Abstract
X-ray crystallography is currently the most successful method for determining the three-dimensional structure of membrane proteins. Nevertheless, growing the crystals required for this technique presents one of the major bottlenecks in this area of structural biology. This is especially true for the α-helical type membrane proteins that are of particular interest due to their medical relevance. To address this problem we have undertaken a detailed analysis of the crystallization conditions from 121 α-helical membrane protein structures deposited in the Protein Data Bank. This information has been analyzed so that the success of different parameters can be easily compared for different membrane protein families. Concurrent with this analysis, we also present the new sparse matrix crystallization screen MemGold.
Keywords: membrane proteins, protein crystallization
Recent successes over the last few years have significantly increased the numbers of high-resolution membrane protein structures using X-ray crystallography (see http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html). A sufficient amount of data is now available on successful α-helical membrane protein crystallization to allow the rational design of a more specific crystallization screen. With this aim in mind, we constructed a database of crystallization information based on detergent solubilized α-helical membrane proteins that were crystallized using the vapor diffusion technique, the most commonly used method for initial crystal screening (Jancarik and Kim 1991; Kimber et al. 2003; Page and Stevens 2004). The conditions were analyzed to establish the current trends in the different crystallization parameters and from this a new sparse-matrix screen was designed. We anticipate that this will greatly facilitate the preliminary crystallization of a range of important α-helical membrane proteins.
Results
Number and types of α-helical membrane proteins used in the analysis
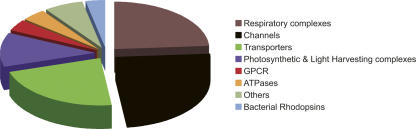
The crystallization conditions from a total of 121 polytopic α-helical membrane proteins (MPs) were used to construct the database. For clarity, these were grouped into eight different families depending on their function (Fig. 1). The largest families are the respiratory proteins and the channels, accounting for 29 entries each, followed by 27 entries for transporters. Of the remaining families, 14 entries are photosynthetic and light harvesting complexes, five entries are for the ATPases and GPCRs, and four for the different bacterial rhodopsins. The remaining eight MPs are not easily ascribed to these seven families and were grouped into a separate category termed “others” (see Supplemental material). At the time this database was compiled, the only available examples for the GPCR family consisted of five entries for bovine rhodopsin. For this reason, the recent structure of the human β2 adrenergic G protein-coupled receptor was not included in the database (Rasmussen et al. 2007).
Figure 1.
MP families. The proportion of structures belonging to each of the eight MP families in the database is shown. Bacterial rhodopsins (blue), GPCR (red), channels (black), transporters (green), photosynthetic and light harvesting complexes (purple), ATPases (orange), respiratory complexes (brown), others (DsbB-DsbA oxidase, intramembrane proteases, membrane-associated proteins in eicosanoid, and gluththione metabolism [MAPEG]) (olive). This color scheme is used throughout.
Detergent selection for crystallization
The choice of detergent is a critical parameter for successful crystallization of MPs. However, the detergent used for the initial extraction and purification will not always be effective in promoting the formation of well diffracting crystals (Iwata 2003). While a great deal of effort has been applied to understanding the role of detergent during crystallization (Rosenow et al. 2002; Iwata 2003), there is no single detergent that can be generally and reliably applied.
