Abstract
The DAF-2 insulin/IGF-1 receptor regulates development, metabolism, and aging in the nematode Caenorhabditis elegans. However, complex differences among daf-2 alleles complicate analysis of this gene. We have employed epistasis analysis, transcript profile analysis, mutant sequence analysis, and homology modeling of mutant receptors to understand this complexity. We define an allelic series of nonconditional daf-2 mutants, including nonsense and deletion alleles, and a putative null allele, m65. The most severe daf-2 alleles show incomplete suppression by daf-18(0) and daf-16(0) and have a range of effects on early development. Among weaker daf-2 alleles there exist distinct mutant classes that differ in epistatic interactions with mutations in other genes. Mutant sequence analysis (including 11 newly sequenced alleles) reveals that class 1 mutant lesions lie only in certain extracellular regions of the receptor, while class 2 (pleiotropic) and nonconditional missense mutants have lesions only in the ligand-binding pocket of the receptor ectodomain or the tyrosine kinase domain. Effects of equivalent mutations on the human insulin receptor suggest an altered balance of intracellular signaling in class 2 alleles. These studies consolidate and extend our understanding of the complex genetics of daf-2 and its underlying molecular biology.
IN mammals, insulin and insulin-like growth factor 1 (IGF-1) receptors are major regulators of energy homeostasis and growth, respectively. In invertebrates such as the nematode Caenorhabditis elegans and the fruit fly Drosophila melanogaster, a single insulin/IGF-1 signaling pathway acts as a major regulator of life history. For example, reduction of insulin/IGF-1 signaling can increase life span in both organisms (Friedman and Johnson 1988; Kenyon et al. 1993; Clancy et al. 2001; Tatar et al. 2001; Ayyadevara et al. 2007). Insulin/IGF-1 signaling may also function as a regulator of aging in mammals, since global reduction of IGF-1 receptor function or of fat-specific reduction of insulin receptor function can increase life span in mice (Bluher et al. 2003; Holzenberger et al. 2003).
In C. elegans, the insulin/IGF-1 receptor is encoded by the gene daf-2 (Kimura et al. 1997). An intracellular pathway similar to the cognate mammalian and Drosophila pathways acts downstream of the DAF-2 receptor. This includes an AAP-1/AGE-1 phosphatidylinositol 3-kinase (PI 3-kinase) (Morris et al. 1996; Wolkow et al. 2002) and PDK-1, AKT-1, AKT-2, and SGK-1 serine/threonine kinases (Paradis and Ruvkun 1998; Paradis et al. 1999; Hertweck et al. 2004).
Mutations reducing activity of many genes in this pathway can cause constitutive formation of dauer larvae (the Daf-c phenotype). The dauer larva is a facultative diapausal form of third-stage larva, which forms in response to several environmental cues linked with conditions that are not propitious for reproduction (Cassada and Russell 1975; Riddle and Albert 1997).
AKT-1, AKT-2, and SGK-1 phosphorylate and inactivate the FOXO transcription factor DAF-16 (Lin et al. 1997; Ogg et al. 1997), whose activity is necessary for the dauer constitutive (Daf-c) and longevity (Age) traits resulting from reduced insulin/IGF-1 signaling (Kenyon et al. 1993; Riddle and Albert 1997). Phosphorylation inactivates DAF-16 at least in part by causing its sequestration in the cytoplasm (Henderson and Johnson 2001; Lee et al. 2001; Lin et al. 2001).
Analysis of genetic epistasis has been an important tool in helping to establish the order in which genes function in pathways in C. elegans (Avery and Wasserman 1992; Huang and Sternberg 1995). In studies of daf-2, epistasis analysis has been complicated by the fact that different daf-2 alleles can behave very differently. For example, mutational loss of function of daf-12, which encodes a nuclear hormone receptor (Antebi et al. 2000), suppresses the Daf-c phenotype of daf-2(m41). Yet when added to daf-2(e1370), daf-12 can result in early larval arrest (Vowels and Thomas 1992; Larsen et al. 1995), even though in terms of Daf-c, e1370 is a weaker allele than m41 (Gems et al. 1998). Moreover, while daf-12(m20) partially suppresses the longevity increase of daf-2(m41), it enhances that of daf-2(e1370) (Larsen et al. 1995).
Such allele-specific effects reflect the presence of distinct phenotypic classes of the daf-2 mutant (Gems et al. 1998). Most daf-2 reduction-of-function (hypomorphic) alleles form a single, approximate allelic series over a range of severity. Weak alleles cause weak temperature-sensitive (ts) Daf-c and Age phenotypes; severe alleles cause strong ts Daf-c and non-ts Age phenotypes, plus other ts pleiotropic traits, such as reduced feeding (Eat) and reduced movement (Unc). However, some daf-2 alleles do not fit within this scheme, exhibiting relatively severe ts Daf-c phenotypes, but not the additional pleiotropic traits. In such alleles, e.g., e1369, m41 and m212, Daf-c is also suppressed by daf-12(m20). On this basis, daf-2 alleles were classified into class 1 (Daf-c, Age: suppressed by daf-12), and class 2 (Daf-c, Age, plus other pleiotropic traits: enhanced by daf-12) (Gems et al. 1998).
daf-2 allele-specific effects of the type seen with interactions with daf-12 are also seen with other components of the daf-12 pathway. These include the daf-9 sterol monooxygenase (Gerisch et al. 2001; Motola et al. 2006), the daf-12 cofactor din-1S (Ludewig et al. 2004), and the dafachronic acid steroid ligands for daf-12 (Motola et al. 2006). Examples of daf-2 allelic differences are summarized in Table 1.
TABLE 1.
daf-2 allele class-specific epistasis and traits
|
daf-2 allele
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class 1
|
Class 2
|
|||||||||||||
| Interacting mutation/treatment | e1371 | e1368 | e1365 | m577 | sa193 | m596 | m41 | m212 | e1369 | m579 | e1370 | e1391 | e979 | Source |
| Daf-c/larval arrest | ||||||||||||||
| daf-12(m20) | — | — | S | S | S | S | S | S | S | — | E | E | C | Gems et al. (1998) |
| din-1S(dh127) | — | S | — | — | — | — | — | — | S | — | Na | Na | — | Ludewig et al. (2004) |
| Δ4-dafachronic acid | — | S | — | — | — | — | — | — | — | — | (S) | — | — | Motola et al. (2006) |
| rop-1(pk93) | — | — | — | — | — | (S) | (S) | — | — | E | E | — | — | Labbé et al. (2000) |
| soc-1(n1789) | — | — | — | S | — | — | — | — | — | — | (S) | — | — | Hopper (2006) |
| Age | ||||||||||||||
| daf-12(m20) | — | — | (S) | (S) | (S) | — | (S) | N | — | — | E | E | E | Gems et al. (1998) |
| soc-1(n1789) | — | — | — | N | — | — | — | — | — | — | E | — | — | Hopper (2006) |
| daf-9(rh50) | — | (S) | — | — | — | — | — | — | — | — | (S)/Eb | — | — | Gerisch et al. (2001) |
| Whole gonad ablation | — | N | — | — | — | E | E | — | — | — | E | — | — | Hsin and Kenyon (1999) |
| Hyp phenotypec | SH | SH | SH | SH | — | + | +d | — | + | ++ | ++ | + | — | Scott et al. (2002) |
daf-2 alleles are listed in approximate order of increasing severity of the Daf-c phenotype within each allele class. S, full suppression. (S), partial suppression. E, enhancement. N, little effect. C, complex interaction. —, not determined.
