Abstract
Myo2p is an essential class V myosin in budding yeast with several identified functions in organelle trafficking and spindle orientation. The present study demonstrates that Myo2p is a component of a large RNA-containing complex (Myo2p–RNP) that is distinct from polysomes based on sedimentation analysis and lack of ribosomal subunits in the Myo2p–RNP. Microarray analysis of RNAs that coimmunoprecipitate with Myo2p revealed the presence of a large number of mRNAs in this complex. The Myo2p–RNA complex is in part composed of the RNA processing body (P-body) based on coprecipitation with P-body protein subunits and partial colocalization of Myo2p with P-bodies. P-body disassembly is delayed in the motor mutant, myo2-66, indicating that Myo2p may facilitate the release of mRNAs from the P-body.
Keywords: RNP, P-body, unconventional myosin, myosin V, Saccharomyces cerevisiae
INTRODUCTION
Myo2p is one of two class V myosins in the budding yeast Saccharomyces cerevisiae; the other is Myo4p. Numerous functions have been identified for Myo2p. Myo2p plays a role in membrane transport; myo2 mutants have defects in several organelle-trafficking pathways including polarized vesicle secretion (Johnston et al. 1991), inheritance of vacuoles (Hill et al. 1996; Catlett and Weisman 1998), peroxisomes (Hoepfner et al. 2001; Fagarasanu et al. 2006), late Golgi (Rossanese et al. 2001), mitochondria (Itoh et al. 2002; Boldogh et al. 2004; Itoh et al. 2004), and spindle pole orientation and resultant nuclear segregation (Gundersen and Bretscher 2003).
Like Myo2p, Myo4p plays a role in organelle inheritance. Myo4p is required for mother to daughter cell inheritance of cortical endoplasmic reticulum (Estrada et al. 2003) and the mitochondrial RNA processing body (Gill et al. 2006). Myo4p is also involved in the transport and localization of a specific set of mRNAs to the bud. This function was first elucidated for the transcript encoding Ash1p. The adaptor protein She3p bridges the interaction between Myo4p and ASH1 mRNA by binding to both the myosin and the ASH1 mRNA-binding protein, She2p (Long et al. 2000; Gonsalvez et al. 2005). In addition to ASH1 mRNA, 23 additional transcripts have been identified that are localized to the daughter cell by a mechanism dependent on Myo4p, She2p, and She3p proteins (Long et al. 1997; Takizawa et al. 1997; Takizawa and Vale 2000; Shepard et al. 2003).
In addition to Myo4p, there have been numerous examples of association of class V myosins with mRNAs. Myosin Va has been shown to associate with peri-axoplasmic ribosomal plaques (PARPS), the discrete ribosome-containing domains distributed along the periphery of axoplasm in myelinated fibers. Translational machinery is spatially organized in PARPS to serve as centers for protein synthesis (Sotelo-Silveira et al. 2006). Myosin Va is thought to accumulate in PARPS as a result of targeted transport of translational machinery and/or mRNAs into these structures (Sotelo-Silveira et al. 2004). Myosin Va is also found to associate with translocation in liposarcoma (TLS), an RNA-binding protein in neurons (Yoshimura et al. 2006). siRNA knockdown of myosin Va expression disrupted accumulation of TLS in dendritic spines (Yoshimura et al. 2006). Moreover, Myosin Va is found to coimmunoprecipitate (IP) with an mRNP complex containing Purα, mStaufen, and fragile X mental retardation protein (FMRP) (Ohashi et al. 2002). Staufen- and FMRP-containing RNP in Drosophila neurons have been recently shown to contain proteins that are present in the RNA-processing (P) body (Barbee et al. 2006), the cytoplasmic site for mRNA storage, decapping, and degradation (Brengues et al. 2005).
In the present study, we show that Myo2p also interacts with mRNAs, although by a mechanism(s) quite distinct from Myo4p–mRNA interactions. Biochemical analyses reveal that Myo2p associates with a large, RNase-sensitive, mRNA-containing particle(s) (Myo2p–RNP) that is distinct from actively translating polysomes and exosomes. In contrast to Myo4p, which as noted above associates with a small set of specified mRNAs, hundreds of mRNAs are associated with Myo2p–RNP. Myo2p co-IPs and partially colocalizes with known components of P-body. In the temperature-sensitive motor mutant myo2-66, there is no defect in P-body formation and distribution; however, the P-bodies in these cells disassemble at a slower rate than in control cells, indicating that Myo2p may play a role in P-body disassembly.
RESULTS
Assessment of Myo2p content in vacuole-, secretory vesicle-, and actin-containing fractions obtained by differential sedimentation and density gradient fractionation
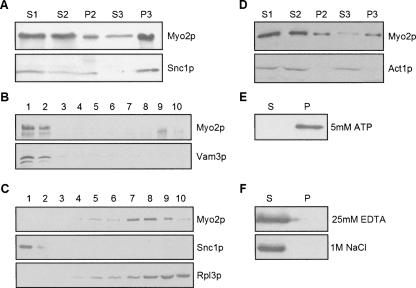
Consistent with our previous studies (Reck-Peterson et al. 1999), fractionation of whole cell lysates by differential sedimentation revealed that Myo2p is present in both intermediate (P2; 30,000g) and high-speed pellet fractions (P3; 100,000g), as well as in the high-speed supernatant/cytosol fraction (S3) (Fig. 1A). Analysis of the P2 fraction on Optiprep (iodixanol) density gradients revealed that Myo2p cofractionates with the vacuole membrane component Vam3p (Fig. 1B), consistent with the known association of Myo2p with the vacuole (Hill et al. 1996). Both Myo2p and secretory vesicles were particularly enriched in P3 (Fig. 1A), based on immunoblot analysis for the secretory vesicle component Snc1p. Although Myo2p plays an essential role in secretory vesicle transport to the bud (Govindan et al. 1995; Pruyne et al. 1998), density gradient fractionation of P3 indicated that Myo2p is absent from low-density, secretory-vesicle-containing fractions (Fig. 1C). Most of the Myo2p was present in much denser fractions that also contained ribosomal subunits based on immunoblot analysis for the ribosomal protein Rpl3p (Fig. 1C). Protein-stained gels of these gradient fractions revealed that the bulk of the proteins present in P3 are found in the dense fractions near and at the bottom of the gradient (data not shown).
FIGURE 1.
Subcellular fractionation of Myo2p. Lysates from wild-type (WT) cells grown on YP dextrose were first spun at low speed to generate S1; P2 and S2 were generated by sedimentation of S1 at 30,000g. S2 was centrifuged at 100,000g, resulting in P3 and S3. Immunoblots of the resultant fractions, loaded stoichiometrically, were probed with antibodies to (A) Myo2p or Snc1p (secretory vesicle marker) or (D) Act1p (actin). (B,C) Optiprep density gradient fractionation of (B) P2 and (C) P3. Gradient fractions (1–10, low to high density) were immunoblotted with antibodies to Myo2p and either (B) Vam3p (vacuole marker) or (C) Snc1p (secretory vesicle marker) or Rpl3p (ribosomal marker). (E,F) Pellet (P) and supernatant (S) fractions after extraction of P3 with either (E) 5 mM ATP or (F) 25 mM EDTA or 1 M NaCl.
The presence of Myo2p was not due to association and cosedimentation with F-actin since actin was absent from P3 (Fig. 1D). Moreover, ATP treatment, which dissociates Myo2p from F-actin (Reck-Peterson et al. 2001), did not release Myo2p (Fig. 1E). Extraction of P3 with either high salt or EDTA (Fig. 1F), conditions commonly used to dissociate protein–protein and protein–nucleic acid complexes, did solubilize Myo2p.
P3-Myo2p exhibits ATP-sensitive actin binding
One obvious concern is that the putative complex with which Myo2p is associated in P3 is actually an artifact of protein aggregation due to denaturation. A stringent test for assessing the native state of myosins is their ATP-sensitive binding to F-actin. The binding of P3-Myo2p to actin was assayed by cosedimentation. Since the high speeds required for sedimentation of F-actin would result in actin-independent sedimentation of Myo2p, a low-speed sedimentation assay with actin bundles formed by addition of high Mg2+ was employed (Fig. 2). Under the sedimentation conditions used, F-actin sedimented (±ATP) in the presence (Fig. 2, lanes 4,6) but not the absence (Fig. 2, lanes 3,5) of high Mg2+. Myo2p alone (with no added actin) did not sediment in the absence (Fig. 2, lane 1) or presence (Fig. 2, lane 2) of high Mg2+. Addition of unbundled filaments (±ATP) did not promote sedimentation of Myo2p (Fig. 2, lanes 3,5). Substantial cosedimentation of Myo2p with actin bundles was observed in the absence (Fig. 2, lane 4) but not the presence (Fig. 2, lane 6) of ATP. These results indicate that P3-associated Myo2p is in a native state and potentially is able to promote the movement of the complex(es) with which it is associated.