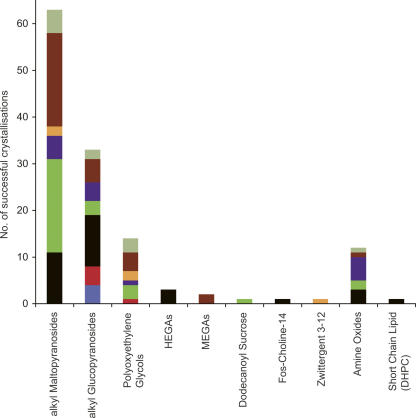
Figure 2 shows the different detergents that have been used to successfully crystallize α-helical MPs to date. These data show a clear preference for the alkyl maltopyranoside detergents, accounting for over 60 entries in the database. The alkyl glucopyranoside detergents were the next most successful with 33 entries. A full breakdown of all the detergents is given in Supplemental Figure S1. Of the alkyl maltopyranoside detergents, n-Dodecyl-β-D-maltopyranoside (DDM) is clearly the most successful, followed by n-Decyl-β-D-maltopyranoside (DM). For the alkyl glucopyranosides, both n-Octyl-β-D-glucopyranoside (OG) and n-Nonyl-β-D-glucopyranoside (NG) have had almost equal success. This finding contrasts with that obtained from an earlier study, which found OG to be the most successful crystallization detergent (Raman et al. 2006). This discrepancy arises because outer membrane proteins (OMP) were included in the latter analysis and so biased the results in favor of OG (one of the most successful detergents for OMP crystallization).
Figure 2.
Detergents. The numbers of successful crystallizations for each detergent class are shown. The bars have been subdivided into the eight MP families. Where more than one detergent type is represented, these have been grouped together.
Interestingly, the distribution of MP families crystallized by these two detergents differs. DDM has been more successful with transporters and the respiratory complexes, whereas OG has been more successful with the channels. The reason for this difference is not clear; one reason could be that ion channels are more stable than transporters, and therefore retain their structure in smaller detergents such as OG (Iwata 2003; Krishnan et al. 2005).
Types of precipitants
Figure 3A illustrates the success of the different types of precipitant within the eight MP families. Of the 121 conditions in the database, 108 were obtained using PEG or another type of polymer as the primary precipitant. Of the remainder, 15 were obtained using salt and two using an organic molecule. At first glance, the results appear quite similar to those obtained for soluble proteins, in which PEGs are the most successful crystallization reagents (Kimber et al. 2003; Page and Stevens 2004). On closer inspection, however, it is clear that small MW PEGs, in particular PEG 400 (Supplemental Fig. S2), have been more successful for MPs, whereas large MW PEGs have been more successful for soluble proteins (Kimber et al. 2003; Wooh et al. 2003; Page and Stevens 2004). To date, the large MW PEGs have been more successful for only one MP family, the respiratory complexes (Fig. 3A). Respiratory complexes usually contain large hydrophilic domains, which are often responsible for the majority of crystal contacts (Jormakka et al. 2002). Due to these large hydrophilic domains, respiratory complexes may behave more like soluble proteins in a crystallization experiment and may explain the success of large MW PEGs in this case.
Figure 3.
(A) Precipitants. The different precipitants used in the successful crystallization of the MP families are shown. Salts, which include ammonium sulphate, sodium chloride, lithium sulphate, sodium phosphate, and tri-sodium citrate (green) and the organic molecule, (+/−)-2-methyl-2,4-pentanediol (MPD) (purple) are indicated. The polyethylene glycols and other polymers, which include jeffamine and pentaerythritol propoxylate (5/4 PO/OH), are grouped into three classes, small MW (blue), medium MW (red), and large MW (yellow). (B) Concentration of PEGs. The concentrations of the polyethylene glycols used for successful MP crystallization are shown. Small MW (blue), medium MW (red), and large MW (yellow).
Salts have been significantly less successful than PEG in crystallizing α-helical MPs. However, of the 15 salt conditions in the database, these are spread across four MP families, showing that these reagents cannot be ignored. For very stable MPs, such as the AmtB–GlnK complex (Conroy et al. 2007) and the magnesium transporter MgtE (Hattori et al. 2007), mild organic solvents can be successful, although these are in general thought too harsh for α-helical MPs (Iwata 2003).
The concentration of PEGs used for crystallizing MPs is also different to those used for soluble proteins (Iwata 2003). Figure 3B shows the optimum concentration range for low MW PEGs lies between 20% and 30%, which is relatively narrow compared to the large MW PEGs, which were used between 5% and 25%.