Tested for suppression, but not for enhancement.
Weakly suppressed at 15°, enhanced at 22.5°.
SH, sensitive to hypoxia. +, resistant to hypoxia, but >10% death. ++, <10% death under hypoxia.
Identified in Scott et al. (2002) as e979 (C. M. Crowder, personal communication). This mistake was due to the m41 allele being misidentified as e979 when initially sent from the Gems laboratory to the Caenorhabditis Genetics Center. Only class 2 alleles show a high level of resistance to hypoxia.
In this study we have investigated the molecular changes that underlie phenotypic differences among daf-2 alleles. We have extended the description of the phenotypes of daf-2 with respect to nonconditional mutants and characterized the molecular lesions in 11 daf-2 mutants, including many unusual alleles. We have also examined the predicted effects of mutations on the DAF-2 receptor using three-dimensional homology modeling (Garza-Garcia et al. 2007) and other approaches. Taken together, our findings provide a clearer and more detailed overview of the genetics of daf-2.
MATERIALS AND METHODS
Nematode culture and strains:
Procedures for growth and manipulation of C. elegans were as published (Brenner 1974; Sulston and Hodgkin 1988). C. elegans strains employed included the following: wild type—N2 Bristol (CGC male stock) (Gems and Riddle 2000), CF1844 fer-15(b26); daf-2(mu150); fem-1(hc17), DR608 daf-2(m212), DR1436 daf-2(m631)/qC1 [dpy-19(e1259ts) glp-1(q339)], DR1468 daf-2(m646)/qC1, DR1563 daf-2(e1370), DR1564 daf-2(m41), DR1566 daf-2(m579), DR1567 daf-2(m577), DR1568 daf-2(e1371), DR1573 daf-2(e1369), DR1942 daf-2(e979), GA302 daf-2(tm1236)/qC1, GR1307 daf-16(mgDf50), JK1438 daf-2(m65)/qC1, JT193 daf-2(sa193), JT6723 daf-2(sa223)/qC1, NS3227 daf-18(nr2037), and TJ356 zIs356 [daf-16∷GFP rol-6(su1006)]. daf-2(m631) and daf-2(m646) were isolated during an EMS screen for nonconditional Daf-c mutants (I. Caldicot and D. L. Riddle, personal communication). daf-2(tm1236) was generated on request by the National Bioresource Project (Tokyo) (http://shigen.lab.nig.ac.jp/c.elegans/).
Strain constructions:
The many multiple mutants created for this study were prepared using standard strain construction methodologies (details available on request from the authors). In construction of all daf-2; daf-18 and daf-16; daf-2 strains involving nonconditional daf-2 alleles, the presence of each mutation was confirmed either by PCR product size or by DNA sequencing. The primers used for PCR to test for the presence of the deletions were as follows: for daf-16(mgDf50)—daf-16F1, gccactttattggaatttgagc; and daf-16R1, atcctcccatagaaggaccatt [PCR from gDNA with this primer pair yields a product with daf-16(+) but not mgDf50]; for daf-18(nr2037)—F3, gattggtgtctacgtggaacgg; or F6, ctattgaaggaggactaacacaggc, and R3, gccaacgaagtgctaaatcgac [PCR with F3 and R3 amplifies a 0.8-kb fragment only from daf-18(+) gDNA; F6 and R3 amplify 0.6- and 1.6-kb fragments from nr2037 and daf-18(+) gDNA], respectively (Mihaylova et al. 1999); for daf-2(tm1236)—F16F, ctggaccggaagccgaatcc; and Ex14F2A, tgacgattcagaagcactgg. In daf-2(+), these primers generate a product of 1.1 kb, and in daf-2(tm1236), they generate a product of 0.6 kb.
Sequencing of mutant alleles:
The entire coding sequence of the daf-2 transcript was amplified as a series of overlapping fragments by RT–PCR. These fragments were then sequenced on both strands; most fragments were sequenced twice, giving fourfold coverage of the region. Sequence was compared to the wild-type sequence reported by Kimura et al. (1997) (GenBank AF012437) and the sequence of the daf-2 predicted gene in WormBase (GenBank NM065249). Fragments that contained identical nucleotide substitutions on both strands compared to the wild-type sequences were taken to be the mutation in the allele. This region was then amplified from genomic DNA isolated from the mutant worms and sequenced to confirm the presence of the lesion.
Differences in the wild-type DAF-2 sequence:
The sequence of the mutant cDNA was translated in silico and then compared to the reported wild-type DAF-2 protein sequences to determine the amino acid change. The two wild-type DAF-2 sequences (Kimura et al. 1997 and WormBase) differ in length by three amino acids (1846 aa vs. 1843 aa); the residues Met–Thr–Arg at the N terminus of the Kimura et al. (1997) sequence are not present in the WormBase sequence, which uses the second in-frame ATG codon as the start of translation. We chose to use the numbering system of Kimura et al. (1997) for the position of the mutations identified in this study. This protein sequence also differs from the WormBase protein sequence at two positions. In the Kimura et al. (1997) DAF-2 sequence, Arg and Gln residues occupy positions 838 and 1313, respectively. In the WormBase DAF-2 sequence, His and Lys residues occupy the equivalent positions. Our DAF-2 sequence proved to be the same as the WormBase sequence.
Irregularities with daf-2 alleles:
The e979 allele was previously reported to have a G383E substitution in the cysteine-rich (CR) domain of the DAF-2 receptor (Scott et al. 2002). However, the m41 allele has also been reported to contain this mutation (Yu and Larsen 2001). The m41 allele is class 1 and e979 is class 2 and the Daf-c phenotype of e979 is more severe than that of m41 (Gems et al. 1998). We sequenced and phenotyped our lab stock of daf-2(e979) and resequenced and phenotyped the e979 allele used by Scott et al. (2002) and found that the mutation in our lab stock of e979 was C146Y, which affects the L1 domain. This stock exhibited a phenotype as previously reported (Gems et al. 1998). Sequencing of the e979 allele used by Scott et al. (2002) confirmed the G383E substitution in this strain. Phenotypic analysis confirmed that this strain was actually m41 and not e979. Scott et al. (2002) obtained the stock from the Caenorhabditis Genetics Center, which initially had an m41 stock that was misidentified as e979. This error was corrected in September 2002.
Oligonucleotide array analysis:
Microarray analysis of sterile, 1-day-old adults of the genotypes glp-4(bn2); daf-2(m577), glp-4(bn2); daf-2(e1370), glp-4(bn2) daf-16(mgDf50); daf-2(m577), and glp-4(bn2) daf-16(mgDf50); daf-2(e1370) was described previously (McElwee et al. 2004). In that study, for each genotype examined, five biological replicates were performed, and data for the two alleles of daf-2 were pooled. For this study, this data set was reanalyzed using newer methods, and the two alleles were treated separately, generating lists of genes that are differentially expressed in each allele.
Raw microarray data (cel files) were normalized, fold-changes between genotypes were determined, and global statistical analysis was performed, using a slightly modified version of the recently described “Goldenspike” methodology implemented in R (version 2.0.1) (Choe et al. 2005) (http://www.r-project.org/). Briefly, this procedure performs eight different normalization routines, which are then used to produce an average fold-change difference and false-discovery rate (q-value) between different genotypes that takes into consideration the variance of probe set intensity across the different normalizations. The Goldenspike methodology has been shown to out-perform most commonly used normalization methods (Choe et al. 2005). The Goldenspike protocol was altered slightly to exclude absent probe sets (those probe sets called “absent” in all hybridizations by MAS5) prior to the final probe-set-level Loess normalization. This alteration was found to reduce the number of false positives associated with the absent probe sets (Schuster et al. 2007).
