Abstract
Nucleoporin Nup98, a 98-kDa protein component of the nuclear pore complex, plays an important role in both protein and RNA transport. During its maturation process, Nup98 undergoes post-translational autoproteolysis, which is critical for targeting to the NPC. Here we present high-resolution crystal structures of the C-terminal autoproteolytic domains of Nup98 (2.3 Å for the wild type and 1.9 Å for the S864A precursor), and propose a detailed autoproteolysis mechanism through an N–O acyl shift. Structural constraints are found at the autocleavage site, and could thus provide a driving force for autocleavage at the scissile peptide bond. Such structural constraints appear to be generated, at least in part, by anchoring a conserved phenylalanine side chain into a highly conserved hydrophobic pocket at the catalytic site. Our high-resolution crystal structures also reveal that three highly conserved residues, Tyr866, Gly867, and Leu868, provide most of the interactions between the autoproteolytic domain and the C-terminal tail. These results suggest that Nup98 may represent a new subtype of protein that utilizes autoprocessing to control biogenesis pathways and intracellular translocation.
Keywords: autoproteolysis mechanism, N–O acyl shift, nucleoporin Nup98, protein autoprocessing, structural constraints
The human nucleoporin 98 (Nup98) is a component of the nuclear pore complexes (NPCs) which provide the only known portals for macromolecule exchange between the nucleus and cytoplasm (Suntharalingam and Wente 2003). Recent studies using a battery of complementary techniques have revealed that the NPC is highly modular on several levels, ranging from the eightfold rotational symmetry of the pore structure down to a limited set of domain structural elements (Schwartz 2005). Thus even though the NPC is a massive complex (sizes ranging from 60 to 125 MDa for the vertebrates), it is built from multiple copies of only approximately 30 different proteins called nucleoporins (Nups). Nup98 is directly involved in the transport of proteins and RNAs (Blevins et al. 2003) and is the target of the vesicular stomatitis (von Kobbe et al. 2000) and influenza viruses (Satterly et al. 2007). Chromosomal rearrangements involving Nup98 have also been implicated in leukemia (Slape and Aplan 2004).
For NPC targeting, an autoproteolytic processing event at the C-terminal domain is required during maturation of Nup98. Nup98 is encoded by two alternatively spliced mRNA transcripts (Fontoura et al. 1999). One transcript is translated into a 98-kDa, 920-amino acid protein which is post-translationally cleaved at the peptide bond between Phe863 and Ser864, into a 92-kDa N-terminal fragment (here termed Nup98-N) and an 6-kDa C-terminal tail (Nup98-C). The other transcript is translated into a 190-kDa polyprotein which is also post-translationally processed between Phe863 and Ser864, to form the 92-kDa N-terminal protein (Nup98-N) and a 96-kDa C-terminal protein (Nup96). Studies have indicated that post-translational cleavage of the Nup98 or Nup98–Nup96 polyprotein is essential for the assembly of both Nup98-N and Nup96 into the NPC (Fontoura et al. 1999). Recently, it has been discovered that human nucleoporins Nup98 and Nup98–Nup96 are both post-translationally processed by intramolecular autoprocessing (Rosenblum and Blobel 1999).
The C-terminal autoproteolytic domain of Nup98 is a unique building module among Nups (Schwartz 2005) but is similar to other proteins capable of intramolecular autoprocessing, such as inteins, pyruvoyl enzymes, and Ntn (N-terminal nucleophile) hydrolases (Paulus 2000). These autoproteolytic processing events are initiated by a nucleophilic attack of a threonine, serine, or cysteine residue at the scissile peptide bond, leading to an N–O or N–S acyl rearrangement of the peptide bond (acyl shift) and resulting in a (thio)ester bond. Usually the equilibrium of an N–O or N–S acyl shift favors the amide bond, thus limiting the peptide bond cleavages. The fact that some proteins can self-process at these scissile peptide bonds suggests that these are not typical peptide bonds. This notion has been supported by our crystallographic analyses of an Ntn precursor, glycosylasparaginase (GA) precursor, which showed the scissile peptide bond to be in distorted trans conformations (Xu et al. 1999). Such an unusual distortion for an N–O or N–S acyl shift has also been found in other autoprocessing proteins (Ditzel et al. 1998; Klabunde et al. 1998; Hewitt et al. 2000; Poland et al. 2000; Kim et al. 2002) and is confirmed by NMR studies (Romanelli et al. 2004). Thus the distorted conformations with higher energy than a regular peptide bond could drive the equilibrium toward ester bond formation. Recently, a 3 Å resolution structure of the Nup98 autoproteolytic domain was reported (Hodel et al. 2002). However, the resolution was insufficient to reveal details of autoproteolysis mechanism and its interactions with Nup96 for NPC targeting. Here, we report a 1.9 Å crystal structure of the autoproteolytic domain to study the detailed autoproteolysis mechanisms of Nup98. The structure also provides a model to explain the dynamic nature of Nup98 and a possible binding site for Nup96.
Results
To determine high-resolution structures of the Nup98 autoproteolytic domain, we searched for new constructs that would produce high-quality crystals. To this end, we screened several constructs of Nup98 C-terminal domains for crystallization. The wild-type coding sequence was used to screen for the mature/autocleaved form (WT), whereas the catalytic Ser864 was replaced with an alanine to prepare the uncleaved precursor form (S864A). The best crystals of S864A precursor, diffracting to 1.9 Å resolution, were obtained using the construct containing residues 716–870 of the Nup98 autoproteolytic domain, which also included a six-histidine tag at its C terminus to facilitate purification. The best crystals of the wild-type domain, diffracting to 2.3 Å resolution, were grown from a construct containing residues 712–870 of the Nup98 autoproteolytic domain, also with a C-terminal His-tag.
Minimal autoproteolytic Nup98 domain
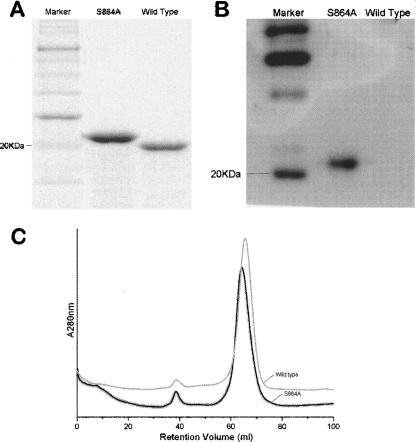
Biochemical analyses were used to study the autoproteolytic activity of the truncated constructs that gave the best diffracting crystals. As mentioned above, crystallization screens had selected a wild-type construct that is four residues longer at the N terminus than the S864A precursor fragment. Nonetheless, a side-by-side comparison on an SDS-PAGE gel (Fig. 1A) shows that the wild-type fragment has a faster mobility than the precursor fragment, consistent with the notion that autoproteolysis has cleaved a 13-residue tail (residues 864–870 followed by a 6-His tag) off the wild-type autoproteolytic domain. To ensure that the C-terminal tail was indeed being cleaved off the wild-type domain, fragments separated on the SDS-PAGE gel were transferred to a nitrocellulose membrane and probed with an antibody against the C-terminal His-tag. As expected, Western blot analysis shows that the C-terminal tail is retained in the S864A precursor construct, but is absent from the wild-type domain (Fig. 1B). Further, it had been shown that even after the autocleavage, the C-terminal tail remains non-covalently associated with the N-terminal domain (Hodel et al. 2002). This has also been confirmed by the fact that we were able to obtain high yield of both the S864A precursor and the wild-type domain using a nickel chelating column, by binding to the C-terminal His-tags. Thus, whether autocleaved or not, the C-terminal tails remain associated with the Nup98-N domain throughout the purification processes. Nonetheless, gel filtration indicates that the S864A and wild-type domains were eluted at slightly different volumes (Fig. 1C), suggesting there are some conformational differences between the wild-type and the S864A precursor domains. Since the difference in molecular mass alone between these two proteins, <500 daltons (four residues), is too small to be resolved by the gel filtration column used (see Materials and Methods), and since gel filtration separates proteins in their native forms by not only molecular size but also shape, the significant difference in elution volume is likely to be a result of the autoproteolysis that cuts a surface loop into two floppy ends with opposite charges. Altogether, these biochemical analyses show that the truncated wild-type domain retains its autoproteolytic activity. High-resolution electron-density maps from crystallographic studies of these two domains provide direct visualization of the backbone cleavage in the minimized wild-type autoproteolytic domain of Nup98 (see below).