Types of buffers and pH ranges
Buffers can have a significant impact on soluble protein crystallization (Newman 2004). Supplemental Figure S3 shows the success of different buffers for crystallization, and Supplemental Figure S4 shows their pH distribution. Supplemental Figure S4 shows a normal distribution around pH 7–8, with 88% of the conditions falling within the range of pH 5–9 and 65% within pH 6–8. Many of the commercially available screen kits are relatively conservative when screening pH, covering usually from 4.5–8.5. While this may be reasonable for soluble proteins, there are a number of examples in this analysis that fall outside this range. The data presented here suggests that screening between pH 3.5 and 10 should cover the broadest crystallization space. Unfortunately, this study shows no strict correlation between different buffer types and successful crystallization.
Types and concentrations of salts
Like buffering agents, polyvalent cations and anions can often be essential for crystallization as they can stabilize proteins within the crystal (Trakhanov and Quiocho 1995). Of the different salts present in the database, both monovalent and divalent salts have been equally successful (Supplemental Fig. S5). The most common salt is sodium chloride, which accounts for 32% of the conditions. Where more than one salt was used, the most common pairing with sodium chloride was magnesium chloride. Our analysis suggests that there is a strong case for screening many different types of counter ions in a MP screen, and that any initial screen should contain five of the most common salts at concentrations between 50 mM and 200 mM.
Additive screening
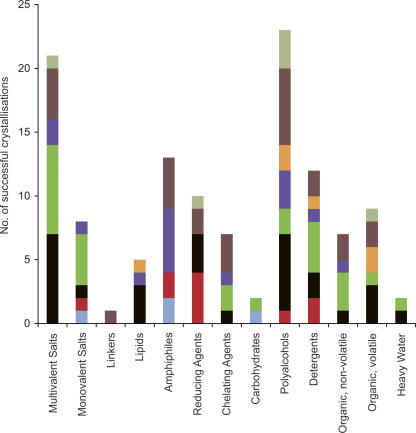
Small organic molecules, additional detergents, multivalent salts, and chemicals can all have a significant impact on the formation and quality of MP crystals (Michel 1982; Schertler et al. 1993; Sørensen et al. 2006). Figure 4 shows the different additive groups used to crystallize α-helical MPs, with the most successful groups being the polyalcohols and the multivalent salts. Within the polyalcohol group, glycerol is the most frequently used additive (Supplemental Fig. S6). However, we cannot rule out the possibility that this observation may be a result of glycerol being used as a cryoprotectant during crystallization. There is no observable trend in the multivalent salt group. Instead, this group is made up of a wide range of different salts and combinations thereof, suggesting that these could be useful in an initial additive screen.
Figure 4.
Additives. The range of additives used for the successful crystallization of the MP families is shown. The different additives have been grouped into more general classes: multivalent salts, monovalent salts, linkers (glycyl-glycyl-glycine), lipids, amphiphiles (1,2,3-heptanetriol, benzamidine hydrochloride), reducing agents, chelating agents, carbohydrates, polyalcohols (glycerol, ethylene glycol), detergents, nonvolatile organic molecules (1,6-hexanediol, MPD, PEG 400), and volatile organic molecules (ethanol, tert-butanol, 1,3-propanediol, isopropanol).
Discussion
This paper presents an in-depth analysis of successful polytopic α-helical MP crystallization conditions and highlights some of the emerging tends. For example, if no prior knowledge is available about the stability of an α-helical MP in different detergents, our analysis suggests that DDM, DM, OG, C12E8, and LDAO are good first candidates. However, one should always take the function and stability of the MP into consideration. For example, we would caution the use of LDAO for transporters, with only one successful transporter structure to date (Conroy et al. 2007). Indeed, a recent screen for the monodispersity of eukaryotic transporters concluded that LDAO caused aggregation for the majority of transporters tested (Newstead et al. 2007). In terms of crystallization, of particular note is the success of small MW PEGs, which are often underrepresented in the more traditional crystallization screens for soluble proteins. This bias in favor of large MW PEGs often makes these screens unsuitable for α-helical MPs. Using the information from this study we developed MemGold (Table 1) to address this problem, and this currently represents the most up-to-date sparse matrix crystallization screen for α-helical MPs.