Life-span measurements:
Animals were raised at 15° and shifted to 22.5° at the L4 stage. In measures of life span, day 0 is the L4 stage. In most trials, the inhibitor of DNA replication fluorodeoxyuridine (FUdR) was added to plates (Gandhi et al. 1980) to prevent death due to internal hatching of eggs.
Microscopy:
For Nomarski (differential interference contrast) imaging of nematodes, we used a Leica RXA2 compound microscope, with a Hamamatsu Orca-ER B/W CCD digital camera.
RESULTS
Incomplete suppression of severe daf-2 mutations by daf-18(0) and daf-16(0):
We began by characterizing four nonconditional daf-2 alleles: m65, m631, m646, and tm1236. In each case, homozygous daf-2 segregants from balanced daf-2/+ hermaphrodites form dauer larvae nonconditionally (data not shown). We performed epistasis tests, initially with m65, m631, and m646, and the dauer defective, suppressor mutations daf-18(nr2037) and daf-16(mgDf50), both of which are putative null alleles. The nr2037 mutation is a 990-bp deletion in daf-18, which results in loss of critical elements of the gene product (Gil et al. 1999; Mihaylova et al. 1999); mgDf50 is a deletion that removes most of the daf-16 coding region (Ogg et al. 1997). Here the aims were to reveal differences in severity among nonconditional daf-2 alleles to identify potential daf-2 null alleles and to understand interactions among daf-2, daf-18, and daf-16.
daf-18 encodes a PTEN phosphatase, which is a negative regulator of signaling via DAF-2 and AGE-1 (Ogg and Ruvkun 1998; Gil et al. 1999; Mihaylova et al. 1999; Rouault et al. 1999). PTEN (phosphatase and tensin homolog deleted on chromosome 10) is a commonly mutated tumor suppressor gene in human cancer. PTEN phosphatases attenuate insulin/IGF-1 signaling by dephosphorylating phosphatidylinositol 3,4,5-trisphosphate (PIP3) on position 3 of the inositol ring. daf-18(nr2037) fully suppresses both the Daf-c and Age phenotypes of daf-2(e1370) (Gil et al. 1999; Mihaylova et al. 1999). Thus, it is thought that DAF-2 acts principally by stimulating PIP3 production and by inhibiting DAF-16 activity (Kenyon et al. 1993; Riddle and Albert 1997; Mihaylova et al. 1999).
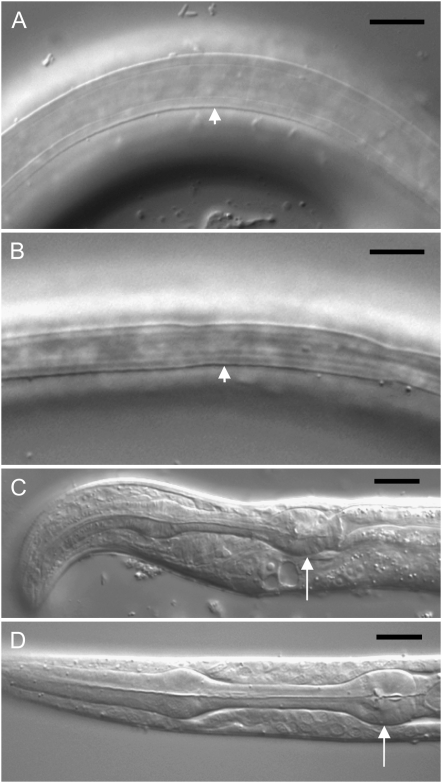
At 25°, daf-18 fully suppressed Daf-c in m631 and m646, but also resulted in 33 and 8% early developmental arrest, respectively (Table 2). This implies that m631 is the more severe allele. In the case of daf-2(m65); daf-18 double mutants, a high proportion of animals arrested development as embryos. At 15°, all three daf-2; daf-18 strains showed some dauer-like arrest at a low frequency (Table 2). This cold-sensitive Daf-c trait implies that the daf-18 mutation is temperature sensitive in its effects. Given that daf-18(nr2037) is a nullimorphic allele, this could imply the presence of a second PIP3 phosphatase that is functionally redundant with DAF-18, but only at lower temperatures. In the case of m631 and m646, these arrested larvae were partial dauers with dark, radially contracted bodies, yet there were also non-dauer alae (Figure 1A) and some pharyngeal pumping. These larvae resumed development after several days. By contrast, daf-2(m65); daf-18 dauers appeared fully differentiated and resumed development only after ∼1 week. Under the dissecting microscope these animals resembled normal dauer larvae with dauer alae (Figure 1B) and no pharyngeal pumping in most cases. Some arrested embryos were also present. These results demonstrate the following order of decreasing mutant allele severity: m65 > m631 > m646.
TABLE 2.
Incomplete suppression of nonconditional Daf-c alleles of daf-2 by daf-16 and daf-18
| Genotype | % L4 adults | % L1, L2a | % dauer | % dead eggs | N |
|---|---|---|---|---|---|
| At 25° | |||||
| + | 99 | 0 | 0 | 1 | 295 |
| daf-18(nr2037) | 100 | 0 | 0 | 0 | 275 |
| daf-16(mgDf50) | 100 | 0 | 0 | 0 | 325 |
| daf-2(m65) | — | — | 92 | — | 1361b |
| daf-2(m65); daf-18 | 24 | 6 | 0 | 70 | 171 |
| daf-2(m631); daf-18 | 67 | 26 | 0 | 7 | 85 |
| daf-2(m646); daf-18 | 92 | 5 | 0 | 3 | 283 |
| daf-16; daf-2(m65) | 0 | 0 | 0 | 100 | 298 |
| daf-16; daf-2(m646) | 100 | 0 | 0 | 0 | 172 |
| At 15° | |||||
| + | 99 | 0 | 0 | 1 | 230 |
| daf-18 | 100 | 0 | 0 | 0 | 189 |
| daf-16 | 100 | 0 | 0 | 0 | 257 |
| daf-2(m65); daf-18 | 65 | 6 | 5 | 24 | 109 |
| daf-2(m631); daf-18 | 78 | 0 | 22c | 0 | 172 |
| daf-2(m646); daf-18 | 89 | 0 | 11c | 0 | 221 |
| daf-16; daf-2(m65) | 43 | 18 | 0 | 39 | 292 |
| daf-16; daf-2(m646) | 100 | 0 | 0 | 0 | 210 |
These larvae are morphologically abnormal at 25°, but not at 15°.
Data were derived from Gems et al. (1998). Seventy-five percent of segregants from m65/+ mothers reached adulthood, 23% formed dauers, and the remainder were a mixture of dead eggs, L1's, or unhealthy, clear-looking arrested L2's.
Partial dauers, recovered within 24 hr.
Figure 1.—
Combined effects of severe daf-2 alleles and daf-18(nr2037) or daf-16(mgDf50) on development. (A) Cuticle of daf-2(m646); daf-18(nr2037) dauer-like larva showing L3 ala (short arrow). (B) Cuticle of daf-2(m65); daf-18(nr2037) dauer larva showing dauer ala (short arrow). (C) Anterior end of morphologically abnormal daf-2(m631); daf-18(nr2037) L1 larva, showing hypertrophy of ventral side of the head and dorsal displacement of pharynx (long arrow). (D) Anterior end of morphologically normal wild-type L1 larva (long arrow). Bars, 10 μm.