Figure 1.
Autoproteolysis activity of the minimized Nup98 domain. (A) Slight migration difference on SDS-PAGE between the wild-type domain (a.a. 712–870 plus six histidines on its C terminus) and the autocleavage-deficient counterpart, the S864A precursor (a.a. 716–870 plus six histidines on its C terminus). The faster mobility of the wild-type fragment suggests an autocleavage has occurred to trim it into a smaller fragment. (B) Western blot results, probed with a His-tag antibody, indicate that the C-terminal His-tagged tail remains attached to the autocleavage-deficient (S864A) fragment but is absent from the wild-type counterpart. (C) FPLC gel filtration curves of the wild-type (gray) and the S864A precursor (black). Significant difference in elution volume is indicative of conformational differences, likely a result of autoproteolysis in the wild-type protein.
Structures of the Nup98 autoproteolytic domains
There is evidence that structural rearrangements subsequent to autoproteolysis are essential for the interaction between Nup98 and Nup96 (for NPC targeting) (Fontoura et al. 1999). In line with this notion, an autoproteolyzed Nup98 domain crystallized in different solution conditions and yielded different crystal forms than its precursor domain. The wild-type domain crystallized in the space group P3121 with unit cell constants a = b = 90.3 Å, and c = 97.6 Å, whereas crystals of the S864A precursor belong to the space group P41212 with unit cell constants a = b = 69.7 Å and c = 91.0 Å (Table 1). To avoid the model bias, the experimental phases of the wild-type structure were obtained at 3.3 Å by three-wavelength MAD method using three selenium sites in a SeMet-derivatized crystal, and the final structure was determined at 2.3 Å and refined to an R free of 26.0% and an R cryst of 22.8% (Table 2). Similarly, the experimental 2.7 Å phases of the S864A precursor were obtained from a single selenium site by three-wavelength MAD method, and the final structure at 1.9 Å resolution has been determined and refined to an R free of 22.9% and an R cryst of 20.6%. As expected, the polypeptide chain of the wild-type sequence is broken at the scissile peptide bond, after Phe863, whereas that of S864A precursor is continuous (see below).
Table 1.
Data-collection statistics of the Nup98 crystals
Table 2.
Statistics of model building and refinement
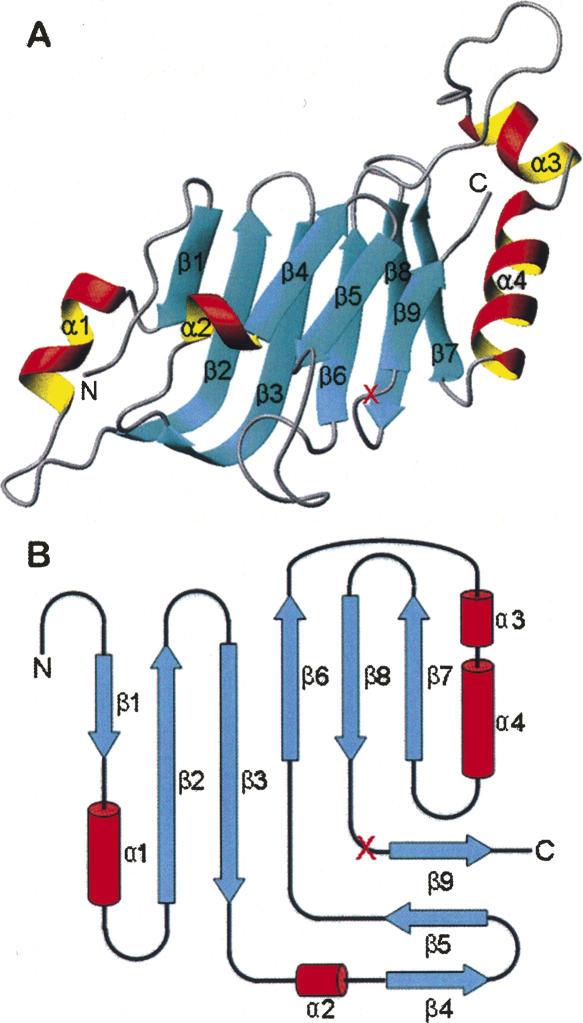
The wild-type autoproteolytic domain of Nup98 adopts a β-sandwich-like fold (Fig. 2), similar to the 3 Å structure (Hodel et al. 2002). The root mean square deviation (RMSD) between the current structure and the previously published low-resolution structure is 0.6 Å for all the equivalent Cα atoms. The structure is rich of β-strands, which form double-layer β-sheets capped with two α-helices on each side (α1, α2 on one side and α3, α4 on the other side). One layer is composed of a three-β-strand sheet (β4, β5, β9), while the other layer contains two side-by-side β-sheets, each with three β-strands (β1, β2, β3, and β6, β8, β7). Additionally, β2 and β3 are two long antiparallel β-strands, forming a tent-like shape capped by helix α2. The autoproteolytic site is located in the loop between the last two strands, β8 and β9.
Figure 2.
Architecture of the autoproteolytic domain of Nup98. The S864A precursor and wild-type domain have very similar overall structures, containing double layers of β-strands with two α-helices capped on each side. (A) Ribbon representation of the S864A precursor structure, with α-helices and β-strands shown as red/yellow and cyan, respectively. The red “X” marks the autocleavage site between Phe863 and Ser864. Figure produced with MOLMOL (Koradi et al. 1996). (B) Schematic diagram showing the topology of the Nup98 autocatalytic domain. The α-helices are represented by red columns, and the β-strands by cyan arrows.
The S864A precursor structure is almost the same as the wild-type domain, except that the polypeptide chain remained connected between Phe863 and Ser864 (Fig. 3). The overall superimposition of two structures gave a RMSD of 0.6 Å between the Cα carbons of the two models, suggesting that the cleavage of peptide bond between Phe863 and Ser864 in the wild-type domain does not have a dramatic effect on the overall structure. However, significant changes as the result of autoproteolysis were observed at the autocatalytic site.
Figure 3.
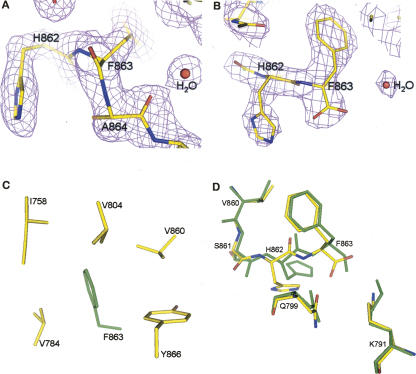
Structural features and comparison at the autocleavage sites of the S864A precursor and the wild-type protein. (A) Electron-density map at the autocleavage site of the S864A precursor. The magenta map is the 2F o–F c map at 1.9 Å resolution and contoured at 1.3σ. Model near the scissile peptide bond, at the carbonyl end of Phe863, is shown by atom type: yellow for carbons, blue for nitrogens, and red for oxygens. The electron density between F863 and A864 is continuous, indicating that S864A is an uncleaved precursor. Also shown are some bound water molecules, but the potential nucleophilic water molecule W97 (discussed in the text) is not shown for clarity. (B) Electron-density map at the autocleavage site of the wild-type domain. Shown is a 2F o–F c map at 2.3 Å resolution of the wild-type structure. The backbone electron density terminates at Phe863, indicating that the C-terminal tail is autocleaved and becomes disordered. Figures produced with PyMOL (http://pymol.sourceforge.net/). (C) A conserved hydrophobic pocket near the autocleavage site. The side chain of Phe863 (colored in green) is deeply buried in a hydrophobic pocket surrounded by side chains of I758, V784, V804, V860, and Y866 (yellow for carbons and red for oxygens); these residues are invariant or highly conserved among mammals, insects, and yeast. Additional contribution for the pocket comes from main-chain atoms of V784, H862, S864, and L798 (not shown). Figure was produced with PyMOL. (D) Structural comparison of the Nup98 autocleavage sites before and after autoproteolysis. Residues at the active sites and common to the S864A precursor and the wild-type structures are shown in green and red/yellow/blue, respectively. Structure superimposition was made by Cα atoms of the major β-sheet (including residues 748–760 [strands β2 and β3], 803–807 [strand β6], and 846–858 [strands β7 and β8], see Fig. 2), with an RMSD of 0.2 Å. Figure produced with PyMOL.