FIGURE 2.
P3-Myo2p exhibits ATP-dependent binding to actin. Resuspended P3 was mixed with or without actin in presence or absence of high Mg2+ (to form actin paracrystal bundles) and in the absence or presence of 4 mM ATP. Samples were then spun at 15,000g to collect the actin bundles. Resulting supernatant and pellet fractions were immunoblotted with antibodies to Myo2p (top panel). (Bottom panel) A Ponceau S-stained membrane to visualize actin.
Myo2p is associated with an RNAase-sensitive complex distinct from ribosomes and the exosome
Sucrose velocity gradient analysis of P3 was performed to assess the size of the Myo2p complex in comparison to the ribosomal components present (Fig. 3). We observed variable levels of Myo2p throughout these gradients including fractions with relatively low sedimentation coefficients (Fig. 3A, -RNase; Fig. 3B). This is likely due to partial disruption of the Myo2p complex either during resuspension of the P3 pellet or upon exposure to high sucrose concentrations since the Myo2p present in fractions pooled from the top of the gradient no longer sediments at 100,000g after dilution out of sucrose (data not shown). Nevertheless, there was also a discrete peak of Myo2p with a sedimentation rate considerably faster than that of the 80S mono-ribosome peak, indicating that Myo2p in P3 was at least in part associated with a relatively large complex(es) that was at least in part distinct from ribosomes. The density gradient results (Fig. 1C) indicated that the complex was sufficiently dense to contain nucleic acid. To determine if the Myo2p complex contains RNA, sucrose gradient fractionation of RNase-treated P3 was performed. As expected, RNase treatment resulted in a loss of polysomes and increase of the 80S monoribosome peak, indicating that the RNase was active (Fig. 3A). In contrast to the discrete shift of the polyribosomes into the 80S peak, RNase treatment resulted in partial shift of Myo2p from the bottom of the gradient toward the top of the gradient with some overlap with the 80S monoribosomes. These results indicate that the P3 Myo2p–RNP complex(es) contain RNAs that are variably accessible to RNase digestion.
FIGURE 3.
Myo2p associates with a large particle distinct from actively translating ribosomes. Sucrose velocity gradients were performed on P3-Myo2p isolated from WT cells (A) with or without RNase treatment or (B) with or without cycloheximide treatment prior to lysis. Immunoblots of gradient fractions were performed with anti-Myo2p tail and anti-Rpl3p (ribosomal marker) antibody. The OD260 of each fraction was read and plotted to generate ribosomal subunit distribution profile. (C) Immunoblot of pellet (P) and supernatant (S) from IP of Myo2p from the resuspended P3 fraction. Blots were probed with antibodies to Myo2p or the large ribosomal subunit protein Rp13p.
Because the Myo2p–RNP exhibits sedimentation properties similar to that of polysomes (Mangus and Jacobson 1999) and RNase digestion results in generation of Myo2p complexes that partially overlap with the 80S monoribosome peak (Fig. 3A), Myo2p could at least in part be associated with polysomes. To assess the possible association state of Myo2p with polysomes, we examined the sedimentation properties of Myo2p from P3 fractions isolated from cells treated with the protein synthesis inhibitor cycloheximide (CHX). Cycloheximide treatment stalls ribosomes on translating RNA, eliminating the 80S peak and resulting in an increase in the polyribosome peak (Wettstein et al. 1964). As expected the ribosomal peak shifted toward the bottom of the gradient with drug treatment (Fig. 3B). In contrast, neither the size nor the amount of Myo2p-containing complex was affected by cycloheximide treatment, suggesting that Myo2p does not associate with actively translating ribosomes. Consistent with this conclusion, IPs of Myo2p from P3 did not contain ribosomes based on immunoblot analysis probed with antibodies to the ribosomal protein Rpl3p (Fig. 3C). Myo2p IPs were performed with antibody directed against the head (motor) domain to minimize the possibility of antibody-dependent disruption of the Myo2–RNP given that the actin cosedimentation studies indicated that the head domains of Myo2p are free to interact with actin.
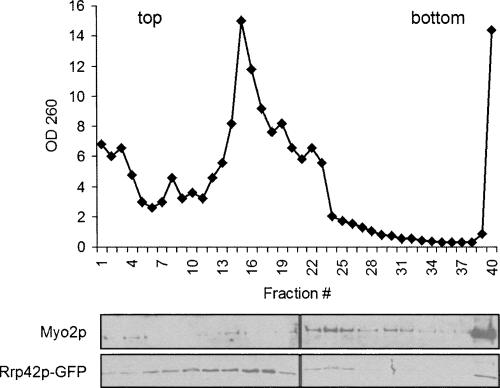
One potential candidate for the RNA-containing Myo2p complex is the RNA-processing complex, the exosome, since several protein subunits of this complex have been identified as potential binding proteins for Myo2p (Gavin et al. 2002). To assess this possibility, we generated a yeast strain expressing a GFP-fusion protein of the exosome subunit, RRP42-GFP, as the sole copy of this gene. Sucrose velocity gradient analysis of P3 isolated from this tagged strain revealed that although a fraction of the exosome sediments with Myo2p, the exosome peak sediments at a slower velocity than the Myo2p complex, indicating that this complex is probably not the exosome (Fig. 4).
FIGURE 4.
P3 Myo2p does not associate with the exosome. Sucrose velocity gradients were performed on P3 isolated from cells expressing a GFP-tagged subunit of the exosome (GFP-RRP42). The OD260 of each fraction was read and plotted to generate a ribosomal subunit distribution profile. Immunoblots of gradient fractions were probed with antibodies to either Myo2p or GFP.
Myo2p–RNP contains many mRNAs
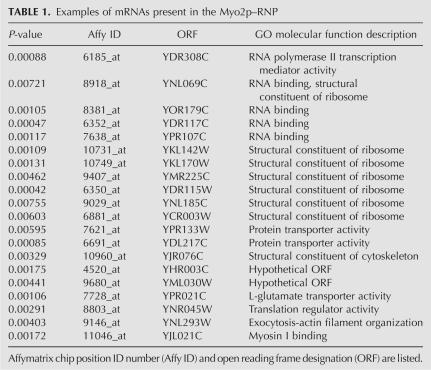
To determine if the RNA associated with the P3 particle included mRNAs, a microarray-based experiment was performed. Myo2p was IPed from P3 with Myo2p-head-specific antibodies; non-immune IgG was used as a negative control. Associated mRNAs were amplified using oligo(dT) and random primers. This cRNA was fluorescently labeled and then hybridized to yeast microarrays. Results show hundreds of transcripts that were specifically IPed by the Myo2p-head antibody. A clustergram of the mRNAs that specifically IP with Myo2p (from two independent experiments) is shown in Figure 5; genes indicated in red represent a physical association of the mRNA with Myo2p. Analysis of the Myo2p IPs revealed nearly 1800 mRNAs with greater than sixfold abundance compared to the control, and more than 2300 transcripts with threefold abundance over control. (A table listing these transcripts is available upon request to the corresponding author.) There was ∼80% (1470/1775) overlap between both experiments. The amount of each transcript present did not correlate with their cellular abundance or with intron content. A short list of the types of mRNAs present is shown in Table 1.
FIGURE 5.