Severe daf-2 alleles causing nonconditional dauer arrest have previously been combined with reduction-of-function alleles of daf-16 (Gottlieb and Ruvkun 1994; Larsen et al. 1995). In daf-2(mg43), (nonconditional) Daf-c is suppressed by daf-16(m27) at 25° (Gottlieb and Ruvkun 1994). The nonconditional Daf-c phenotype of daf-2(m65) was also suppressed by the reduction-of-function allele daf-16(m26) at 15°; however, at 25.5°, the double mutant was embryonic lethal (Larsen et al. 1995). We examined the developmental effects of daf-16(mgDf50) on daf-2(m65) and daf-2(m646). In both daf-2 alleles, dauer formation was fully suppressed at 15° and in m646 at 25° (Table 2). In the case of daf-16(mgDf50); daf-2(m65) at 25°, 100% embryonic arrest occurred, similar to previous observations of daf-16(m26); daf-2(m65) (Larsen et al. 1995). In addition, daf-16(mgDf50); daf-2(tm1236) animals developed normally at 25°, demonstrating that tm1236 is a weaker allele than m65 (data not shown).
daf-2; daf-18 and daf-16; daf-2 mutants exhibited a range of other mutant phenotypes. All three daf-2; daf-18 strains and daf-16; daf-2(m65)-arrested first-stage larvae exhibited variable and sometimes severe morphological abnormalities, including ventral swelling in the region of the terminal bulb of the pharynx, resulting in dorsal displacement of the pharynx (Figure 1, C and D) and, in some cases, kinks in the anterior intestine. In this region, disorganization of the cuticular alae was sometimes seen. A small proportion of adult animals also showed a weak uncoordinated phenotype, with reduced movement, and a tendency to kink behind the pharynx when backing. These defects suggest that insulin/IGF-1 signaling plays a role in morphogenesis and early development.
Nonconditional daf-2 alleles and aging:
We tested the requirement for daf-18 and daf-16 gene function of the daf-2 life extension (Age) phenotype in severe daf-2 mutants. By itself, daf-18(nr2037) shortened life span relative to wild type, as previously seen. The Age phenotype of the weak mutant daf-2(e1370) was largely, but not fully, suppressed by daf-18 (Table 3). Consistent with their greater loss of function, m65, m631, and m646 all resulted in larger increases in life span when added to daf-18. In two of three trials, daf-2(m65); daf-18 animals were also longer lived than wild-type controls. In a direct comparison of daf-2; daf-18 strains (Table 3, trial 4), daf-2(m65); daf-18 animals were longer lived than daf-2(m631); daf-18 animals (P < 0.0001, log rank test), and daf-2(m631); daf-18 than daf-2(m646); daf-18 (P = 0.03). The implied order of mutant severity is consistent with that inferred from effects on development. Given its relative phenotypic severity and molecular identity, daf-2(m65) is a strong candidate for a null allele.
TABLE 3.
Effects of daf-2, daf-18, and daf-16 on life span
| Genotype | Mean (days) ±SE | Median (days) | Maximum (days) | Na | Pb | Pc |
|---|---|---|---|---|---|---|
| Trial 1 | ||||||
| + | 15.5 ± 0.5 | 15 | 22 | 55 (84) | — | — |
| daf-18(nr2037) | 8.2 ± 0.3 | 7 | 13 | 63 (102) | <0.0001 | — |
| daf-16(mgDf50) | 10.6 ± 0.3 | 9 | 17 | 68 (80) | <0.0001 | — |
| daf-2(e1370) | 29.9 ± 2.4 | 35 | 42 | 21 (77) | <0.0001 | — |
| daf-2(e1370); daf-18(0) | 9.9 ± 0.3 | 10 | 13 | 77 (94) | <0.0001 | <0.0001 |
| daf-2(m646); daf-18(0) | 13.2 ± 0.4 | 13 | 21 | 79 (102) | <0.0083 | <0.0001 |
| daf-16(0); daf-2(m646) | 9.8 ± 0.3 | 9 | 16 | 84 (84) | <0.0001 | 0.1423 |
| Trial 2 | ||||||
| + | 16.2 ± 0.5 | 18 | 21 | 62 (75) | — | — |
| daf-18(nr2037) | 9.2 ± 0.1 | 10 | 10 | 87 (100) | <0.0001 | — |
| daf-16(mgDf50) | 11.2 ± 0.1 | 11 | 11 | 100 (100) | <0.0001 | — |
| daf-2(m65); daf-18(0) | d 8.2 ± 0.5 | 7 | 14 | 72 (75) | <0.0001 | 0.6757 |
| daf-16(0); daf-2(m65) | 9.7 ± 0.4 | 11 | 14 | 95 (98) | <0.0001 | 0.6486 |
| Trial 3 | ||||||
| + | 16.3 ± 0.6 | 16 | 26 | 60 (60) | — | — |
| daf-18(nr2037) | 10.4 ± 0.3 | 12 | 14 | 89 (100) | <0.0001 | — |
| daf-16(mgDf50) | 13.4 ± 0.2 | 14 | 14 | 93 (100) | <0.0001 | — |
| daf-2(m65); daf-18(0) | 28.9 ± 0.5 | 29 | 37 | 95 (100) | <0.0001 | <0.0001 |
| daf-16(0); daf-2(m65) | 14.2 ± 0.2 | 14 | 16 | 97 (100) | <0.0001 | <0.0002 |
| Trial 4 | ||||||
| + | 13.4 ± 0.3 | 13 | 17 | 72 (75) | — | — |
| daf-18(nr2037) | 8.4 ± 0.2 | 10 | 11 | 77 (100) | <0.0001 | — |
| daf-2(m65); daf-18(0) | 17.6 ± 0.6 | 17 | 26 | 90 (100) | <0.0001 | <0.0001 |
| daf-2(m631); daf-18(0) | 14.9 ± 0.3 | 17 | 19 | 90 (100) | <0.0001 | <0.0001 |
| daf-2(m646); daf-18(0) | 13.6 ± 0.3 | 14 | 14 | 96 (100) | 0.0284 | <0.0001 |
| Trial 5 | ||||||
| + | 19.4 ± 0.4 | 18 | 28 | 99 (100) | — | — |
| daf-16(mgDf50) | 12.9 ± 0.1 | 12 | 16 | 100 (100) | <0.0001 | — |
| daf-16(0); daf-2(m65) | 12.3 ± 0.3 | 12 | 20 | 90 (100) | <0.0001 | 0.3922 |
| daf-16(0); daf-2(m646) | 12.2 ± 0.1 | 12 | 14 | 99 (100) | <0.0001 | <0.0001 |
| daf-16(0); daf-2(tm1236) | 12.4 ± 0.1 | 12 | 16 | 96 (100) | <0.0001 | 0.0041 |
| Trial 6 | ||||||
| + | 16.2 ± 0.2 | 16 | 20 | 98 (100) | — | — |
| daf-16(mgDf50) | 12.2 ± 0.1 | 12 | 16 | 99 (100) | <0.0001 | — |
| daf-16(0); daf-2(m65) | 11.1 ± 0.3 | 12 | 18 | 96 (100) | <0.0001 | 0.0552 |
| daf-16(0); daf-2(m646) | 12.2 ± 0.2 | 12 | 20 | 99 (100) | <0.0001 | 0.5758 |
| daf-16(0); daf-2(tm1236) | 12.6 ± 0.1 | 12 | 16 | 96 (100) | <0.0001 | 0.0138 |
FUdR was used for trials 2–4 to prevent death due to internal hatching of eggs, which affected some of the double-mutant strains. Maximum life span is the last day on which live worms were observed.