Autocatalytic sites
One major difference between electron-density maps of S864A precursor and the wild-type domain is at the scissile peptide bond, between Phe863 and Ser864 (Ala864 in the mutant) (Fig. 3). As expected, the S864A precursor, the non-cleavable construct, has continuous electron densities at the autocleavage site (Fig. 3A), while the wild-type main chain is broken between Phe863 and Tyr866 (Fig. 3B). The lack of electron density for Ser864 and Lys865 indicates structural disorder for these two residues in the wild-type construct. However, with a density gap of 8.5 Å (for two residues) between the well-ordered Phe863 and Tyr866, it is most likely that the disorder is caused by main chain autocleavage rather than flexibility of a linked polypeptide chain.
In the S864A precursor structure, an unusual conformation was found at the scissile peptide bond between Phe863 and Ala864 (Fig. 3A). A cis peptide bond fit best into the omit electron density, but we cannot completely rule out the possibility of a distorted trans peptide bond. Furthermore, the refinement software tends to minimize the bond distortion at the expense of local density fit (Fig. 3A). However, it is certain that the scissile peptide bond takes an unusual conformation, similar to those found in other autoprocessing proteins (Ditzel et al. 1998; Klabunde et al. 1998; Xu et al. 1999; Hewitt et al. 2000; Poland et al. 2000). These unusual, constrained peptide bonds have higher energy than their trans counterparts and could thus shift the equilibrium toward ester bond formation and break the energetically unfavorable amide bond (the N–O acyl shift). We were unable to compare this constrained peptide bond with the previously reported low-resolution structure (Hodel et al. 2002), due to the omission of this segment in the deposited coordinates (PDB ID: 1KO6).
To initiate the N–O acyl shift, the side chain of Ser864 acts as the necessary nucleophile to attack the backbone carbonyl carbon of Phe863 (Rosenblum and Blobel 1999). As demonstrated above, substitution of the Ser864 with an alanine (S864A mutant) completely abrogates the autoproteolysis activity and thus results in the precursor domain. When a serine was modeled into the precursor structure, the side-chain hydroxyl group interacts with His862 by a hydrogen bond and is also positioned 3.4 Å from the carbonyl carbon of Phe863, the target of the nucleophilic attack to initiate an N–O acyl shift. Although the distance is not ideal for the nucleophilic attack, it is possible that in the actual wild-type precursor a dynamic motion of the side chain could bring the nucleophile to an optimal distance and geometry for the attack.
In addition, several other conserved residues are also positioned around the autocatalytic sites to play various roles during the autocleavage. The imidazole ring of the highly conserved His862 is stabilized by a hydrogen bond to the side-chain oxygen of Ser861 (2.9 Å). Moreover, when Ser864 was modeled into the precursor structure with the most favored side-chain rotamer, it is properly positioned to form a 2.4 Å hydrogen bond with the side chain of His862. This peculiar structure is apparently stabilized by anchoring the side chain of the conserved Phe863 into a deep hydrophobic pocket (Fig. 3C), thus exposing the imidazole ring of His862 to act as the general base for activation at the nucleophile on Ser864. Side-chain nitrogen of the conserved Lys791 forms a 2.6 Å hydrogen bond with the carbonyl oxygen of Phe863. In addition, side chain of the conserved Asn799 forms a 2.9 Å hydrogen bond with the main-chain nitrogen of Phe863, and makes another hydrogen bonding (3.2 Å apart) to the carbonyl oxygen of Phe863. Thus both Lys791 and Asn799 are positioned to serve as the oxyanion hole to stabilize the negatively charged reaction intermediate (see proposed mechanism below).
Our high-resolution data provide further structural explanations for previous mutagenesis results. Single mutations with non-hydroxyl- or non-thiol-containing side chains at Ser864 completely abolish the autocleavage activity (Rosenblum and Blobel 1999). Similarly, point mutations at His862 also substantially reduce the level of autoproteolysis (Rosenblum and Blobel 1999; Hodel et al. 2002). Importantly, a F863W mutant does not affect much the autoproteolysis activity, while F863V or F863M mutants retain partial autoproteolysis activity. However, when Phe863 was mutated to amino acids with hydrophilic side chains, such as histidine, arginine, and glutamine, the autoproteolysis activity is completely destroyed. These results support the notion that tight fitting of a large hydrophobic side chain into the deep hydrophobic pocket (Fig. 3C) is critical to maintain the constrained scissile peptide bond for the Nup98 autoproteolysis. Further, single mutations at Lys791 or Asn799 also resulted in uncleaved precursors (Hodel et al. 2002), consistent with their role as the oxyanion hole in stabilizing the autoproteolysis intermediate.
Conformational rearrangements after autocleavage
Although autocleavage did not result in a dramatic conformational change, there were significant structural movements at the autocatalytic site (Fig. 3D). First, after autocleavage, the hydrogen bond between the side chains of Ser 861 and His862 was lost. This resulted in a 120° rotation at Ser861's χ1 angle, away from the side chain of His862. In addition, break of the scissile peptide bond also causes the side chain of His862 to lose another hydrogen bond with Ser864. These movements, however, allow formation of a new hydrogen bond: between the carbonyl oxygen of Ser861 and the main-chain nitrogen of Lys801 (2.6 Å). Furthermore, despite the stability of the overall structure, significant movement is observed at residues 861–863. The Cα's move away from the autocleavage site by 0.61, 0.95, and 0.88 Å for Ser861, His862, and Phe863, respectively (Fig. 3D), while the RMSD of the residues around the active site is only 0.2 Å. These changes in hydrogen bond interactions and main-chain shifts allow the side chain of His862 to tilt about 21° around the main-chain direction, resulting in movement in excess of 2 Å for the imidazole ring. Moreover, after cleavage, the aromatic side chain of Phe863 inserts deeper (by about 0.4 Å) into the hydrophobic pocket.