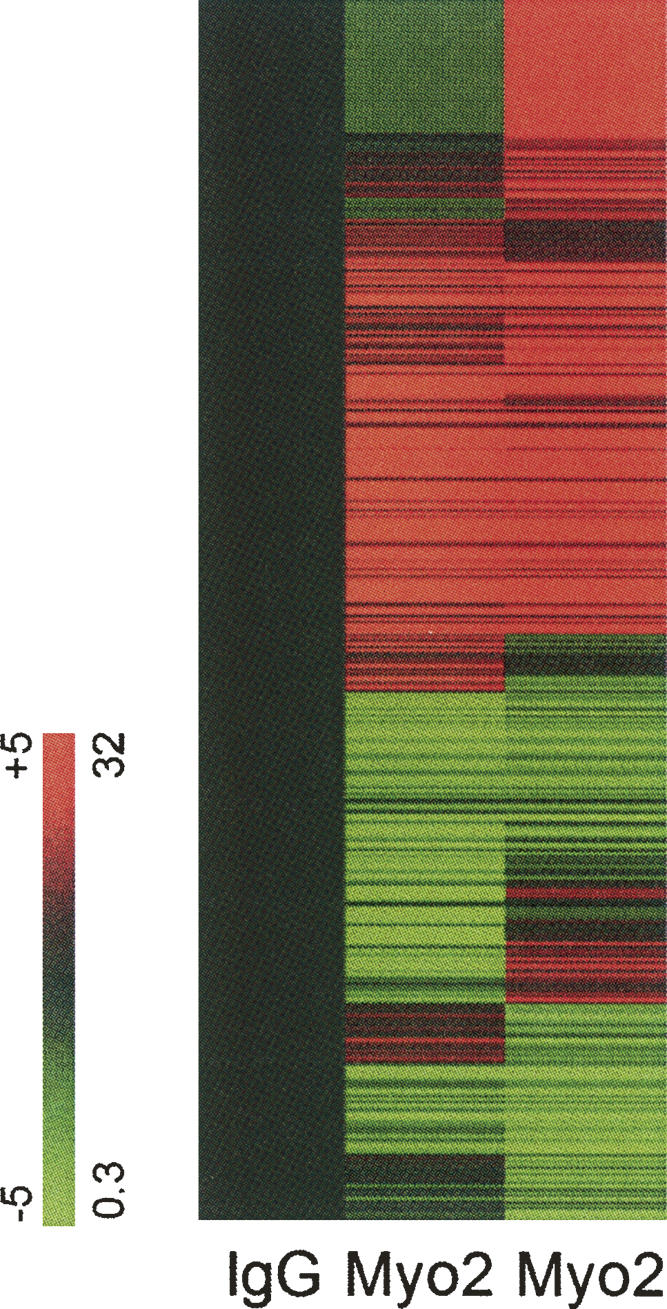
Microarray hybridization of mRNAs that coimmunoprecipitate with Myo2p. Hybridization data reveals a large number of mRNAs that co-IP with Myo2p. Each row is a gene; each column is an IP sample. To facilitate visualization of enriched mRNAs, the expression level of each gene is normalized to the control (non-immune IgG) IP hybridization values. Hybridization values above (red) and below (green) the non-immune IgG IP. These fold differences range from 32-fold to 0.03-fold relative to mock IP hybridization values (+5 to −5 logs on log base 2 scale). (Black) Normalized hybridization value.
TABLE 1.
Examples of mRNAs present in the Myo2p–RNP
Myo2p associates and partially colocalizes with the P-body
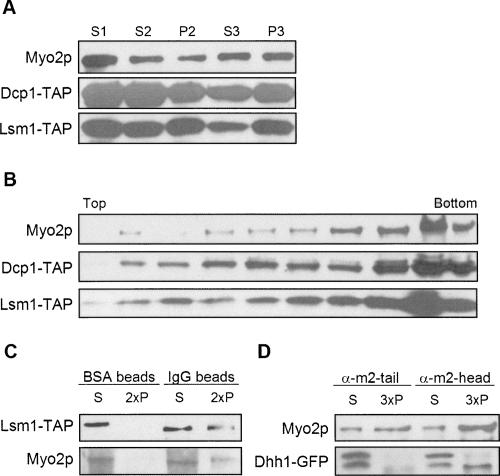
P-bodies are dynamic eukaryotic cytoplasmic granules that contain translationally inactive mRNAs and proteins involved in mRNA degradation and translation initiation (Coller and Parker 2004). mRNAs in P-bodies can be degraded or can return to translation (Brengues et al. 2005). The P-body shares properties with the Myo2p-P3 RNA particle. These include the following: (1) P-bodies contain a broad spectrum of mRNAs. (2) In cells grown in YPD, P-bodies components (Dcp1p, Lsm1p, Pat1p, Dhh1p, Xrn1p) were found in P3 and partially cofractionated with Myo2p on iodixanol gradients (Fig. 6A,B) as well as sucrose velocity gradients (Dhh1p was tracked in these studies) (data not shown). However, since P-body components were also present in fractions not enriched in Myo2p, such overlap may be independent of any association. To test whether Myo2p associates with P-bodies, P3 or S2 fractions were prepared from yeast strains that were expressing TAP- or GFP-tagged proteins and analyzed by IP. Myo2p was IPed by the P-body component Lsm1p-TAP (Fig. 6C) using IgG-coated magnetic beads and conversely the anti-Myo2p-head antibody IPed Dhh1-GFP (Fig. 6D); note that only the lower-molecular-weight (∼80 kDa) Dhh1-GFP immunogen was IPed by anti-Myo2p-head antibody. The upper, ∼100-kDa band has an apparent molecular weight greater than predicted (84 kDa), and its molecular relationship to the presumed bona fide Dhh1-GFP in the lower band is not known. A similar higher-molecular-weight immunogen has been seen in a different Dhh1-GFP strain (Jang et al. 2006). Interestingly, the Myo2p-tail antibody did not coIP Dhh1p, suggesting that the antibody may disrupt Myo2p associations with the P-body or that tail epitopes are occluded. Lsm1p, along with Lsm2p-Lsm7p, form a cytoplasmic heteroheptameric complex that interacts with proteins involved in decapping-dependent mRNA degradation and is required for efficient decapping (Beggs 2005). Dhh1p is a DExD/H-box helicase which promotes both mRNA decapping and translational repression (Coller and Parker 2005).
FIGURE 6.
Myo2p associates with P-body components. (A) Differential sedimentation of lysates from cells expressing TAP-tagged P-body subunits Dcp1p and Lsm1p. (B) Optiprep density gradient fractionation of P3 isolated from cells expressing TAP-tagged Dcp1p or Lsm1p. (C) Coprecipitation of Myo2p with TAP-tagged Lsm1p. P3 from cells expressing TAP-tagged Lsm1p was mixed with either BSA- or mouse IgG-coated magnetic beads. The pellet and supernatant fractions were analyzed by immunoblotting. (D) Dhh1-GFP coprecipitates with Myo2p pulled down by anti-myo2-head but not anti-myo2-tail antibody. Gradient and IP fractions were immunoblotted with anti-Myo2p or anti-GFP antibodies or rabbit IgG to visualize the TAP tag. The pellet fractions were loaded at twofold (2×) or threefold (3×) concentrations of the supernatant fractions.
In cells expressing both Myo2p-GFP and Dcp2-RFP, Myo2p was also found to partially colocalize with Dcp2p (Fig. 7). To enhance the P-body fluorescence signal, cells were grown to OD > 1 and fixed prior to LM observation (Teixeira et al. 2005). Approximately 50% of the Dcp2p-RFP structures (65 of 134 examined) were also Myo2p-GFP-positive. These results indicate that a fraction of Myo2p associates with the P-body and that not all the P-bodies contain Myo2p, indicating that the interaction between the two might be regulated. It is also important to note that the percentage of Myo2p-GFP particles that also contain Dcp2p-DFP is much lower, consistent with the idea that Myo2p associates with multiple cellular components. Time-lapse imaging of Dcp2p-GFP-tagged puncta in live cells revealed that most exhibit only random movements in wild-type (WT) cells, suggesting that Myo2p does not effect long-range directed movement of P-bodies in contrast to the role of Myo2p in organelle trafficking and spindle pole orientation.
FIGURE 7.