Deaths scored (initial sample size).
Probability that life span is identical to that of wild type with same temperature history (log rank test).
Probability that life span is identical to that of single Daf-d mutant (log rank test).
In trial 2, daf-2(m65); daf-18(0) is not longer lived than daf-18(0), in contrast to trials 3 and 4. While the reason for this discrepancy is unknown, we believe that the latter results are correct, given that daf-2(m631) and daf-2(m646) also increase daf-18(0) life span and that the severity ranking of the effects of these three daf-2 alleles is the same for both life span and larval arrest.
We examined the requirement for daf-16 gene function of the daf-2 life extension (Age) phenotype in three severe daf-2 alleles: m646, m65, and tm1236. In no case was the life span of daf-16(mgDf50); daf-2 animals consistently different from that of daf-16 alone (Table 3). Thus, the effect of daf-2 on aging is fully dependent on daf-16.
In humans, many mutations affecting the insulin receptor are semidominant (Taylor et al. 1992). This may reflect wild-type proreceptors combining with mutant proreceptors to form nonfunctional receptor dimers. By contrast, the life span of daf-2/+ hermaphrodites using the most severe daf-2 allele, m65, was not increased relative to wild-type controls (two trials at 22.5°; data not shown). Thus, in contrast to the mammalian insulin receptor, mutations affecting the DAF-2 receptor appear to be fully recessive.
Sequence analysis of nonconditional daf-2 alleles:
DNA sequencing was used to identify the mutant lesions in the nonconditional alleles. Three are nonsense alleles: m65 (W1449*, amber), m631 (R281*, opal), and m646 (Q1223*, ochre) (Figure 2B, Table 4). Although m65 is phenotypically the most severe allele, it is predicted to result in the least truncated protein (see discussion). W1449 is in the tyrosine kinase domain and the region following it is necessary for kinase function, so m65 is predicted to result in a truncated receptor without kinase activity. tm1236 contains an in-frame 561-bp deletion in exon 14, which is predicted to result in a Y1234S substitution and the loss of 187 amino acids, 1235–1421, from the kinase domain. This deletion removes the catalytic and activation loops of the kinase domain, but leaves intact the unusual C-terminal extension that DAF-2 possesses.
Figure 2.—
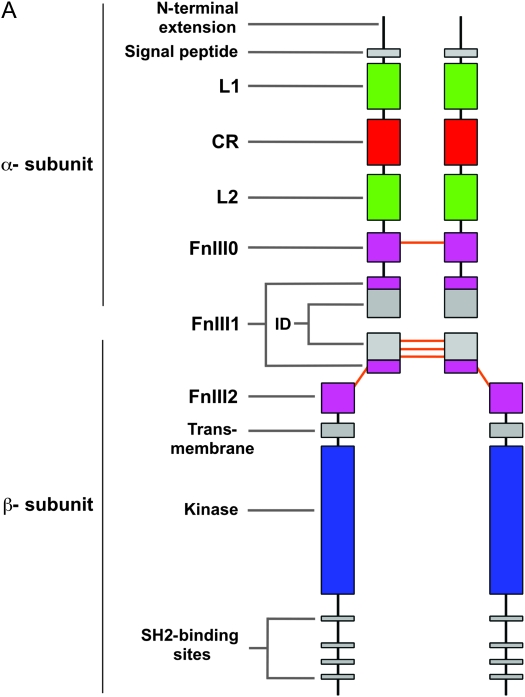
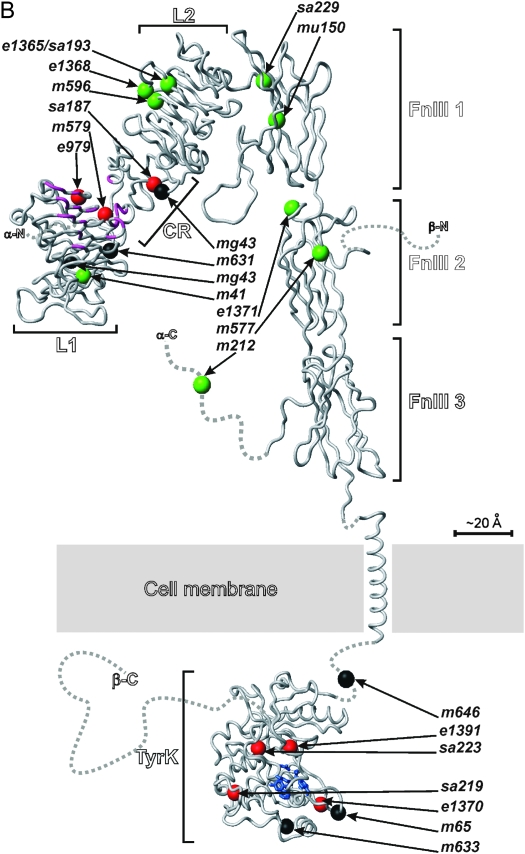
Position and nature of mutant lesions affecting the DAF-2 protein. (A) DAF-2 domain structure. The N-terminal extension and signal peptide are predicted to be cleaved off during processing of the proreceptor. The C-terminal extension is the region C-terminal of the kinase domain. The orange lines represent cysteine residues involved in interchain disulfide bonds. The α- and β-subunits of each heterodimer are linked by a predicted bond between C827, located in the insert domain of FnIII2, and C1126, located within the FnIII3 domain. The holoreceptor is formed by disulfide bonds between equivalent cysteines in adjacent α-subunits at positions C706 (FnIII1), C882, C883, and C886 (insert domain of FnIII2). (B) Backbone representation of homology models of the domains of DAF-2, showing the location of the mutations in the alleles studied. Green, class 1 mutations; red, class 2 mutations; black, nonconditional mutations. The regions of L1 and CR that are thought to interact with human IGF-1 and/or Ins are in purple (note the clustering of class 2 and nonconditional mutations in this region). The active site of the tyrosine kinase domain is in blue. Dashed lines represent regions that are intrinsically unfolded or without known structure. The model was constructed using the atomic coordinates of the extracellular domain of hInsR (PDB code: 2DTG) and the kinase domain of hInsR (PDB code: 1IRK).
TABLE 4.