Association of the C-terminal tail after autocleavage
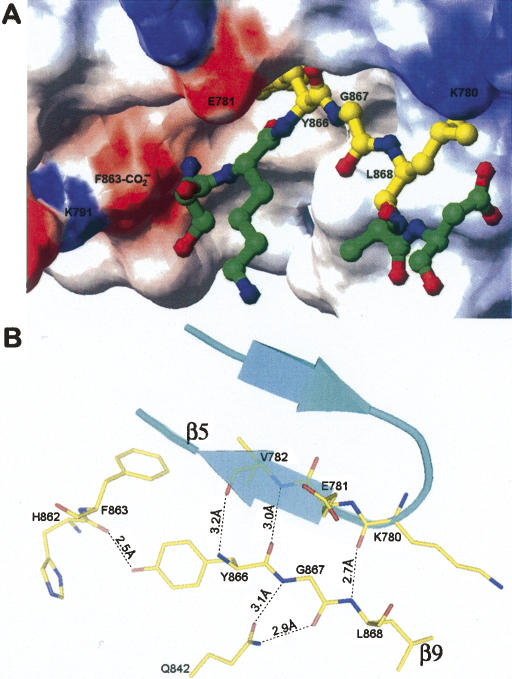
In agreement with the proposal of Hodel et al. (2002), our data show that the C-terminal tail remained associated with the N-terminal domain after autocleavage. In the wild-type structure, the autocleaved C-terminal tail is visible and remained associated with the Nup98-N domain (Fig. 4A). However, a relatively weak electron density of the tail also suggests a weak association between the Nup98-N domain and the autocleaved tail. The electron-density maps of the wild-type structure only allowed us to build three residues with confidence for the C-terminal tail (out of a total of seven residues followed by a 6-His tag). In our wild-type autocleaved model, only Tyr866, Gly867, and Leu868 are included, whereas Ser864, Lys865, Asn869, and Asp870 appear to be disordered. Comparison of the S864A precursor and the wild-type structures indicates that the interactions of these three residues with neighboring residues are very similar before and after autoproteolysis. In the S864A precursor structure, the portion corresponding to the C-terminal tail forms the strand β9 which runs antiparallel with strand β5 (residues 780–784) and makes numerous contacts with neighboring residues (Fig. 4B). The three residues, Tyr866, Gly867, and Leu868, assume most of the interactions with the Nup98-N domain and thus form the most stable segment of the strand β9. These interactions also appear to correlate well with the backbone stability of the C-terminal tail after autocleavage. In the wild-type autocleaved structure, these three residues remain bound to the N-terminal domain, whereas the rest of C-terminal tail becomes disordered. There are several hydrogen bonds formed between these three residues and the Nup98-N domain, including the ones connecting the main-chain nitrogen of Tyr866 with the carbonyl oxygen of Val782 (3.2 Å), the hydroxyl oxygen of Tyr866 with carbonyl oxygen of His862 (2.5 Å), the carbonyl oxygen of Tyr866 with the main-chain nitrogen of Val782 (3.0 Å), the main-chain nitrogen and the carbonyl oxygen of Gly867 with the side chain of Gln842 (3.1 Å and 2.9 Å, respectively), and the main-chain nitrogen of Leu868 with the carbonyl oxygen of Lys780 (2.7 Å). In addition, the hydrophobic side chain of Leu868 is buried in a bipolar pocket, one side of which is built by hydrophobic side chains and the other side is built by hydrophilic side chains. On the other hand, in the precursor structure, the other four residues on the C-tail, Ser864, Lys865, Gln869, and Asp870, only make hydrogen-bond contacts with water molecules, with one exception of the hydrogen bond between the nucleophile at Ser864 and the base at His862. After cleavage at the scissile peptide bond, these residues become disordered and thus no electron densities were observed. Importantly, sequence alignments indicate that the three residues 866–868 are highly conserved among all Nup98 autoproteolytic homologs (Fig. 5): The position of Tyr866 is always filled by a large hydrophobic residue, such as Tyr or Trp, and the other two residues, Gly867 and Leu868, are invariant in all known homologs. Conservation of these three residues is likely to preserve the critical function of associating the Nup98-N domain with the C-terminal tail or Nup96 for NPC targeting. At the same time, the disordered features for the rest of the C-tail also suggest that it could readily dissociate from the N-terminal domain, such that the binding site for the tail is opened for Nup96 binding. Since the N-terminal sequence of Nup96 (the first 50 residues), which is autoprocessed from the 190-kDa polyprotein, has the identical sequence as the Nup98 C-terminal tail, specific yet dynamic binding of Nup96 onto Nup98-N domain are most likely through the same binding residues Tyr866–Leu868.
Figure 4.
Structural details of the interactions between Nup98-N and the C-terminal tail. (A) Binding of the C-terminal tail to a cleft on the Nup98-N domain. Figure was generated based on the wild-type autocleaved structure. The solvent-accessible surfaces of the Nup98-N domain are colored according to electrostatic potential (blue, positively charged; red, negatively charged; and white, uncharged). Indicated areas are the charged surfaces contributed by the carboxylate terminus of Nup98-N domain (labeled F863-CO2 −) and by residues Lys791, Glu781, and Lys780. The structure of the C-terminal tail is displayed with a ball-and-stick model, using the stable fragment (residues Tyr866–Leu868) from the wild-type structure, whereas the disordered sections (a.a. 864–865 and 869–870) are modeled with the help of the corresponding sections from the precursor structure). The C-terminal tail is colored by atom types: nitrogens in blue, oxygens in red, and carbons in yellow and green for the stable and disordered sections, respectively. The figure was produced by MOLMOL (Koradi et al. 1996). (B) Associations between the C-terminal tail and the Nup98-N domain. The well-ordered section of C-terminal tail (residues Tyr866, Gly867, and Leu868) corresponds to the last strand, β9, in the precursor structure, which forms an antiparallel sheet with strand β5 (residues 780–784) through backbone hydrogen-bond interactions (distances in Å are indicated). In addition, Gly867 also interacts with the side chain of Gln842. Thus these three residues remain stable after autocleavage. Figure was produced with the S864A precursor model with PyMOL.
Figure 5.
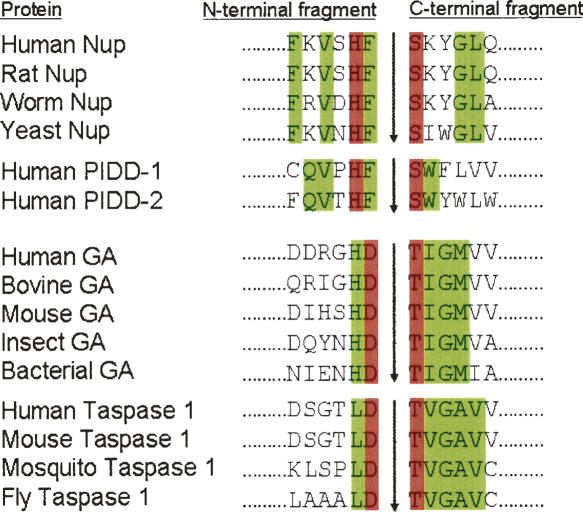
Amino acid sequence alignments at the autoproteolytic sites of Nups, GAs, and related proteins. Amino acid sequences are from Saarela et al. (1998), Rosenblum and Blobel (1999), Hsieh et al. (2003), and Tinel et al. (2007). Residues directly involved in catalysis are shown in red. Other conserved residues within subfamilies of homologs are shown in green.
Discussion
In this report, we provide high-resolution structures to examine the autoproteolysis mechanism of Nup98. Similar to other Ntn enzymes, there are no dramatic conformational changes as the result of autocleavage. However, autocleavage does produce local conformational rearrangements near the cleavage site, which are required for NPC targeting of Nup98 and Nup96. Here, we compare the mechanistic similarities and differences of autoproteolysis between Nup98 and other autoprocessing proteins.
Autoproteolytic mechanism
Structural constraints near the scissile peptide bond are essential for autoproteolysis to break the peptide linkage through an N–O or N–S acyl shift and an ester intermediate (Xu et al. 1999). However, it is worth noting that in aspartate decarboxylase, no conformational strain was found at the scissile peptide bond linking a glycine and the nucleophilic serine (Schmitzberger et al. 2003). In the latter case, the oxyanion hole appears to play the dominant role in facilitating the autoprocessing event. For other cases, almost all of the constrained scissile peptide bonds were found to be in distorted trans conformations, with one exception of a cis conformation in the GyrA intein (Klabunde et al. 1998). Here we report another example of Nup98 adopting a cis-peptide bond at the autoprocessing site. All these distorted trans or cis conformations at the scissile peptide bonds would have higher energy than regular trans bonds and therefore would drive the equilibrium of an N–O acyl shift toward ester bond formation to break the peptide linkage. As we have previously argued (Xu et al. 1999), since the peptide bond is naturally synthesized in the trans conformation, the fact that the cis bond is found in crystal structures indicates that it is favored over a trans bond, presumably distorted, at such location. Interestingly, both structures with a cis-peptide bond at the autocatalytic sites have a substitution of alanine at the defective scissile peptide bonds. It thus remains a possibility that the relatively small side chain of alanine at these two autocatalytic-deficient mutants might have allowed the distorted backbone to convert from a naturally strained trans bond to the observed cis bond, in order to minimize the energetically unfavorable geometry at these locations.