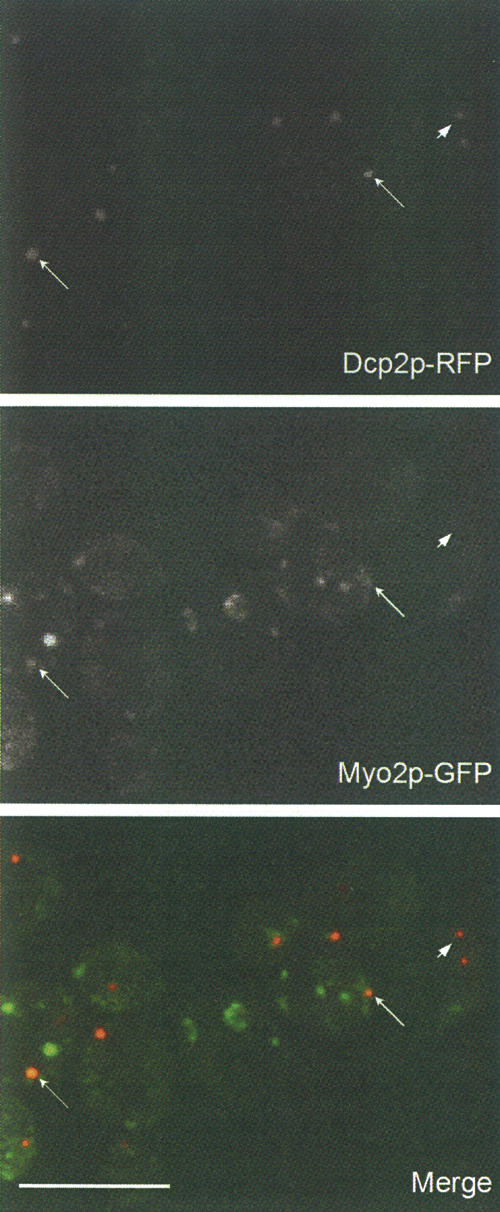
Myo2p partially colocalizes with Dcp2p. Spinning disk confocal images of cells expressing both Myo2p-GFP and Dcp2p-RFP. OD > 1 cultures were used to enhance the P-body florescence signal. (Arrows) Particles containing both Myo2p and Dcp2p; (arrowhead) a particle that contains Dcp2p but not Myo2p. Bar, 10 μm.
Myo2p may play a role in P-body disassembly
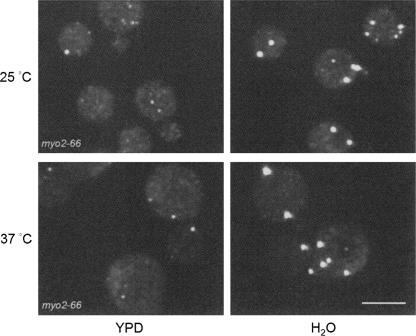
P-bodies are dynamic aggregates of translation repression complexes, and their number and size are affected by the environment and the cellular state (Teixeira et al. 2005). P-bodies increase in number and small P-bodies aggregate to form large P-bodies in response to cellular stress. In contrast, inhibition of translation elongation or growing cells at low density cause P-bodies to fall apart and eventually are no longer detectable by fluorescence microscopy in cells expressing tagged P-body subunits (Teixeira et al. 2005). Whether there are functional differences between large and smaller P-bodies is not known, although it is clear that small P-body aggregates are sufficient for a basal level of translation repression and/or decapping (Parker and Sheth 2007). Myo2p, through interactions with P-body proteins, may be playing a role in P-body assembly and/or disassembly. To test if Myo2p plays a role in P-body assembly, the amount of Dcp2p-GFP in P3 was analyzed in both WT and myo2-66 (a temperature-sensitive motor mutant of Myo2p) (Johnston et al. 1991) cells. Shifting cells to 37°C (restrictive temperature) for 2 h resulted in a slight increase in the amount of P-bodies in both myo2-66 and WT cells, based on the florescence signal of Dcp2p-GFP puncta (data not shown), indicating that there is not a defect in P-body assembly in the mutant cells. Because it is possible that the P-bodies observed were formed before all the Myo2p motors were inactivated, we further tested P-body formation by hypotonic shock (Teixeira et al. 2005). Cells grown in YPD were spun down and resuspended in H2O or YPD as control. Both WT and myo2-66 cells were able to assemble P-bodies, and shifting cells for 2 h to 37°C before hypotonic shock did not have an effect (Fig. 8). These results indicate that the motor activity of Myo2p is not required for P-body assembly.
FIGURE 8.
Myo2p is not required for P-body formation. myo2-66-expressing Dcp2p-GFP cells were incubated for 2 h at 25°C or 37°C. Cells were then spun down and resuspended in either YPD or H2O for 10 min to induce P-body formation. P-body formation was not affected by the loss of Myo2p motor activity. Bar, 10 μm.
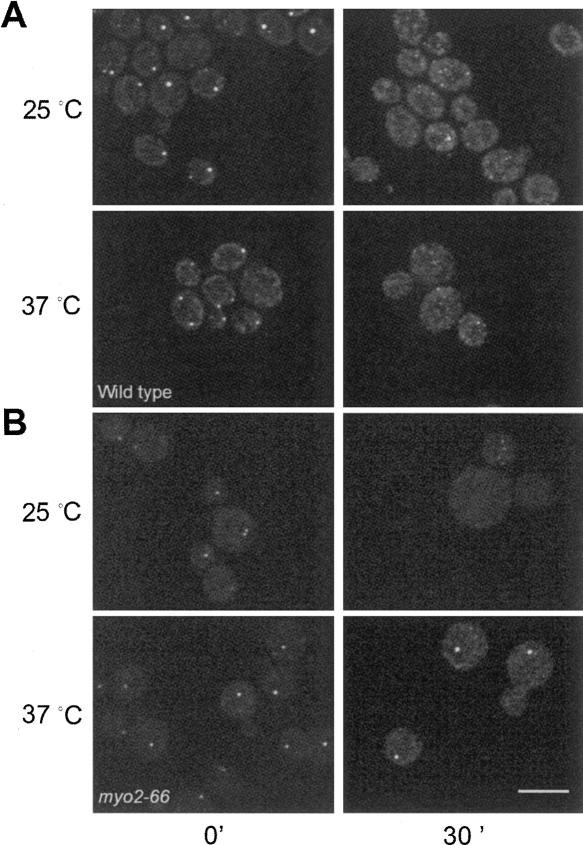
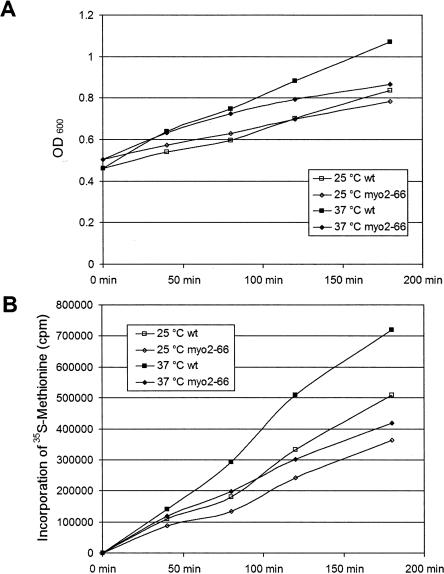
To test whether Myo2p is required for P-body disassembly, myo2-66 cells and WT cells were grown to high OD in YPD to allow P-bodies to form, followed by dilution to induce disassembly of the P-bodies, at permissive (25°C) or restrictive (37°C) temperature. In the WT cells, large P-bodies disassembled into smaller aggregates at both temperatures (Fig. 9A), while P-body disassembly was delayed at the restrictive temperature in myo2-66 cells as shown by the persistence of large P-bodies (Fig. 9B). Similarly, there was a delay in P-body disassembly induced by cycloheximide treatment in myo2-66 cells grown at the restrictive temperature (data not shown). A delay in P-body disassembly may be a result of translational defects (Brengues et al. 2005). To assess this possibility, rates of 35S-methionine incorporation into protein by WT and myo2-66 cells were examined (Fig. 10). myo2-66 cells exhibited linear rates of methionine incorporation into protein over a 3-h time course at both the permissive and nonpermissive temperatures. The incorporation rate in myo2-66 cells was faster at 37°C than at 25°C. However, the rates of methionine incorporation by WT cells at both temperatures were significantly faster than in myo2-66 presumably indicative of overall metabolic impairment resulting from loss of polarized cell growth. These results are comparable to those reported by Johnston et al. (1991) and indicate that the delay in P-body disassembly is not due to a translational arrest at the restrictive temperature.
FIGURE 9.