Table 1.
MemGold: A targeted sparse matrix α-helical MP crystallization screen
Materials and Methods
A database was built by collecting the crystallization information from all of the available unique α-helical MP structures in the PDB (α-MP-database.xls, Supplementary material). Only conditions from proteins crystallized using the vapor diffusion technique were recorded in the database, as this method is the most commonly used method for screening. To avoid biasing the analysis, conditions from the same MP were excluded if they were identical. This task was greatly facilitated by the Membrane Protein Data Bank (MPDB) (www.mpdb.ul.ie) (Raman et al. 2006) and the “Membrane Proteins of Known 3D Structure” Web site from the Stephen White laboratory at UC Irvine (http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html).
Crystallization conditions were then divided into the different components, precipitant, buffer, pH, salt(s), additives, and detergents along with their respective concentrations. Chemicals were considered additives if their concentration was <20 mM, for example, zinc sulphate at 5 mM was counted as an additive and not as a salt. The crystallization conditions were then analyzed by constructing a series of stacked bar charts showing the number of successful crystallizations for each MP family against the individual chemicals for each component of the crystallization experiments.
The MemGold screen was designed by selecting 96 conditions from this database. The conditions were chosen to be as nonredundant as possible. The concentration of the precipitant was increased by 10%, such that 20% PEG would become 22%, to promote nucleation and the formation of crystals. The MemGold screen is currently commercially available through Molecular Dimensions Ltd.
Electronic supplemental material
The electronic supplemental material consists of Figures S1–S6 and a copy of the database used for the analysis in Excel format: α-MP-database.xls.
Acknowledgments
We thank Bernadette Byrne, Liz Carpenter, and David Drew (Imperial College London, UK) for discussions concerning this work. This project was supported by the Biotechnology and Biological Sciences Research Council, the Membrane Protein Structure Initiative consortium (www.mpsi.ac.uk), and the EU-PF6 E-MEP European Membrane Protein Consortium (www.e-mep.org).
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: So Iwata, MPC Group, Division of Molecular Biosciences, Imperial College London, Wolfson Laboratory, London SW7 2AZ, UK; e-mail: s.iwata@imperial.ac.uk; fax: 0044 20-759-43022.
Abbreviations: C12E8, polyoxyethylene(8)dodecyl ether; LDAO, lauryldimethylamine-N-oxide; DDM, n-Dodecyl-β-D-maltopyranoside; DM, n-Decyl-β-D-maltopyranoside; NG, n-Nonyl-β-D-glucopyranoside; OG, n-Octyl-β-D-glucopyranoside; HEGA, (N-hydroxyethylglucamide); MEGA, (N-methylglucamide); DHPC, (1,2-diheptanoyl-sn-glycero-3-phosphocholine); MPD, (+/−)-2-Methyl-2,4-pentanediol; KMES, 2-(N-morpholino)ethanesulfonic acid potassium salt; MME, monomethylether.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073263108.