The proposed autoproteolytic mechanism of Nup98 is similar to that of GA through an N–O acyl shift with the side chain of Ser864 acting as the nucleophile (Rosenblum and Blobel 1999). Sequence alignments at the autocleavage sites are shown in Figure 5 for GAs and Nups. Interestingly, the only identical residue across GA and Nup is a histidine positioned two residues N-terminal from the nucleophilic side chain. This conserved histidine was initially proposed to act as the base in GA, through a mechanism similar to serine proteases. Nonetheless, both structural and mutational studies have later shown that this conserved histidine plays only a structural role to stabilize the backbone distortion in GA but is not in a position to act as the base (Guan et al. 1998). Instead, the conserved aspartic acid at the N-terminal side of the scissile peptide bond (colored red in Fig. 5) is the base for GA to activate the nucleophile, as well as holding the backbone distortion required to trigger autoproteolysis (Qian et al. 2003). Interestingly, a conserved phenylalanine is found in Nup98 at the corresponding position of the general base for GA autoproteolysis. Furthermore, neither the conserved residues (colored green in Fig. 5) near the cleavage sites nor the pattern of conservation are similar between GA and Nup. Apparently, the observed differences in sequence requirements would originate from differences in their precursor structures. Based on the present structural analysis, we found that the two residues immediately N-terminal to the scissile peptides bonds of Nup98 and GA have switched their roles in autoproteolysis. For GA precursor, the general base is provided by the aspartate at the P1 position (Asp151), whereas this functional group was carried by the histidine at the P2 position of Nup98 precursor (His862). Meanwhile, both the P2 histidine of GA (His150) and the P1 phenylalanine of Nup98 (Phe863) provide the structural anchors to maintain the distorted backbone conformation, by fitting their side chains into complementary pockets. Even though neither the conserved residues near the cleavage sites, nor the patterns of conservation are similar between GA and Nup, these two autoprocessing proteins apply a similar strategy to break up the scissile bonds.
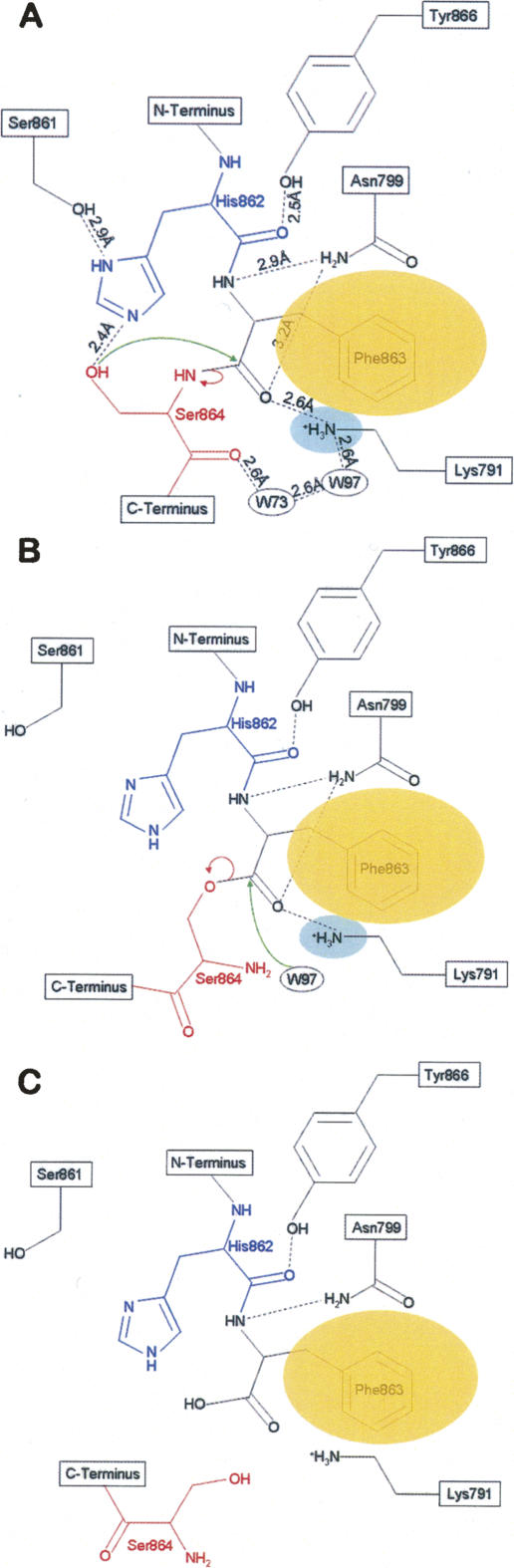
Based on the high-resolution structures of the autoproteolytic domain, we propose the detailed mechanism of Nup98 autoproteolysis through an N–O acyl shift. To this end, we have modeled a wild-type precursor structure by changing the Ala864 in the S864A structure to a serine with the most-favored side-chain rotamer. The three key elements of the proteolytic process are the nucleophile, the base, and the oxyanion hole. Based on the wild-type precursor model, we propose that His862 acts as the general base, Ser864 as the nucleophile, and Lys791 together with Asn799 serve as the oxyanion hole. His862 polarizes the hydroxyl oxygen of Ser864 to enhance its nucleophilicity through a hydrogen bond contact (Fig. 6A), thereby activating the nucleophilic attack. This critical polarization is made possible by Phe863, whose aromatic side chain is buried in a deep hydrophobic pocket, thus placing the side chain of His862 in an appropriate geometry to act as the base. In addition, the side chain of Tyr866 also appears to constrain the scissile backbone via a hydrogen bond to the main-chain carbonyl oxygen of His862 (Fig. 4B). Nucleophilic attack of the hydroxyl oxygen of Ser864 at the carbonyl carbon of Phe863 would result in a tetrahedral intermediate with a negatively charged carbonyl oxygen stabilized by an oxyanion hole. This oxyanion hole is formed by the positively charged side chain of Lys791, and the side chain of the conserved Asn799. Collapse of the tetrahedral intermediate results in an N–O acyl shift, that changes linkage from an amide to an ester bond (Fig. 6B). Finally, a nearby water molecule nucleophilically attacks the same carbonyl carbon at Phe863 to form another tetrahedral intermediate, which later collapses to produce the autocleaved Nup98 (Fig. 6C). This second tetrahedral intermediate is likely to be stabilized by the same oxyanion hole as the first intermediate. According to current high-resolution structures, water W97 is a potential candidate for the second nucleophilic attack to hydrolyze the ester linkage. Although in the precursor structure this water is 3.7 Å away from the carbonyl carbon of Phe863, conformational rearrangements during the N–O acyl shift could bring this water molecule to an optimal position for the second nucleophilic attack.
Figure 6.