Myo2p may function in P-body disassembly. WT and myo2-66 cells expressing Dcp2p-GFP were grown to high OD to allow P-body formation, followed by dilution of cells to induce P-body disassembly. (A) WT cells immediately before dilution (left panels) and 30 min after dilution at 25°C and 37°C (right panels). P-body disassembly was essentially complete 30 min after dilution (right panels) at both 25°C and 37°C. (B) myo2-66 cells prior to dilution (left panels) and 30 min after dilution (right panels). P-bodies were able to disassemble in myo2-66 cells at the permissive temperature, but disassembly was delayed in cells at the restrictive temperature. Bar, 10 μm.
FIGURE 10.
myo2-66 cells at restrictive temperature incorporate methionine. myo2-66 cells at both permissive and restrictive temperatures (A) continued to grow (OD600) and (B) synthesize proteins (35S-methionine incorporation) at similar rates over a 3-h time course, although the rates at both temperatures were lower compared to the wild-type cells.
DISCUSSION
The results presented here demonstrate that a significant fraction of the total Myo2p in the cell is associated with a large, mRNA-containing particle that is distinct from the exosome and actively translating polysomes. Several lines of evidence indicate that the Myo2p–RNP is composed at least in part of the P-body. This is based on the partial colocalization of Myo2p with the P-body component, Dcp2p, and the coprecipitation of Myo2p with the P-body subunits Lsm1p-TAP and Dhh1-GFP. Myo2p–RNP and the P-body share similar properties:
Similar to P-bodies that are most prominent in cells at G0 phase, the amount of P3 Myo2p is cell-cycle-dependent (data not shown), with the greatest levels (70%–80% total Myo2p) observed in glycerol-grown and G-phase-arrested cells (cdc28-4); conversely, P3 Myo2p is significantly reduced (∼20% total) in M-phase-arrested cells (cdc15-4).
Both have similar sedimentation profiles.
They both associate with bulk cytoplasmic transcripts and not a specific set of mRNAs. However, P3 Myo2p may be associated with RNP complexes in addition to the P-body since only a small fraction of Myo2p present in P3 co-IPs with P-body components and only a low percentage of Myo2p-GFP particles contain Dcp2p-DFP.
There are several possible functions for Myo2p association with the P-body. Myo2p may function in the movement of mRNAs from and/or to P-bodies. However, the P-body does assemble in myo2-66 mutant cells, indicating that the Myo2p motor activity is not essential for P-body formation. In this regard, it is important to note that recent studies have shown that no single known protein subunit of the P-body is essential for assembly (Teixeira and Parker 2007). Interestingly, we observed that P-body disassembly induced either by a release of G0-arrested cells or cycloheximide treatment is slowed in myo2-66 cells at the nonpermissive temperature, indicating that Myo2p may promote disassembly of P-bodies to release transcripts for either degradation or translation. If so, disassembly apparently does not involve long-range directed movement of P-body components. However, perhaps Myo2p applies short-range torque on P-body components (e.g., via interaction with cortical actin filaments) that, in turn, facilitates dissociation. In this regard, it is interesting to note that disruption of actin with latrunculin A results in loss of P-bodies from the cell cortex and resultant aggregation of P-bodies in the central cytoplasm (data not shown). This suggests that P-bodies interact with cortical actin and that these cortical filaments could be accessible to interaction with P-body-associated Myo2p. However, localization of P-bodies to the cortex is not dependent on Myo2p anchoring since such a central collapse of P-bodies is not observed in myo2-66 cells.
Although Myo2p coprecipitates with two P-body proteins tested, these interactions could be indirect. Additional components, such as mRNAs or other adaptor proteins, may affect this association. It is likely that the tail domain of Myo2p effects its association with P-body components since the head domain is free to interact with actin. Moreover, only the antibody to the head domain pulls down Dhh1p, suggesting that tail epitopes are either occluded from antibody access in the Myo2p–RNP or that the antibody disrupts Myo2p association with P-bodies. Future studies to identify the molecular basis for the association of Myo2p with the P-body should provide critical insights into the role of this myosin in P-body functions.
Several examples of class V myosin–RNP interactions have been reported, including the transport and localization of mRNAs to the bud by yeast Myo4p (Takizawa et al. 1997), the association of mammalian myosin Va to the Purα/mStaufen/FMRP complex (Ohashi et al. 2002), the accumulation of class V myosin in PARP domains in mammalian and mauthner myelinated fibers (Sotelo-Silveira et al. 2006), and the spinal accumulation of TLS and its target mRNA Nd1-L facilitated by mammalian Myosin Va (Yoshimura et al. 2006). However, in all these instances, a small and specific subset of mRNAs associate with these myosins (Shepard et al. 2003). In contrast, Myo2p–RNP is unique in that based on the IP data, much of the yeast mRNA genome is present in this RNP, suggesting, as has been shown for the P-body, that it is involved in bulk sequestration/storage of transcripts. While best characterized in budding yeast, P-bodies and related structures (stress granules, germinal granules, and neuronal granules) are also found in higher animals (Anderson and Kedersha 2006). Even though both stress granules and neuronal granules have been reported to contain ribosomal subunits, no evidence for this has been described for P-bodies (Anderson and Kedersha 2006). Consistent with this, we failed to detect coprecipitation of ribosomal proteins with Myo2p.
Staufen, an RNA-binding protein initially found in Drosophila germinal granules can be recruited into stress granules upon cellular stress (Thomas et al. 2005) and has been recently demonstrated to partially colocalize with P-body components (Barbee et al. 2006). Similar to our observation that P-body distribution is not affected by the head mutant myo2-66, the localization of Staufen1 is not affected by overexpression of the dominant-negative tail domain of myosin-Va, even though overexpression of the same domain inhibits the translocation of TLS (Yoshimura et al. 2006). Given that Myosin Va is reported to interact with Staufen (Ohashi et al. 2002) and the Myo2p–P-bodies association reported here, it will be of much interest to assess whether class V myosins play a similar role with P-bodies in other cell types.
MATERIALS AND METHODS
Yeast strains and growth conditions
Yeast cells were grown in YP medium (1% Bacto yeast extract, 2% Bacto peptone; Difco) containing 2% dextrose or YP 2% glycerol for the growth-restricted experiments. The yeast strains used in this study are NY 580 (MATa pep4Δ∷URA3 leu2-3,112, ura3-52), NY 1002 (MATa ura3-52 leu2-3,112 myo2-66), yRP1824 (MATa leu2-3,112 trp1 ura3–52 his4-539 DHH1-GFP), and yRP1825 (MATα leu2-3,112 trp1 ura3-52 his4-539 DCP2-GFP). yRP1824 and yRP1825 (Teixeira et al. 2005) are generous gifts from R. Parker (University of Arizona, Tucson, AZ). Dcp2-RFP/myo2-66 and Dcp2-RFP/Myo2-GFP were constructed as follows: the mRFP sequence was PCRed from pmRFP1 (Campbell et al. 2002) using the following primers: 5′-primer: CCTTAATTAACGCCTCCTCCGAGGACGTCATC; and 3′-primer: TTGCGCGCTTAGGCGCCGGTGGAGTGGC. Underlined sequences depict restriction sites. The PCR product and pFA6a-GFP (S65T)-kanMX6 were digested by 5′-PacI and 3′-BssHI, and ligated to form pRFP-KanMX6. Stable yeast strains expressing Dcp2-GFP (myo2-66) and Dcp2p-RFP (Myo2-GFP) were generated by homologous recombination, using plasmids pFA6a-GFP (S65T)-kanMX6 and pRFP-KanMX6, as described previously (Longtine et al. 1998). The primer sequences used were as follows: Forward, TTCAGGGTCTAATGAATTATTAAGCATTTTGCATAGGAAGCGGATCCCCGGGTTAATTAA; Reverse, CATTTACAGTGTGTCTATAAAACGTATAACACTTATTCTTGAATTCGAGCTCGTTTAAAC. Underlined sequences indicate GFP/RFP homology regions. Strain MMY147 (Myo2-GFP-HIS3; leu2; URA3; his3d200) is a sporulation product of YJC1454, a generous gift from J. Cooper (Washington University, St. Louis, MO). MMY147 was used to generate a stable Myo2-GFP/Dcp2-RFP strain. TAP-tagged strains are described in Ghaemmaghami et al. (2003). NY580 was used to generate the strain expressing a C-terminal GFP tag of the exosome subunit, RRP42-GFP as the sole copy of this gene, by homologous recombination as described previously (Longtine et al. 1998). The primers used were as follows: Forward primer F2, CAATGGTGGAGAAATACGCCCCCGACGTAGTACGATCACTGGAGAATTTACGGATCCCCGGGTTAATTAA; reverse primer R1, TTTTTAGTATGTAAACTAATTTGTCTTTTTATCTAATTGTGACTATATTTGAATTCGAGCTCGTTTAAAC. Underlined sequences depict GFP homology regions.
