, S., Bruell, C., Zarivach, R., Yonath, A., Bottger, E.C. 23S rRNA base-pair 2057-2611 determines ketolide susceptibility and fitness cost of the macrolide resistance mutation 2058A→G. Proc. Natl. Acad. Sci. 2005;102:5180–5185. doi: 10.1073/pnas.0501598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poehlsgaard, J., Douthwaite, S. The bacterial ribosome as a target for antibiotics. Nat. Rev. Microbiol. 2005;3:870–881. doi: 10.1038/nrmicro1265. [DOI] [PubMed] [Google Scholar]
- Polacek, N., Mankin, A.S. The ribosomal peptidyl transferase center: structure, function, evolution, inhibition. Crit. Rev. Biochem. Mol. Biol. 2005;40:285–311. doi: 10.1080/10409230500326334. [DOI] [PubMed] [Google Scholar]
- Schlunzen, F., Zarivach, R., Harms, J., Bashan, A., Tocilj, A., Albrecht, R., Yonath, A., Franceschi, F. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- Sigmund, C.D., Ettayebi, M., Morgan, E.A. Antibiotic resistance mutations in 16S and 23S ribosomal RNA genes of Escherichia coli . Nucleic Acids Res. 1984;12:4653–4664. doi: 10.1093/nar/12.11.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenson, T., Lovmar, M., Ehrenberg, M. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J. Mol. Biol. 2003;330:1005–1014. doi: 10.1016/s0022-2836(03)00662-4. [DOI] [PubMed] [Google Scholar]
- Teraoka, H. A reversible change in the ability of Escherichia coli ribosomes to bind to erythromycin. J. Mol. Biol. 1970;48:511–515. doi: 10.1016/0022-2836(70)90062-8. [DOI] [PubMed] [Google Scholar]
- Tu, D., Blaha, G., Moore, P.B., Steitz, T.A. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121:257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Vester, B., Douthwaite, S. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 2001;45:1–12. doi: 10.1128/AAC.45.1.1-12.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai, H.H., Vu, L., Oakes, M., Nomura, M. Complete deletion of yeast chromosomal rDNA repeats and integration of a new rDNA repeat: Use of rDNA deletion strains for functional analysis of rDNA promoter elements in vivo. Nucleic Acids Res. 2000;28:3524–3534. doi: 10.1093/nar/28.18.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner, J.R., Gorenstein, C. The ribosomal proteins of Saccharomyces cerevisiae . Methods Cell Biol. 1978;20:45–60. doi: 10.1016/s0091-679x(08)62008-7. [DOI] [PubMed] [Google Scholar]
- Weisblum, B. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann, H.G., Stoffler, G., Apirion, D., Rosen, L., Tanaka, K., Tamaki, M., Takata, R., Dekio, S., Otaka, E. Biochemical and genetic studies on two different types of erythromycin resistant mutants of Escherichia coli with altered ribosomal proteins. Mol. Gen. Genet. 1973;127:175–189. doi: 10.1007/BF00333665. [DOI] [PubMed] [Google Scholar]
- Zaman, S., Fitzpatrick, M., Lindahl, L., Zengel, J. Novel mutations in ribosomal proteins L4 and L22 that confer erythromycin resistance in Escherichia coli . Mol. Microbiol. 2007;66:1039–1050. doi: 10.1111/j.1365-2958.2007.05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]