Proposed autoproteolysis mechanism of Nup98. The nucleophilic Ser864 and the general base His862 are shown in red and blue, respectively. The side chain of Phe863 is buried in a conserved hydrophobic pocket (shown as an orange ellipse) to hold the strained scissile peptide bond between Phe863 and Ser864. Nucleophilic attacks are indicated by green curved arrows, and the electron-pair transfers are indicated by red curved arrows. Dashed lines indicate hydrogen bonds. Two bound water molecules near the autocleavage center are displayed in small ellipses (labeled as W73 and W97). The three key elements involved in the autoproteolysis are the nucleophile (the side-chain hydroxyl group of Ser864 for the first nucleophilic attack, and possibly water W97 for the second nucleophilic attack), the oxyanion hole (the positively charged side chain of Lys791, indicated by the cyan ellipse, and possible contribution from the side chain of Asn799), and the general base (side chain of His 862 for the first step). (A) Side chain of Phe863 is anchored into a conserved hydrophobic pocket to maintain the critical structural constraints at the scissile peptide bond and also positions the side chain of His862 to act as the general base and to deprotonate the hydroxyl group of Ser 864 for the first nucleophilic attack. Such a nucleophilic attack at the carbonyl carbon of Phe863 results in a negatively charged tetrahedral transition state, that is stabilized by the oxyanion hole formed by the side chain of Lys791 and possibly the side chain of Asn799. Subsequent collapse of this tetrahedral intermediate results in an N–O acyl arrangement, forming an ester intermediate. (B) The ester intermediate is further hydrolyzed to break the linkage. A bound water molecule (W97) is the likely candidate to make the second nucleophilic attack at the same carbonyl carbon of Phe863, forming the second tetrahedral transition state that is also stabilized by the same oxyanion hole as the first step. Collapse of this second tetrahedral transition state breaks the ester bond and results in the autoproteolyzed Nup98 protein. (C) The autoproteolyzed (mature) form of Nup98, in which the peptide bond between Phe863 and Ser864 is broken, caused conformational changes around the autocatalytic center (see text for details).
Minimal C-terminal tail required to support the Nup98 autoproteolysis
For different autoproteolytic proteins, the fragment length requirement on either side of the scissile peptide bond varies substantially. The smallest autoproteolytically active domain of Nup98 previously reported contains 149 residues upstream to the cleavage site and only 10 residues C-terminal of the scissile peptide bond (Rosenblum and Blobel 1999). However, GA requires almost the entire sequence to maintain its autoproteolysis activity, likely due to its unique three-dimensional structure of an interwoven αββα sandwich (Guo et al. 1998; Xu et al. 1999; Oinonen and Rouvinen 2000). Natural deletions from the GA C terminus found in aspartylglucosaminuria (AGU) patients results from loss of the autoproteolytic activity. An AGU-causing mutation of an American patient results in premature termination of translation, with the expected mutant GA shortened by 28 amino acids at the C-terminal end (Ikonen et al. 1991). Another AGU patient was found to carry a C-terminal deletion of 84 amino acids in GA (Park et al. 1996). The latter truncated GA contains complete sequence (205 amino acids) upstream of the autocleavage site, and 57 of the complete 141 amino acids downstream of the autocleavage site. When it was expressed in COS-1 cells, this C-terminally deleted protein remained as a single-chain precursor form, suggesting a loss of the autoproteolytic activity. These observations indicate that GA is vulnerable to deletion, likely to be caused by misfolding of deleted mutants. To the contrary, a PI-SceI truncated precursor containing 10- and four-residue extensions at the N- and C-terminal sides of the scissile peptide bond, respectively, was able to capture the intein in distorted trans conformations (Poland et al. 2000). However, to block protein splicing for crystallization, two critical residues were mutated to alanines in that truncated PI-SceI precursor. It is thus unclear whether such a minimal PI-SceI precursor without mutation is fully active in the N–S acyl shift and splicing.
In this report, we have further shortened the C-terminal fragment of Nup98 autoproteolytic domain to contain only seven residues downstream of the autocleavage site, three amino acids shorter than the previously published minimal sequence (Rosenblum and Blobel 1999). Biochemical and structural analyses demonstrate that this further minimized domain still retains native-like autoproteolytic activity (Figs. 1, 3). Furthermore, only three residues, Tyr866 to Leu868, in the C-terminal tail make substantial contacts with the N-terminal domain. It thus appears that a C-terminal tail containing only five residues (Ser864 to Leu868) might be sufficient to hold the structural constraints necessary for the Nup98 autoproteolysis.
Nup98 as the founder of a new subtype of autoprocessing proteins
Intein-independent protein autoprocessing was first discovered in prokaryotes. Now there is emerging evidence that these reactions also occur in several eukaryotic and human proteins. These include human GA involved in disease AGU (Guan et al. 1998), proteasomes in protein/antigen processing (Seemuller et al. 1996), γ-glutamyltranspeptidase in glutathione metabolism (Suzuki and Kumagai 2002), Taspase1 in MLL regulation (Hsieh et al. 2003), PCSK9 involved in hypercholesterolemia (Benjannet et al. 2004), and G protein-coupled receptor in cell signaling (Lin et al. 2004), as well as the maturation of viral proteins (McCann III et al. 1994; Zlotnick et al. 1994; Wu et al. 1998). Furthermore, autoprocessing has also recently been found in Clostridium difficile toxins (Reineke et al. 2007).
Nup98 represents a new subtype of autoprocessing proteins. Previously four subtypes of autoprocessing proteins have been described: intein, hedgehog, pyruvoyl enzymes, and Ntn hydrolases. To resolve the ester linkage during autoprocessing, a second nucleophilic attack is made by a cis- (intramolecular) side chain, a cholesterol molecule, or by a β-elimination for intein, hedgehog, and pyruvoyl enzyme, respectively. Since Nup98 uses a water molecule for the second nucleophilic attack to finish the autocleavage of the peptide backbone, its autoproteolysis most closely resembles that of Ntn hydrolases. However, significant differences exist between the autoprocessings of Nup98 and the well-studied GA, an Ntn member. In GA precursor, the residue immediately N-terminal to the scissile peptide bond, Asp151, acts as the general base to deprotonate Thr152, whereas the residue further upstream, His150, acts as a structural anchor to maintain the critical distorted geometry at the scissile peptide bond. In contrast, Nup98 uses the residue next to the scissile peptide bond, Phe863, as the structural anchor, but uses the penultimate residue of Nup98-N, His862, as the general base to activate the nucleophilic Ser864. Further, GA autoproteolysis results in conformational rearrangements to open up a substrate site with the newly generated N-terminal threonine (Thr152) as the catalytic residue (Wang and Guo 2007). To the contrary, no enzymatic activities have been detected for the autocleaved Nup98 or Nup96. Thus Nup98 autoproteolysis appears to be mainly involved in separating the linkage between Nup98-N and Nup98-C tail or Nup96. Nonetheless, such a cleavage is necessary for correct translocations of Nup98 and Nup96, so that they could take different biogenesis pathways for distinct functions. Recently, autocleavage has also been found to be essential in translocations of PIDD (p53-induced protein with a death domain) (Tinel et al. 2007). Sequence alignments also indicate that Nup98 and PIDD have closely related autocleavage sites that are distinct from the GA/Taspase1 subtype (Fig. 5). Thus it appears that Nup98 represents a new subtype of autoprocessing proteins that utilize backbone autocleavage to control cellular translocations and signalings.
Materials and Methods
Cloning of C-terminal domain of Nup98
To create the autoproteolytic domains for crystallographic study, various lengths of the Nup98 C-terminal domain with the wild-type sequences or an autoproteolysis-deficient mutation (S864A) were subcloned into the pET-21a vector (Novagen), followed with a hexahistidine (6-His) tag to facilitate protein purification. The constructs were flanked by NdeI and XhoI restriction sites, and were inserted into the pET-21a vector. In the recombinant constructs, the NdeI site at the 5′ end provided an ATG start codon to initiate the expression, whereas two stop codons were added to the 3′ end of the cloned sequences. The final sequences of the constructs were confirmed by primer-extended sequencings.