References
- Conroy, M.J., Durand, A., Lupo, D., Li, X.D., Bullough, P.A., Winkler, F.K., Merrick, M. The crystal structure of the Escherichia coli AmtB-GlnK complex reveals how GlnK regulates the ammonia channel. Proc. Natl. Acad. Sci. 2007;104:1213–1218. doi: 10.1073/pnas.0610348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori, M., Tanaka, Y., Fukai, S., Ishitani, R., Nureki, O. Crystal structure of the MgtE Mg2+ transporter. Nature. 2007;448:1072–1075. doi: 10.1038/nature06093. [DOI] [PubMed] [Google Scholar]
- Iwata, S. Methods and results in crystallization of membrane proteins. International University Line; La Jolla, CA: 2003. [Google Scholar]
- Jancarik, J., Kim, S.-H. Sparse Matrix sampling: A screening method for crystallisation of proteins. J. Appl. Crystallogr. 1991;24:409–411. [Google Scholar]
- Jormakka, M., Tornroth, S., Byrne, B., Iwata, S. Molecular basis of proton motive force generation: Structure of formate dehydrogenase-N. Science. 2002;295:1863–1868. doi: 10.1126/science.1068186. [DOI] [PubMed] [Google Scholar]
- Kimber, M.S., Vallee, F., Houston, S., Necakov, A., Skarina, T., Evdokimova, E., Beasley, S., Christendat, D., Savchenko, A., Arrowsmith, C.H., et al. Data mining crystallization databases: Knowledge-based approaches to optimize protein crystal screens. Proteins. 2003;51:562–568. doi: 10.1002/prot.10340. [DOI] [PubMed] [Google Scholar]
- Krishnan, M.N., Bingham, J.P., Lee, S.H., Trombley, P., Moczydlowski, E. Functional role and affinity of inorganic cations in stabilizing the tetrameric structure of the KcsA K+ channel. J. Gen. Physiol. 2005;126:271–283. doi: 10.1085/jgp.200509323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, H. Three-dimensional crystals of a membrane protein complex. The photosynthetic reaction centre from Rhodopseudomonas viridis . J. Mol. Biol. 1982;158:567–572. doi: 10.1016/0022-2836(82)90216-9. [DOI] [PubMed] [Google Scholar]
- Newman, J. Novel buffer systems for macromolecular crystallization. Acta Crystallogr. D Biol. Crystallogr. 2004;60:610–612. doi: 10.1107/S0907444903029640. [DOI] [PubMed] [Google Scholar]
- Newstead, S., Kim, H., von Heijne, G., Iwata, S., Drew, D. High-throughput fluorescent-based optimization of eukaryotic membrane protein overexpression and purification in Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. 2007;104:13936–13941. doi: 10.1073/pnas.0704546104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, R., Stevens, R.C. Crystallization data mining in structural genomics: Using positive and negative results to optimize protein crystallization screens. Methods. 2004;34:373–389. doi: 10.1016/j.ymeth.2004.03.026. [DOI] [PubMed] [Google Scholar]
- Raman, P., Cherezov, V., Caffrey, M. The Membrane Protein Data Bank. Cell. Mol. Life Sci. 2006;63:36–51. doi: 10.1007/s00018-005-5350-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, S.G., Choi, H.J., Rosenbaum, D.M., Kobilka, T.S., Thian, F.S., Edwards, P.C., Burghammer, M., Ratnala, V.R., Sanishvili, R., Fischetti, R.F., et al. Crystal structure of the human β2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- Rosenow, M.A., Magee, C.L., Williams, J.C., Allen, J.P. The influence of detergents on the solubility of membrane proteins. Acta Crystallogr. D Biol. Crystallogr. 2002;58:2076–2081. doi: 10.1107/s0907444902016736. [DOI] [PubMed] [Google Scholar]
- Schertler, G.F., Bartunik, H.D., Michel, H., Oesterhelt, D. Orthorhombic crystal form of bacteriorhodopsin nucleated on benzamidine diffracting to 3.6 Å resolution. J. Mol. Biol. 1993;234:156–164. doi: 10.1006/jmbi.1993.1570. [DOI] [PubMed] [Google Scholar]
- Sørensen, T.L., Olesen, C., Jensen, A.M., Møller, J.V., Nissen, P. Crystals of sarcoplasmic reticulum Ca2+-ATPase. J. Biotechnol. 2006;124:704–716. doi: 10.1016/j.jbiotec.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Trakhanov, S., Quiocho, F.A. Influence of divalent cations in protein crystallization. Protein Sci. 1995;4:1914–1919. doi: 10.1002/pro.5560040925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooh, J.W., Kidd, R.D., Martin, J.L., Kobe, B. Comparison of three commercial sparse-matrix crystallization screens. Acta Crystallogr. D Biol. Crystallogr. 2003;59:769–772. doi: 10.1107/s0907444903002919. [DOI] [PubMed] [Google Scholar]