Cell fractionation
Log-phase WT (NY580) cells (100–600 mL) were spun down and washed with either lysis Buffer A (40 mM Tris at pH 7, 2 mM MgCl2, 1 mM EGTA, 2 mM DTT), Buffer B (10 mM imidazole at pH 7.2, 75 mM KCl, 2.5 mM MgCl2, 1 mM EGTA, 2 mM DTT), or Buffer C for microarray experiments (20 mM HEPES at pH 7.2, 150 mM KCl, 1 mM EGTA at pH 7.2, 0.5% TX-100, 2 mM MgCl2, 2 mM DTT) plus 2 mM Pefabloc (Roche Scientific) and 1 mM benzamidine (Calbiochem). We found that lysing the cells in the lower-ionic-strength Buffers A and B resulted in an equivalent amount of Myo2p in the ultraspeed pellet fraction, but an increase in the amount in the 30,000g pellet fraction compared to lysis in the detergent containing higher ionic strength Buffer C. Buffer C was used for lysis of preparations to be used for IP of Myo2p from the ultraspeed pellet fraction as this lysis condition resulted in more efficient IP. Cells were lysed in 1.5-mL aliquots containing 2 g of zirconium/silica beads (Biospec Products) using a Mini-BeadBeater-8 (BioSpec Products). Lysis was achieved by three pulses of 45 sec at 4°C with 1 min on ice between pulses. The resulting lysate was spun at 2000g for 10 min. The following supernatant (S1) was spun at 30,000g for 30 min. The 30,000g supernatant (S2) was then spun at 100,000g for 1 h onto a 100-μL 60% sucrose (in lysis buffer) cushion resulting in a soluble pool S3 and the ultraspeed pellet P3. The relative amount of Myo2p in each of these fractions was determined by immunoblot analysis.
Further fractionation of P3 was performed by density gradient sedimentation using Optiprep (iodixanol) density gradient medium (Sigma D-1556). P3 (including the sucrose cushion) was resuspended in one-half original lysis volume, mixed 1:1 with Optiprep density gradient medium and spun at 287,000g for 2 h in a fixed-angle TLA 120.2 rotor (Beckman Instruments). Fractions (100 μL) were collected from the top of the gradient. Immunoblots were performed with appropriate markers.
For the velocity gradients, RNA content in P3 was determined spectrophotometrically and 40–50 OD260 units were loaded on a continuous 20% to 47% (w/v) sucrose (in lysis buffer) gradient (Mangus and Jacobson 1999). Samples were spun in an SW-41 rotor at 36,000 rpm (160,000g) for 5 h. Identical gradients were run on cells treated (10 min prior to lysis) with 100 μg/mL cycloheximide (Sigma C-7698); 100 μg/mL cycloheximide was also included in the gradients. To test the effects of RNase treatment, P3 was digested with 5 units/μL S7 Micrococcal nuclease (Roche Scientific 107,921) in the presence of 3 mM CaCl2 for 10 min on ice followed by 10 min at room temperature prior to loading on the gradient. Fractions (300 μL) were collected and the OD260 was read to generate plots of ribosomal subunit distribution (Nelson et al. 1992). Samples were then loaded on an SDS-PAGE gel, transferred to nitrocellulose, and immunoblotted for the appropriate markers. The antibodies used were anti-Myo2p-tail antibody (0.2 μg/mL) (Reck-Peterson et al. 1999), Snc1p polyclonal serum (Rossi et al. 1997) (1:1000) as a vesicle marker, and monoclonal anti-ribosomal protein L3 (1:5000) (generous gift of Jonathon Warner, Albert Einstein College of Medicine, New York, NY).
High-salt, EDTA, and ATP treatment was performed on P3. P3 was obtained as described above, and resuspended to half the original lysis volume in Buffer A that contained either 1 M NaCl, 25 mM EDTA, or 5 mM ATP; samples were placed for 10 min at room temperature then spun at 100,000g for 1 h. The amount of Myo2p present in the supernatant and pellet fractions was determined by immunoblot analysis.
Actin cosedimentation assays
Actin cosedimentations were performed on Myo2p from P3. P3 was obtained as described above in Buffer B with 1 mM Pefabloc and was resuspended to the original lysis volume. Chicken skeletal muscle F-actin (7 μM) was mixed with P3 in the presence of 20 mM MgCl2 (to form actin paracrystals that would pellet at low speeds) (Hanson 1973), 25 μM phalloidin (Sigma P-2141) in Buffer B with or without 4 mM ATP. The reaction mixture was incubated for 10 min on ice and then spun at 15,000g for 10 min to pellet the F-actin paracrystals. The resultant supernatant and pellet fractions were separated by SDS-PAGE. F-actin is visualized by Ponceau S staining, and Myo2p was detected by immunoblot analysis.
Immunomagnetic separation
To isolate complexes from cells expressing tandem affinity tag (TAP)-tagged P-body subunits, 0.4 mg of magnetic beads (Dynabeads M-450 uncoated; Dynal Biotech) were coated, according to the manufacturer's instructions, with BSA (Sigma) or non-immune mouse IgG (Jackson Immunoresearch) at a concentration of 0.2 mg of antibody for 15 mg of Dynabeads. Coated beads were then resuspended in 150 μL of Buffer B (containing 0.5% BSA) and added to 50 μL of P3 prepared from TAP-tagged strains and incubated for 30 min at 4°C with continuous end-over-end rotation. The supernatants were collected, and the beads were washed three times with Buffer A containing 0.5% BSA followed by an additional wash with Buffer B (without BSA). The supernatant and pellet fractions were analyzed by SDS-PAGE and immunoblotted for TAP-tag and Myo2p using rabbit non-immune IgG and anti-Myo2p-tail, respectively.
Immunoprecipitation assay
Yeast cells expressing Dhh1-GFP (yRP1824) were grown at 25°C to OD5 and S2 fraction were prepared in Buffer B. S2 was precleared with protein A-Sepharose (1:10 volume; Amersham Biosciences) for 45 min at 4°C and incubated with 30 μg/mL non-immune IgG or antibodies to myo2p-head or myo2p-tail for 1 h at 4°C. Protein A-Sepharose was then added (1:10), and the samples were incubated for 45 min at 4°C. Pellets were washed (five times for 2 min) with Buffer B + 0.1% TX and processed for SDS-PAGE and immunoblot analysis.
Immunopurification of Myo2p, RNA isolation, and microarray analysis
Immunopurification of the Myo2p-associated mRNAs was performed according to Takizawa et al. (2000). The ultraspeed pellet, P3, was prepared from WT NY580; P3 was resuspended with Buffer C to half the original lysis volume and incubated with protein A-Sepharose CL-4B (Amersham Pharmacia Biotech) for 30 min at 4°C as a pre-clear step. Beads were then removed, samples were divided into two aliquots, and 100 μg/mL rabbit anti-Myo2p-head antibody (Reck-Peterson et al. 1999) or an equivalent amount of rabbit non-immune IgG (Jackson Immunoresearch) as a control was added. This mixture was incubated with gentle agitation for 5 h at 4°C. Protein A-Sepharose beads were then added and incubated for 30 min at 4°C. The beads were then washed five times with 1 mL of Buffer B. Aliquots of IP supernatants and pellets were analyzed for Myo2p content by immunoblot to assess IP efficiency, and RNA was extracted from the remainder of the IP.
For RNA isolation, the immunobeads were suspended in 100 μL of 1% SDS in TE (10 mM Tris at pH 8.0, 1 mM EDTA at pH 8.0) and then boiled for 1 min. An equal volume of phenol-chloroform was added, vortexed, and spun; the aqueous layer containing RNA was treated with DNase I (Roche scientific 776,785) for 30 min at 37°C. Following DNase I treatment RNA was extracted with phenol-chloroform; the aqueous layer was topped with 3 vol of ethanol, 300 mM (NH4)2SO4, and 1 μL of 5 mg/mL glycogen as carrier and stored overnight at −80°C to pellet RNA. RNA was spun and resuspended in RNase free water and stored at −80°C.