Mutant lesions in daf-2 alleles
| Allele | Classa | Mutation location location (domain) | DNA change | Amino acid change | Reference | Predicted outcome of mutationb |
|---|---|---|---|---|---|---|
| e979 | 2D | L1 | TGC-TAC | C146Y | This study | a |
| m631 | NC | L1 | CGA-TGA | R281STOP | This study | b |
| m41 | 1B | Cys-Rich | GGA-GAA | G383E | Yuet al. (2002) | c |
| mg43c | NC | Cys-Rich | TGT-TAT | C401Y | Kimuraet al. (1998) | a |
| m579 | 2B | Cys-Rich | CGT-TGT | R437C | Scott et al. (2002) | d, h, i |
| sa187 | 2C | Cys-Rich | TGT-AGT | C469S | Kimuraet al. (1998) | a |
| mg43c | NC | Cys-Rich | CCC-CTC | P470L | Kimuraet al. (1998) | d |
| m596d | 1E | L2 | GGC-AGC | G547S | Scott et al. (2002) | e, h |
| e1368 | 1A | L2 | TCA-TTA | S573L | Kimuraet al. (1998) | e |
| e1365/sa193e,f | 1A | L2 | TGC-TAC | A580T | Kimuraet al. (1998); this study | f |
| sa229 | 1A | FnIII1 | GAT-AAT | D648N | Kimuraet al. (1998) | g |
| mu150 | 1A | FnIII1 | GGC-GAC | G682D | This study | f |
| e1371 | 1A | FnIII2α | GGA-GAA | G803E | This study | d |
| m212 | 1D | FnIII2ID | TGT-TAT | C883Y | This study | a, h |
| m577f | 1C | FnIII2β | TGC-TAC | C1045Y | This study | a, h |
| m646 | NC | Prekinase | CAA-TAA | Q1223STOP | This study; K. Kimura and G. Ruvkun (personal communication) | b |
| sa219 | 2C | Kinase | GAT-AAT | D1374N | Kimuraet al. (1998) | d |
| sa223 | 2E | Kinase | CGA-CAA | R1430Q | This study | d, h, i |
| e1391 | 2D | Kinase | CCC-CTC | P1434L | Kimuraet al. (1998) | d, i |
| m65 | NC | Kinase | TGG-TAG | W1449STOP | This study | b |
| e1370 | 2C | Kinase | CCA-TCA | P1465S | Kimuraet al. (1998) | d |
| m633 | NC | Kinase | CGT-CAT | R1510H | K. Kimura and G. Ruvkun (personal communication) | d |
| tm1236 | NC | Kinase | Deletion (see text) | This study |
Classes as defined in Gems et al. (1998). NC, nonconditional Daf-c.
a: Residue affected is a disulfide-bonded cysteine. Substitution is likely to disrupt fold stability. b: Nonsense mutation implying truncation of the polypeptide chain. Mutant protein is expected not to be functional. c: Residue affected is in a part of the sequence present only in nematodes. We were unable to predict the effect of substitution on the basis of homology to mammalian receptors. d: Residue affected is highly conserved in all phyla. Substitution is likely to affect stability of the structure or aspects of the receptor mechanism. e: Residue affected is solvent inaccessible and not highly conserved across phyla. The conservative nature of the substitution means that it is not predicted to have major detrimental effects. f: Residue affected is solvent inaccessible and not highly conserved across phyla. The nonconservative nature of the substitution suggests that it is likely to disrupt fold stability. g: Residue affected is solvent accessible and not highly conserved across phyla. Currently unable to predict effect of substitution. h: A nonidentical, naturally occurring, or engineered mutation in the equivalent hInsR position has been reported. i: An identical, naturally occurring, or engineered mutation in the equivalent hInsR position has been reported. For further details, see http://www.biochem.ucl.ac.uk/rilm.
The mg43 allele has two lesions, both affecting the CR domain.
Previously defined as class 2A (Gems et al. 1998). Given that this allele is suppressed by daf-12(m20), we have redesignated it as a class 1 allele.
The e1365 and sa193 alleles were isolated in different labs but contain identical sequence changes.
Sequence analysis revealed that the allele identified as e1365 in Gems et al. (1998) was in fact daf-2(m577). The real e1365 (Kimura et al. 1997) is a class 1A allele.
Analysis of conditional alleles of daf-2:
Next, we conducted an analysis and survey of conditional daf-2 alleles. Our overall aim was to build up a more detailed picture of the phenotypic range of daf-2 mutants and to understand it in terms of genotype and of changes in receptor structure and function. We first probed the difference in terms of DAF-16 function and gene expression between daf-2(m577), a class 1 allele, and daf-2(e1370), a class 2 allele.
Allele differences in effects on DAF-16 localization:
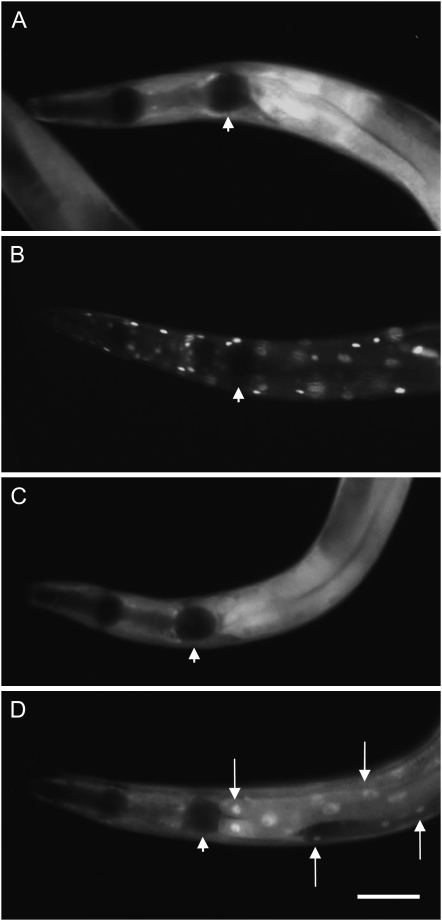
daf-2(e1370) results in loss of cytoplasmic retention of the DAF-16 transcription factor and its concentration in the nucleus (Henderson and Johnson 2001; Lee et al. 2001; Lin et al. 2001). We compared nuclear localization of GFP-tagged DAF-16 in daf-2(m577) and daf-2(e1370) mutant backgrounds. In daf-2(e1370) animals raised at 15° and shifted as L3 larvae to 25° for 4 hr, full nuclear localization was seen (Figure 3B). By contrast, under the same conditions, no nuclear localization was seen in daf-2(+) or daf-2(m577) animals (Figure 3, A and C). After 24 hr at 25°, and more so after 48 hr, a small degree of nuclear localization was visible in daf-2(m577) but not in daf-2(+) adults (not shown). In daf-2(e1370) adults raised and maintained at 15°, weak nuclear localization was observed (Figure 3D). This implies greater activation of DAF-16 in daf-2(e1370).
Figure 3.—
Effects of two daf-2 mutations on DAF-16 localization. (A) daf-2(+) at 25°. (B) daf-2(e1370) at 25°. (C) daf-2(m577) at 25°. (D) daf-2(e1370) at 15°. Anterior of animal to the right. Arrows indicate nuclear localization of DAF-16∷GFP. Arrowheads indicate position of the grinder of the pharynx. Bar, 50 μm.
We then asked whether this difference in DAF-16∷GFP localization is typical of class 1 vs. class 2 alleles by examining four more daf-2 alleles: m41 (weak class 1), m212 (severe class 1), m579 (weak class 2), and e979 (severe class 2). The pattern of DAF-16∷GFP localization was the same in m41 as m577 and in m579 as e1370 (data not shown). Unfortunately, in the case of m212 and e979, the presence of the daf-16∷gfp transgene caused nonconditional dauer formation. These results suggest that DAF-16 is typically more active in weak class 2 alleles than weak class 1 alleles. It is notable that DAF-16 showed little nuclear localization in m41 despite the fact that in terms of Daf-c it is a more severe allele than e1370.
daf-16-dependent differences in gene expression in daf-2(m577) and daf-2(e1370) mutants:
The different effects of daf-2 alleles on DAF-16 localization may differentially affect DAF-16-mediated gene expression, which could contribute to phenotypic differences among daf-2 mutants. To explore this, we used whole-genome oligonucleotide microarrays (Affymetrix) to compare DAF-16-dependent transcription in daf-2(m577) and daf-2(e1370). An analysis of summed data from these two alleles has been described previously (McElwee et al. 2004). Our expectations were that in daf-2(e1370) there would be more differentially expressed genes, on the basis of the greater nuclear localization of DAF-16, and also the additional mutant phenotypes of class 2 daf-2 mutants (Gems et al. 1998). Comparisons were made between daf-2 (long lived, DAF-16 ON) and daf-16(mgDf50); daf-2 (not long lived, DAF-16 OFF) for daf-2(m577) and daf-2(e1370) (five biological replicates per genotype). Reproduction was blocked using the glp-4(bn2ts) mutation, and age-synchronized young adults were studied (McElwee et al. 2004).