Protein expression and purification
Wild-type and S864A precursor proteins were expressed in bacterial BL21 (Gold) cells for 3 h at 37°C or 23°C, and by induction with 0.1 mM IPTG at mid-log growth phase. Selenomethionine-labeled proteins were expressed in the same cell line in M9 media supplemented with 100 mg/L of lysine, phenylalanine, threonine, 50 mg/L of isoleucine, leucine, valine, and 60 mg/L of l-selenomethionine added 15 min before IPTG induction at 23°C for 5 h. To release the target proteins, cell walls of harvested bacteria were broken by incubation with lysozyme and DNAse I followed by sonication and then clarified by high-speed centrifugation. The proteins were then purified by nickel chelating affinity purification, followed by a gel filtration column (Amersham Pharmacia, Sephacryl S-200 HR column) in 10 mM Tris, 10 mM NaCl, pH 7.5. For selenomethionine-labeled proteins, 10 mM dithiothreitol (DTT) was included in the entire purification process to prevent oxidation. All of the purified native and selenomethionine-labeled proteins were concentrated to 5–8 mg/mL for crystallization.
Crystallization and diffraction data collection
All crystals were grown by a vapor diffusion method in hanging-drops at 23°C. Hampton Crystal Screen Kits I and II were used for initial crystallization screens. The best diffracting crystals of wild-type domain belong to the space group P3121 (a = b = 90.3 Å, c = 97.6 Å, α = β = 90°, γ = 120°), which was grown at 23°C above a well solution containing 0.1 M Tris (pH 8.1), 0.2 M magnesium acetate, and 22% PEG 8000. Macroseeding was used to improve crystal quality. Crystals of the S864A precursor grew at 23°C against a well solution containing 0.1 M sodium citrate (pH 4.6) and 4.5 M sodium formate, and belong to the space group P41212 (a = b = 69.7 Å, c = 91.0 Å). All of the selenomethionine-labeled crystals were obtained under the same condition as the native crystals.
Data for all the native and SeMet-labeled crystals were collected at the National Synchrotron Light Source (NSLS) beamline X12C, Brookhaven National Laboratory. The flash-freezing method was used during data collection. Before freezing, crystals were soaked in the well solution supplemented with 20% glycerol. Three-wavelength MAD experiments were performed using SeMet-labeled crystals to provide phases for both the wild-type protein and the precursor. Data were processed using the program HKL (Otwinowski and Minor 1997) and were converted to structure factors using TRUNCATE from the CCP4 suite.
Structure determination and refinement
To avoid model bias, the experimental phases were calculated from data of SeMet MAD experiments. Parameters of selenium sites found were refined and the phases were calculated at 3.3 Å for the wild-type and 2.7 Å for the S864A precursor structures. An overall FOM (figure of merit) of 0.49 and a Z score of 10.9 were obtained for the wild-type structure, whereas an overall FOM of 0.52 and a Z score of 63.6 were found for the precursor structure. The first electron-density map was calculated using the experimental phases. There is one molecule in an asymmetric unit of the S864A precursor crystal, and two molecules in an asymmetric unit of the wild-type crystals. After density modification (including a noncrystallographic-symmetry averaging for the wild-type crystals), clear electron-density maps were obtained. Later in model building, a combination of the experimental and calculated phases was used to further improve the structures. After a few rounds of model rebuilding, stepwise resolution extension, and automated refinement using CNS (Brunger et al. 1998), the wild-type structure was determined at 2.3 Å resolution. One monomer (A) in the final wild-type model covers 155 out of 159 residues (missing residues 864, 865, 869, and 870), and the side chains of nine residues (D731, E739, K740, K753, E761, K814, K841, Q845, and Y866) were disordered. The second monomer (B) of the wild-type model covers 151 out of 159 residues (missing residues 787–790, 864, 865, 869, and 870), with no density for the side chains of four residues (E739, S746, K814, and Y866). The final precursor model has 151 out of 155 residues built (missing residues 787–790), and the side chains of three residues (E796, K814, and Q869) were disordered. The statistics of the final structures are shown in Table 2.
Accession numbers
Atomic coordinates and structure-factor amplitudes have been deposited in the Protein Data Bank under the accession codes 2Q5Y and 2Q5X for the wild-type and the S864A mutant, respectively.
Acknowledgments
We thank Drs. Günter Blobel and Jonathan S. Rosenblum for Nup98 plasmids. We also thank Jeffrey Brown for assistance on manuscript preparation and Anand Saxena, Yeming Wang, and Xiaofeng Qian for assistance with data collection at the National Synchrotron Light Source, Brookhaven National Laboratory. This research was supported in part by grants from the National Institutes of Health (DK053893 and DK075294).
Footnotes
Reprint requests to: Hwai-Chen Guo, Department of Physiology and Biophysics, Boston University School of Medicine, 715 Albany Street, W302, Boston, MA 02118-2526, USA; e-mail: hcguo@bu.edu; fax: (617) 638-4041.
Abbreviations: AGU, aspartylglucosaminuria; GA, glycosylasparaginase; Ntn, N-terminal nucleophile; RMSD, root mean square deviation.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073311808.
References
- Benjannet, S., Rhainds, D., Essalmani, R., Mayne, J., Wickham, L., Jin, W., Asselin, M.C., Hamelin, J., Varret, M., Allard, D., et al. NARC-1/PCSK9 and its natural mutants: Zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J. Biol. Chem. 2004;279:48865–48875. doi: 10.1074/jbc.M409699200. [DOI] [PubMed] [Google Scholar]
- Blevins, M.B., Smith, A.M., Phillips, E.M., Powers, M.A. Complex formation among the RNA export proteins Nup98, Rae1/Gle2, and TAP. J. Biol. Chem. 2003;278:20979–20988. doi: 10.1074/jbc.M302061200. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Ditzel, L., Huber, R., Mann, K., Heinemeyer, W., Wolf, D.H., Groll, M. Conformational constraints for protein self-cleavage in the proteasome. J. Mol. Biol. 1998;279:1187–1191. doi: 10.1006/jmbi.1998.1818. [DOI] [PubMed] [Google Scholar]
- Fontoura, B.M., Blobel, G., Matunis, M.J. A conserved biogenesis pathway for nucleoporins: Proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J. Cell Biol. 1999;144:1097–1112. doi: 10.1083/jcb.144.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, C., Liu, Y., Shao, Y., Cui, T., Liao, W., Ewel, A., Whitaker, R., Paulus, H. Characterization and functional analysis of the cis-autoproteolysis active center of glycosylasparaginase. J. Biol. Chem. 1998;273:9695–9702. doi: 10.1074/jbc.273.16.9695. [DOI] [PubMed] [Google Scholar]
- Guo, H.-C., Xu, Q., Buckley, D., Guan, C. Crystal structures of Flavobacterium glycosylasparaginase: An N-terminal nucleophile hydrolase activated by intramolecular proteolysis. J. Biol. Chem. 1998;273:20205–20212. doi: 10.1074/jbc.273.32.20205. [DOI] [PubMed] [Google Scholar]
- Hewitt, L., Kasche, V., Lummer, K., Lewis, R.J., Murshudov, G.N., Verma, C.S., Dodson, G.G., Wilson, K.S. Structure of a slow processing precursor penicillin acylase from Escherichia coli reveals the linker peptide blocking the active-site cleft. J. Mol. Biol. 2000;302:887–898. doi: 10.1006/jmbi.2000.4105. [DOI] [PubMed] [Google Scholar]