Microarray Affymetrix (Affymetrix) gene analysis was performed in the Singer laboratory (Albert Einstein) and by the Keck center (Yale University); the results described are a combination of experiments performed in both laboratories. Preparation of labeled cRNA for hybridization onto Affymetrix GeneChips was performed according to the recommended Affymetrix protocol: double-stranded cDNA was synthesized from 5 μg of RNA by using a Superscript Choice System (Invitrogen), with an HPLC-purified oligo(dT)24 primer containing a T7 RNA polymerase promoter sequence at its 5′-end (5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG-(dT)24-3′) (Proligo LLC). The second cDNA strand was synthesized by using Escherichia coli DNA polymerase I, RNase H, and DNA ligase. After second-strand synthesis, cDNA was cleaned using the GeneChip sample cleanup module (Affymetrix). Labeled cRNA was generated from cDNA by in vitro transcription using a GeneChip IVT labeling kit (Affymetrix) that incorporates biotinylated synthetic analog (pseudouridine) mediated by MEGAscript T7 RNA Polymerase. Biotin-labeled cRNA was then purified using the GeneChip cleanup module prior to fragmenting to a size of 35–200 bases by incubating for 35 min at 94°C in fragmentation buffer (40 mM Tris-acetate at pH 8.1, 100 mM potassium acetate, 30 mM magnesium acetate). The IVT yield criteria are at least fivefold of the starting total RNA amount. If the yield is less than fivefold, the sample is not processed further. A Test-3 Gene Array was performed to assess the quality of the biotin-labeled cRNA prior to using the standard arrays. Array hybridization buffer (100 mM MES, 1 M [Na+], 20 mM EDTA, 0.01% Tween 20) was used to prehybridize the Test-3 Gene array for 10–15 min at 45°C. The prehybridized solution was then removed and replaced with 80 μL of hybridization mixture containing hybridization buffer, fragmented cRNA (0.05 μg/μL), and herring sperm DNA (0.1 mg/mL; Promega). Also included in the hybridization buffer are acetylated BSA (0.5 mg/mL; Invitrogen) and four control bacterial and phage cRNAs (1.5 pM BioB, 5 pM BioC, 25 pM BioD, and 100 pM Cre) to serve as internal controls for hybridization efficiency. The arrays were hybridized for 16 h at 45°C in a rotisserie oven. After hybridization, arrays were washed using the Affymetrix fluidics station and stained with streptavidin-phycoerythrin (10 μg/mL; Molecular Probes) according to the Affymetrix technical manual. Washed arrays were then scanned on an Affymetrix GeneChip scanner 3000. Scanned output files were visually inspected for hybridization artifacts and then analyzed by using Affymetrix GCOS software; the GeneSpring cross-gene error model function was used to estimate variance.
Cell imaging
WT and myo66 cells were pipetted onto a pad of 2% YPD-agarose and observed using an Ultraview RS spinning disk confocal microscope (PerkinElmer). Images from multiple focal planes were collected, and the stacks of images were collapsed to single images using the maximal projection. For examination of the colocalization of Myo2p with P-bodies, cells expressing Myo2p-GFP and Dcp2p-RFP were grown to OD > 1 in YPD, fixed by addition of paraformaldehyde (PFA) to 3.7% from a fresh ampoule of 16% PFA (Electron Microscopy Sciences), and washed three times with PBS by sedimentation at 700g before imaging. In order to determine if P-bodies exhibit directed, presumably Myo2p-dependent movements, cells expressing Dcp2-GFP were pipetted directly onto a YPD-agarose pad, and continuous image stacks were recorded as above at time intervals of 0.2–1 sec for 1–10 min. As noted in the Results, almost all P-body movements recorded appeared random.
P-body assembly and disassembly assays
P-body assembly assay
WT (yRP1825) and myo2-66 cells expressing Dcp2p-GFP were grown at 25°C to OD of 0.1, and half of the cells were shifted for 2 h to 37°C. The cells were spun down (700g for 2 min), resuspended in pre-warmed H2O or YPD, and incubated for 10 min at 25°C or 37°C. Cells were then fixed by addition of paraformaldehyde solution to a final concentration of 3.7% and prepared for fluorescence microscopy as described above.
P-body disassembly assay
WT (yRP1825) and myo2-66 cells expressing Dcp2p-GFP were grown at 25°C to an OD of 1, and half of the cells were shifted for 2 h to 37°C. P-body disassembly was induced by two different protocols: (1) 9× volumes of YPD were added to dilute the cells, and cells were then fixed at different time points; (2) 100 μg/mL cycloheximide was added to the cells, and cells were fixed after 10 min and then viewed as described above.
35S-methionine incorporation assay
WT (yRP1825) and myo2-66 cells expressing Dcp2p-GFP were grown at 25°C in synthetic complete medium containing 50 μM cold methionine to OD about 0.5. One μCi/mL 35S-labeled methionine (PerkinElmer) was added to each culture, and half of the cells were shifted to 37°C. ODs were measured, and 1-mL aliquots were removed from each culture at different time points. The aliquots were mixed with 0.2 mL of 50% (w/v) ice-cold trichloroacetic acid (TCA), incubated on ice for 10 min, heated for 20 min to 70°C, and filtered through Whatman GF/C filters. Filters were washed with 10 mL of 5% ice-cold TCA and 10 mL of 95% ethanol, dried, and counted in a scintillation counter.
ACKNOWLEDGMENTS
We thank Dr. Roy Parker, University of Arizona, for his extensive and very helpful advice. This work was supported by NIH grants RO-1 DK-25387 (to M.S.M.), GM073823 (to M.S.M.), PO-1-DK-55389 (Jon Morrow, Yale School of Medicine; MSM PI of project 3), and GM57071 (to R.H.S.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.665008.