We identified genes (represented by probe sets on the oligonucleotide arrays) where transcript abundance is significantly different between daf-2 and daf-16; daf-2 (q < 0.1). There were 1252 and 581 such probe sets for daf-2(m577) and daf-2(e1370), respectively, including 356 probe sets in common. The smaller number of genes with altered expression in daf-2(e1370) was unexpected, but is likely to reflect greater variance observed in gene expression in daf-2(e1370) strains. The average standard deviation of probe sets for the five replicates of each genotype, for a typical data normalization, were as follows: daf-2(m577), 0.59; daf-16; daf-2(m577), 0.50; daf-2(e1370), 0.75; and daf-16; daf-2(e1370), 0.78.
A total of 525 genes showed a significant change in expression in daf-2 relative to daf-16; daf-2 for m577 (q < 0.1) but not for e1370 (q > 0.5) (data not shown). Although some of these genes may actually show m577-specific regulation, for others this may be an artifact of the difference in variance, described above. More interesting, therefore, were the 56 genes showing altered expression in daf-2 relative to daf-16; daf-2 for e1370 but not for m577 (listed in Table 5). Potentially, these genes contribute to the more pleiotropic character of e1370 and to its epistatic interactions (Table 1).
TABLE 5.
Genes showing differential expression between daf-2 and daf-16; daf-2 in e1370 but not in m577
| daf-2 vs daf-16; daf-2 mean log2 fold-change | q | Gene sequence name | Protein |
|---|---|---|---|
| Upregulated in daf-2 | |||
| 6.13 | 0.000961 | Y39H10A.1 | Similarity to Bradyrhizobium sp. Hypothetical protein TR:Q35RI1 |
| 5.318 | 0.00204 | F08H9.4 | IPR002068 heat-shock protein Hsp20 |
| 3.707 | 0.0101 | F08H9.3 | IPR002068 heat-shock protein Hsp20 |
| 3.754 | 0.00795 | gst-41 | IPR004045 glutathione S-transferase, N-terminal |
| 3.572 | 0.0276 | T20D4.12 | IPR002542 protein of unknown function DUF19 |
| 3.48 | 0.0209 | F20A1.6 | Similarity to Dictyostelium discoideum TR:Q55ED2 |
| 3.417 | 0.00252 | cyp-13B1 | IPR001128 cytochrome P450 |
| 3.22 | 0.0233 | C45G7.3 | Similarity to Interpro domain IPR008597 (Destabilase) |
| 3.07 | 0.0732 | F46F5.15 | Similarity to Saccharomyces cerevisiae GAP; negatively regulates RAS |
| 3.03 | 0.0414 | tyr-3 | Predicted tyrosinase |
| 2.982 | 0.00838 | T20G5.8 | IPR003582 metridin-like ShK toxin |
| 2.908 | 0.0514 | C34B2.4 | IPR001781 LIM, zinc binding |
| 2.852 | 0.0126 | ins-22 | IPR004825 insulin/IGF/relaxin |
| 2.823 | 0.0749 | F35E8.6 | IPR003582 metridin-like ShK toxin |
| 2.81 | 0.0734 | F09E10.10 | Probable noncoding RNA |
| 2.77 | 0.0621 | W08A12.4 | Similarity to Oryctolagus cuniculus trichohyalin; SW:P37709 |
| 2.73 | 0.0317 | F36D1.7 | Similarity to Clostridium cellulovorans hydrophobic protein A |
| 2.71 | 0.0480 | C08A9.3 | Similarity to Mus musculus SID1; SW:Q8CIF6 |
| 2.645 | 0.0611 | F49E11.6 | IPR001283 allergen V5/Tpx-1 related |
| 2.615 | 0.0321 | M162.5 | IPR007114 major facilitator superfamily |
| 2.54 | 0.0180 | T23F2.4 | Similarity to Pfam domain PF01679 |
| 2.38 | 0.0248 | F19F10.3 | Similarity to Burkholderia thailandensis TR:Q2SYY4 |
| 2.284 | 0.0352 | Y38E10A.11 | IPR009853 protein of unknown function DUF1412 |
| 2.18 | 0.0249 | C50D2.6 | Similarity to D. melanogaster CG9896-PA |
| 1.97 | 0.0850 | B0563.5 | Similarity to Desulfovibrio desulfuricans metal-dependent phosphohydrolase |
| 1.93 | 0.0433 | Y43C5A.3 | Similarity to Ixodes scapularis putative secreted salivary gland peptide |
| 1.76 | 0.0669 | F20A1.10 | Similarity to H. hepaticus Seryl-tRNA synthetase |
| 1.703 | 0.0731 | nhr-206 | IPR000324 vitamin D receptor |
| Downregulated in daf-2 | |||
| −7.084 | 0.000238 | F22A3.6 | IPR008597 destabilase |
| −3.542 | 0.0323 | C52E2.5 | IPR001810 cyclin-like F-box |
| −3.527 | 0.00422 | T16G12.1 | IPR001930 peptidase M1, membrane alanine aminopeptidase |
| −3.494 | 0.0216 | R03G8.6 | IPR001930 peptidase M1, membrane alanine aminopeptidase |
| −3.128 | 0.00234 | C17H12.8 | IPR003366 protein of unknown function DUF141 |
| −2.826 | 0.0272 | F52F10.4 | IPR002656 acyltransferase 3 |
| −2.755 | 0.00648 | R09H10.5 | IPR006582 MD |
| −2.653 | 0.00878 | T01D3.6 | IPR001846 von Willebrand factor, type D |
| −2.553 | 0.0176 | amt-4 | IPR010256 Rh-like protein/ammonium transporter |
| −2.496 | 0.0277 | F28B4.3 | IPR006209 EGF-like |
| −2.349 | 0.0173 | C41A3.1 | IPR008262 lipase, active site |
| −2.307 | 0.0188 | K11H12.4 | IPR005071 protein of unknown function DUF274 |
| −2.297 | 0.025 | T25B6.2 | IPR002052 N-6 adenine-specific DNA methylase |
| −2.18 | 0.0252 | K09C4.1 | IPR007114 major facilitator superfamily |
| −2.055 | 0.0831 | R193.2 | IPR002035 von Willebrand factor, type A |
| −2.042 | 0.0354 | glc-1 | IPR006201 neurotransmitter-gated ion channel |
| −1.985 | 0.0415 | C12D8.5 | IPR005806 Rieske [2Fe-2S] region |
| −1.932 | 0.0323 | fat-7 | IPR005804 fatty acid desaturase |
| −1.910 | 0.0372 | C29F7.2 | IPR004119 protein of unknown function DUF227 |
| −1.889 | 0.0693 | ugt-43 | IPR002213 UDP-glucuronosyl/UDP-glucosyltransferase |
| −1.870 | 0.0541 | K08D8.6 | IPR003366 protein of unknown function DUF141 |
| −1.83 | 0.0611 | C11E4.7 | Similarity to Trichodesmium erythraeum aminotransferase, class I and II |
| −1.809 | 0.0547 | ckb-2 | IPR002573 choline/ethanolamine kinase |
| −1.74 | 0.00950 | F08G5.6 | IPR003366 protein of unknown function DUF141 |
| −1.66 | 0.0695 | C18H9.6 | Similarity to Geobacter metallireducens replication initiation factor |
| −1.54 | 0.0482 | F15E11.12 | Similarity to Wolinella succinogenes hypothetical protein, TR:Q7MR69 |
| −1.393 | 0.0947 | ugt-22 | IPR002213 UDP-glucuronosyl/UDP-glucosyltransferase |
| −1.117 | 0.0182 | asp-1 | IPR001461 peptidase A1, pepsin |
Of the 56 genes listed here, half are upregulated and half downregulated. Among the most strongly upregulated genes are F08H9.3 and F08H9.4, which are neighboring genes regulated by distinct promoters and which encode small heat-shock proteins (smHSPs) of the HSP-16 type. Several lines of evidence implicate smHSPs in longevity assurance in C. elegans (Walker et al. 2001; Hsu et al. 2003; Walker and Lithgow 2003). Both F08H9.3 and F08H9.4 are constitutively expressed, the former in the pharynx and the latter in the excretory canal and ventral nerve-cord neurons (Shim et al. 2003). Also of note is the upregulation of ins-22, a neuronally expressed type α insulin-like peptide and potential ligand for DAF-2 (Pierce et al. 2001), and nhr-206, an orphan nuclear hormone receptor.