- Hodel, A.E., Hodel, M.R., Griffis, E.R., Hennig, K.A., Ratner, G.A., Xu, S., Powers, M.A. The three-dimensional structure of the autoproteolytic, nuclear pore-targeting domain of the human nucleoporin Nup98. Mol. Cell. 2002;10:347–358. doi: 10.1016/s1097-2765(02)00589-0. [DOI] [PubMed] [Google Scholar]
- Hsieh, J.J., Cheng, E.H., Korsmeyer, S.J. Taspase1: A threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell. 2003;115:293–303. doi: 10.1016/s0092-8674(03)00816-x. [DOI] [PubMed] [Google Scholar]
- Ikonen, E., Aula, P., Gron, K., Tollersrud, O., Halila, R., Manninen, T., Syvanen, A.C., Peltonen, L. Spectrum of mutations in aspartylglucosaminuria. Proc. Natl. Acad. Sci. 1991;88:11222–11226. doi: 10.1073/pnas.88.24.11222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y., Kim, S., Earnest, T.N., Hol, W.G. Precursor structure of cephalosporin acylase. Insights into autoproteolytic activation in a new N-terminal hydrolase family. J. Biol. Chem. 2002;277:2823–2829. doi: 10.1074/jbc.M108888200. [DOI] [PubMed] [Google Scholar]
- Klabunde, T., Sharma, S., Telenti, A., Jacobs W.R., Jr, Sacchettini, J.C. Crystal structure of GyrA intein from Mycobacterium xenopi reveals structural basis of protein splicing. Nat. Struct. Biol. 1998;5:31–36. doi: 10.1038/nsb0198-31. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., Wuthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Lin, H.H., Chang, G.W., Davies, J.Q., Stacey, M., Harris, J., Gordon, S. Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J. Biol. Chem. 2004;279:31823–31832. doi: 10.1074/jbc.M402974200. [DOI] [PubMed] [Google Scholar]
- McCann P.J., III, O'Boyle D.R., II, Deckman, I.C. Investigation of the specificity of the herpes simplex virus type 1 protease by point mutagenesis of the autoproteolysis sites. J. Virol. 1994;68:526–529. doi: 10.1128/jvi.68.1.526-529.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oinonen, C., Rouvinen, J. Structural comparison of Ntn-hydrolases. Protein Sci. 2000;9:2329–2337. doi: 10.1110/ps.9.12.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z., Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Park, H., Rossiter, M., Fensom, A.H., Winchester, B., Aronson N.N., Jr Single base deletion in exon 7 of the glycosylasparaginase gene causes a mild form of aspartylglycosaminuria in a patient of Mauritian origin. J. Inherit. Metab. Dis. 1996;19:76–83. doi: 10.1007/BF01799351. [DOI] [PubMed] [Google Scholar]
- Paulus, H. Protein splicing and related forms of protein autoprocessing. Annu. Rev. Biochem. 2000;69:447–496. doi: 10.1146/annurev.biochem.69.1.447. [DOI] [PubMed] [Google Scholar]
- Poland, B.W., Xu, M.Q., Quiocho, F.A. Structural insights into the protein splicing mechanism of PI-SceI. J. Biol. Chem. 2000;275:16408–16413. doi: 10.1074/jbc.275.22.16408. [DOI] [PubMed] [Google Scholar]
- Qian, X., Guan, C., Guo, H.-C. A dual role for an aspartic acid in glycosylasparaginase autoproteolysis. Structure. 2003;11:997–1003. doi: 10.1016/s0969-2126(03)00150-3. [DOI] [PubMed] [Google Scholar]
- Reineke, J., Tenzer, S., Rupnik, M., Koschinski, A., Hasselmayer, O., Schrattenholz, A., Schild, H., von Eichel-Streiber, C. Autocatalytic cleavage of Clostridium difficile toxin B. Nature. 2007;446:415–419. doi: 10.1038/nature05622. [DOI] [PubMed] [Google Scholar]
- Romanelli, A., Shekhtman, A., Cowburn, D., Muir, T.W. Semisynthesis of a segmental isotopically labeled protein splicing precursor: NMR evidence for an unusual peptide bond at the N-extein–intein junction. Proc. Natl. Acad. Sci. 2004;101:6397–6402. doi: 10.1073/pnas.0306616101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum, J.S., Blobel, G. Autoproteolysis in nucleoporin biogenesis. Proc. Natl. Acad. Sci. 1999;96:11370–11375. doi: 10.1073/pnas.96.20.11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarela, J., Laine, M., Tikkanen, R., Oinonen, C., Jalanko, A., Rouvinen, J., Peltonen, L. Activation and oligomerization of aspartylglucosaminidase. J. Biol. Chem. 1998;273:25320–25328. doi: 10.1074/jbc.273.39.25320. [DOI] [PubMed] [Google Scholar]
- Satterly, N., Tsai, P.L., van Deursen, J., Nussenzveig, D.R., Wang, Y., Faria, P.A., Levay, A., Levy, D.E., Fontoura, B.M. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc. Natl. Acad. Sci. 2007;104:1853–1858. doi: 10.1073/pnas.0610977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitzberger, F., Kilkenny, M.L., Lobley, C.M., Webb, M.E., Vinkovic, M., Matak-Vinkovic, D., Witty, M., Chirgadze, D.Y., Smith, A.G., Abell, C., et al. Structural constraints on protein self-processing in l-aspartate-α-decarboxylase. EMBO J. 2003;22:6193–6204. doi: 10.1093/emboj/cdg575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, T.U. Modularity within the architecture of the nuclear pore complex. Curr. Opin. Struct. Biol. 2005;15:221–226. doi: 10.1016/j.sbi.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Seemuller, E., Lupas, A., Baumeister, W. Autocatalytic processing of the 20S proteasome. Nature. 1996;382:468–471. doi: 10.1038/382468a0. [DOI] [PubMed] [Google Scholar]
- Slape, C., Aplan, P.D. The role of NUP98 gene fusions in hematologic malignancy. Leuk. Lymphoma. 2004;45:1341–1350. doi: 10.1080/10428190310001659325. [DOI] [PubMed] [Google Scholar]
- Suntharalingam, M., Wente, S.R. Peering through the pore: Nuclear pore complex structure, assembly, and function. Dev. Cell. 2003;4:775–789. doi: 10.1016/s1534-5807(03)00162-x. [DOI] [PubMed] [Google Scholar]
- Suzuki, H., Kumagai, H. Autocatalytic processing of γ-glutamyltranspeptidase. J. Biol. Chem. 2002;277:43536–43543. doi: 10.1074/jbc.M207680200. [DOI] [PubMed] [Google Scholar]
- Tinel, A., Janssens, S., Lippens, S., Cuenin, S., Logette, E., Jaccard, B., Quadroni, M., Tschopp, J. Autoproteolysis of PIDD marks the bifurcation between pro-death caspase-2 and pro-survival NF-κB pathway. EMBO J. 2007;26:197–208. doi: 10.1038/sj.emboj.7601473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kobbe, C., van Deursen, J.M., Rodrigues, J.P., Sitterlin, D., Bachi, A., Wu, X., Wilm, M., Carmo-Fonseca, M., Izaurralde, E. Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol. Cell. 2000;6:1243–1252. doi: 10.1016/s1097-2765(00)00120-9. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Guo, H.C. Crystallographic snapshot of a productive glycosylasparaginase–substrate complex. J. Mol. Biol. 2007;366:82–92. doi: 10.1016/j.jmb.2006.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Z., Yao, N., Le, H.V., Weber, P.C. Mechanism of autoproteolysis at the NS2–NS3 junction of the hepatitis C virus polyprotein. Trends Biochem. Sci. 1998;23:92–94. doi: 10.1016/s0968-0004(98)01180-3. [DOI] [PubMed] [Google Scholar]
- Xu, Q., Buckley, D., Guan, C., Guo, H.-C. Structural insights into the mechanism of intramolecular proteolysis. Cell. 1999;98:651–661. doi: 10.1016/s0092-8674(00)80052-5. [DOI] [PubMed] [Google Scholar]
- Zlotnick, A., Reddy, V.S., Dasgupta, R., Schneemann, A., Ray W.J., Jr, Rueckert, R.R., Johnson, J.E. Capsid assembly in a family of animal viruses primes an autoproteolytic maturation that depends on a single aspartic acid residue. J. Biol. Chem. 1994;269:13680–13684. [PubMed] [Google Scholar]