REFERENCES
- Anderson, P., Kedersha, N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbee, S.A., Estes, P.S., Cziko, A.M., Hillebrand, J., Luedeman, R.A., Coller, J.M., Johnson, N., Howlett, I.C., Geng, C., Ueda, R., et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs, J.D. Lsm proteins and RNA processing. Biochem. Soc. Trans. 2005;33:433–438. doi: 10.1042/BST0330433. [DOI] [PubMed] [Google Scholar]
- Boldogh, I.R., Ramcharan, S.L., Yang, H.C., Pon, L.A. A Type V myosin (Myo2p) and a Rab-like G-protein (Ypt11p) are required for retention of newly inherited mitochondria in yeast cells during cell division. Mol. Biol. Cell. 2004;15:3994–4002. doi: 10.1091/mbc.E04-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues, M., Teixeira, D., Parker, R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, R.E., Tour, O., Palmer, A.E., Steinbach, P.A., Baird, G.S., Zacharias, D.A., Tsien, R.Y. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catlett, N.L., Weisman, L.S. The terminal tail region of a yeast myosin-V mediates its attachment to vacuole membranes and sites of polarized growth. Proc. Natl. Acad. Sci. 1998;95:14799–14804. doi: 10.1073/pnas.95.25.14799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller, J., Parker, R. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- Coller, J., Parker, R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada, P., Kim, J., Coleman, J., Walker, L., Dunn, B., Takizawa, P., Novick, P., Ferro-Novick, S. Myo4p and She3p are required for cortical ER inheritance in Saccharomyces cerevisiae . J. Cell Biol. 2003;163:1255–1266. doi: 10.1083/jcb.200304030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasanu, A., Fagarasanu, M., Eitzen, G.A., Aitchison, J.D., Rachubinski, R.A. The peroxisomal membrane protein Inp2p is the peroxisome-specific receptor for the myosin V motor Myo2p of Saccharomyces cerevisiae . Dev. Cell. 2006;10:587–600. doi: 10.1016/j.devcel.2006.04.012. [DOI] [PubMed] [Google Scholar]
- Gavin, A.C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J.M., Michon, A.M., Cruciat, C.M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami, S., Huh, W.K., Bower, K., Howson, R.W., Belle, A., Dephoure, N., O'Shea, E.K., Weissman, J.S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Gill, T., Aulds, J., Schmitt, M.E. A specialized processing body that is temporally and asymmetrically regulated during the cell cycle in Saccharomyces cerevisiae . J. Cell Biol. 2006;173:35–45. doi: 10.1083/jcb.200512025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalvez, G.B., Urbinati, C.R., Long, R.M. RNA localization in yeast: Moving towards a mechanism. Biol. Cell. 2005;97:75–86. doi: 10.1042/BC20040066. [DOI] [PubMed] [Google Scholar]
- Govindan, B., Bowser, R., Novick, P. The role of Myo2, a yeast class-V myosin, in vesicular transport. J. Cell Biol. 1995;128:1055–1068. doi: 10.1083/jcb.128.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen, G.G., Bretscher, A. Cell biology: Microtubule asymmetry. Science. 2003;300:2040–2041. doi: 10.1126/science.1084938. [DOI] [PubMed] [Google Scholar]
- Hanson, J. Evidence from electron microscope studies on actin paracrystals concerning the origin of the cross-striation in the thin filaments of vertebrate skeletal muscle. Proc. R. Soc. Lond. B. Biol. Sci. 1973;183:39–58. doi: 10.1098/rspb.1973.0003. [DOI] [PubMed] [Google Scholar]
- Hill, K.L., Catlett, N.L., Weisman, L.S. Actin and myosin function in directed vacuole movement during cell division in Saccharomyces cerevisiae . J. Cell Biol. 1996;135:1535–1549. doi: 10.1083/jcb.135.6.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner, D., van den Berg, M., Philippsen, P., Tabak, H.F., Hettema, E.H. A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae . J. Cell Biol. 2001;155:979–990. doi: 10.1083/jcb.200107028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, T., Watabe, A., Toh, E.A., Matsui, Y. Complex formation with Ypt11p, a rab-type small GTPase, is essential to facilitate the function of Myo2p, a class V myosin, in mitochondrial distribution in Saccharomyces cerevisiae . Mol. Cell. Biol. 2002;22:7744–7757. doi: 10.1128/MCB.22.22.7744-7757.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, T., Toh, E.A., Matsui, Y. Mmr1p is a mitochondrial factor for Myo2p-dependent inheritance of mitochondria in the budding yeast. EMBO J. 2004;23:2520–2530. doi: 10.1038/sj.emboj.7600271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, L.T., Buu, L.M., Lee, F.J. Determinants of Rbp1p localization in specific cytoplasmic mRNA-processing foci, P-bodies. J. Biol. Chem. 2006;281:29379–29390. doi: 10.1074/jbc.M601573200. [DOI] [PubMed] [Google Scholar]
- Johnston, G.C., Prendergast, J.A., Singer, R.A. The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J. Cell Biol. 1991;113:539–551. doi: 10.1083/jcb.113.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, R.M., Singer, R.H., Meng, X., Gonzalez, I., Nasmyth, K., Jansen, R.P. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- Long, R.M., Gu, W., Lorimer, E., Singer, R.H., Chartrand, P. She2p is a novel RNA-binding protein that recruits the Myo4p–She3p complex to ASH1 mRNA. EMBO J. 2000;19:6592–6601. doi: 10.1093/emboj/19.23.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie A., III, Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., Pringle, J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae . Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Mangus, D.A., Jacobson, A. Linking mRNA turnover and translation: Assessing the polyribosomal association of mRNA decay factors and degradative intermediates. Methods. 1999;17:28–37. doi: 10.1006/meth.1998.0704. [DOI] [PubMed] [Google Scholar]
- Nelson, R.J., Ziegelhoffer, T., Nicolet, C., Werner-Washburne, M., Craig, E.A. The translation machinery and 70 kDa heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- Ohashi, S., Koike, K., Omori, A., Ichinose, S., Ohara, S., Kobayashi, S., Sato, T.A., Anzai, K. Identification of mRNA/protein (mRNP) complexes containing Puralpha, mStaufen, fragile X protein, and myosin Va and their association with rough endoplasmic reticulum equipped with a kinesin motor. J. Biol. Chem. 2002;277:37804–37810. doi: 10.1074/jbc.M203608200. [DOI] [PubMed] [Google Scholar]
- Parker, R., Sheth, U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Pruyne, D.W., Schott, D.H., Bretscher, A. Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J. Cell Biol. 1998;143:1931–1945. doi: 10.1083/jcb.143.7.1931. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson, S.L., Novick, P.J., Mooseker, M.S. The tail of a yeast class V myosin, Myo2p, functions as a localization domain. Mol. Biol. Cell. 1999;10:1001–1017. doi: 10.1091/mbc.10.4.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck-Peterson, S.L., Tyska, M.J., Novick, P.J., Mooseker, M.S. The yeast class V myosins, Myo2p and Myo4p, are nonprocessive actin-based motors. J. Cell Biol. 2001;153:1121–1126. doi: 10.1083/jcb.153.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossanese, O.W., Reinke, C.A., Bevis, B.J., Hammond, A.T., Sears, I.B., O'Connor, J., Glick, B.S. A role for actin, Cdc1p, and Myo2p in the inheritance of late Golgi elements in Saccharomyces cerevisiae . J. Cell Biol. 2001;153:47–62. doi: 10.1083/jcb.153.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi, G., Salminen, A., Rice, L.M., Brunger, A.T., Brennwald, P. Analysis of a yeast SNARE complex reveals remarkable similarity to the neuronal SNARE complex and a novel function for the C terminus of the SNAP-25 homolog, Sec9. J. Biol. Chem. 1997;272:16610–16617. doi: 10.1074/jbc.272.26.16610. [DOI] [PubMed] [Google Scholar]
- Shepard, K.A., Gerber, A.P., Jambhekar, A., Takizawa, P.A., Brown, P.O., Herschlag, D., DeRisi, J.L., Vale, R.D. Widespread cytoplasmic mRNA transport in yeast: Identification of 22 bud-localized transcripts using DNA microarray analysis. Proc. Natl. Acad. Sci. 2003;100:11429–11434. doi: 10.1073/pnas.2033246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotelo-Silveira, J.R., Calliari, A., Cardenas, M., Koenig, E., Sotelo, J.R. Myosin Va and kinesin II motor proteins are concentrated in ribosomal domains (periaxoplasmic ribosomal plaques) of myelinated axons. J. Neurobiol. 2004;60:187–196. doi: 10.1002/neu.20015. [DOI] [PubMed] [Google Scholar]
- Sotelo-Silveira, J.R., Calliari, A., Kun, A., Koenig, E., Sotelo, J.R. RNA trafficking in axons. Traffic. 2006;7:508–515. doi: 10.1111/j.1600-0854.2006.00405.x. [DOI] [PubMed] [Google Scholar]
- Takizawa, P.A., Vale, R.D. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc. Natl. Acad. Sci. 2000;97:5273–5278. doi: 10.1073/pnas.080585897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa, P.A., Sil, A., Swedlow, J.R., Herskowitz, I., Vale, R.D. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- Takizawa, P.A., DeRisi, J.L., Wilhelm, J.E., Vale, R.D. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000;290:341–344. doi: 10.1126/science.290.5490.341. [DOI] [PubMed] [Google Scholar]
- Teixeira, D., Parker, R. Analysis of P-body assembly in Saccharomyces cerevisiae . 2007;18:2274–2287. doi: 10.1091/mbc.E07-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira, D., Sheth, U., Valencia-Sanchez, M.A., Brengues, M., Parker, R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, M.G., Martinez Tosar, L.J., Loschi, M., Pasquini, J.M., Correale, J., Kindler, S., Boccaccio, G.L. Staufen recruitment into stress granules does not affect early mRNA transport in oligodendrocytes. Mol. Biol. Cell. 2005;16:405–420. doi: 10.1091/mbc.E04-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettstein, F.O., Noll, H., Penman, S. Effect of cycloheximide on ribosomal aggregates engaged in protein synthesis in vitro. Biochim. Biophys. Acta. 1964;87:525–528. doi: 10.1016/0926-6550(64)90131-8. [DOI] [PubMed] [Google Scholar]
- Yoshimura, A., Fujii, R., Watanabe, Y., Okabe, S., Fukui, K., Takumi, T. Myosin-Va facilitates the accumulation of mRNA/protein complex in dendritic spines. Curr. Biol. 2006;16:2345–2351. doi: 10.1016/j.cub.2006.10.024. [DOI] [PubMed] [Google Scholar]