A comparison of gene expression in daf-16(0); daf-2(m577) and daf-16(0); daf-2(e1370) strains revealed no genes showing a significant difference in expression level (q < 0.1). Thus, all differences in gene expression between these two alleles appear to be daf-16 dependent.
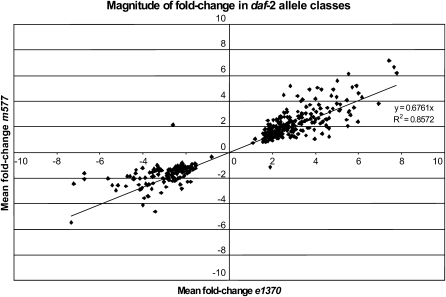
Next we performed a global comparison between m577 and e1370 of the magnitude of mean fold-change of genes that showed significant (q < 0.1) differences in expression between daf-2 and daf-16; daf-2 (Figure 4). There is a clear trend toward a greater magnitude of transcriptional change in daf-2(e1370) than in daf-2(m577). Taken together, these studies of DAF-16 localization and transcript abundance support the view that in daf-2(e1370) there is a lower level of insulin/IGF-1 signaling, leading to more nuclear localization of DAF-16 and greater DAF-16-dependent changes in gene expression.
Figure 4.—
Magnitude of mean fold-change of genes (in two alleles of daf-2), which showed significant (q < 0.1) differences in expression between daf-2 and daf-16; daf-2. (356 probe sets were expressed significantly differently in both comparisons). There is a strong trend toward a greater magnitude of transcriptional change in daf-2(e1370) than in daf-2(m577).
Sequencing of eight more daf-2 alleles:
To gain further insight into the basis of daf-2 allele differences, we sequenced eight more daf-2 alleles that have been studied previously (Gems et al. 1998; Garigan et al. 2002; Murphy et al. 2003; Nanji et al. 2005): e979, sa193, mu150, e1371, m212, m577, sa223, and e1369. Apart from e1369, the lesions were identified for all alleles (Figure 2B, Table 4). e1369 is an unusual allele, exhibiting a severe ts Daf-c phenotype, yet suppressed by daf-12. Although we verified that e1369 fails to complement daf-2(m577) and maps very close to the daf-2 locus at −9.61 ± 0.27, we found no mutation in the coding sequence in the 5.4-kb first intron or in the first 5.2 kb upstream of the putative translational start. Possibly the e1369 mutation lies in a distant cis regulatory region; however, using RT–PCR we saw no change in daf-2 mRNA levels in this mutant (data not shown).
We applied several data-mining approaches to gain insight into the possible mode of action of 24 daf-2 alleles (Figure 2B, Table 4). The alleles include the 11 sequenced in this study, plus 13 daf-2 alleles sequenced previously (Kimura et al. 1997; Yu and Larsen 2001; Scott et al. 2002). First, we looked at the predicted effects of each mutation on homology models of the individual DAF-2 domains. Second, we surveyed reports on mutations in other insulin-like receptors for equivalent or similar changes to those in daf-2 alleles for any phenotypic, biochemical, and/or structural information available. The resulting information was collated into an online web resource, Receptors for Insulin and Insulin-Like Molecules (RILM) (http://www.biochem.ucl.ac.uk/rilm/). Details of the construction and utilities of this resource have been reported elsewhere (Garza-Garcia et al. 2007).
Clustering of phenotypically defined classes of daf-2 alleles:
A survey of all sequenced daf-2 alleles reveals that the site of the lesion in the receptor is a predictor of its phenotypic consequences. Class 1 mutations occur in the CR, L2, and fibronectin type III (FnIII) domains of the extracellular part of the receptor (Figure 2B). By contrast, class 2 and nonconditional alleles cluster around the ligand-binding cleft in the L1 and CR domains or in the tyrosine kinase domain. Thus, the phenotype-based class 1/class 2 distinction corresponds to classes of defect in the DAF-2 receptor protein. In the following sections, we survey daf-2 mutations in more detail, emphasizing the more notable alleles.
Mutations in the receptor L domains and CR region:
e979 is the only conditional allele with a lesion in the L1 domain. This allele is also unique in that at 25.5° it causes 100% embryonic lethality and L1 arrest (Vowels and Thomas 1992; Gems et al. 1998). e979 results in a C146Y substitution. From the structure of the human receptors, this amino acid residue is predicted to be involved in an intradomain disulfide bond with C181. Given that it affects a structurally important residue, e979 is likely to destabilize the protein fold. Possibly, the ts early arrest resulting from this mutation reflects resulting thermolability of the mutant receptor. In hInsR, C8 (equivalent to DAF-2 C146) is also very close to a ligand-binding motif (Whittaker and Whittaker 2005), which is conserved in DAF-2. Thus, e979 may affect binding of ligand to receptor. Four daf-2 alleles have mutations in the L2 domain: e1365, e1368, m596, and sa193. e1365 and sa193 have identical sequence changes (Figure 2, Table 4).
Four alleles have mutations in the CR region: m41, mg43, m579, and sa187. m41 is a distinctive allele in that it is suppressed by daf-12 yet relatively severe in terms of Daf-c, and Daf-c shows some maternal rescue (Gems et al. 1998). This allele has a G383E substitution (Yu and Larsen 2001) (reconfirmed by us). G383 is unusual in that it is conserved in all four nematode DAF-2 proteins examined, but absent from most other lineages. m41 is the only mutation in the CR region that is not in the ligand-binding cleft (Figure 2B).
Mutations in the FnIII domains:
Five daf-2 alleles are located in the FnIII region: e1371, m212, m577, mu150, and sa229. m577 has a Cys-to-Tyr mutation that disrupts an intradomain disulfide bond in the β-subunit region of the FnIII2 domain and likely destabilizes the polypeptide fold. The equivalent positions have been experimentally substituted to Ser in both hInsR and hIGF-1R, and this disulfide bond was found to be essential for correct processing and transport to the plasma membrane when the mutant receptor was expressed in COS1 cells (Maggi and Cordera 2001). mu150 has a Gly-to-Asp substitution in the third β-strand of the FnIII1 domain. Although this Gly is not fully conserved, it is usually a small residue (Gly or Ser) in other insulin/IGF-1 receptors; possibly the bulkier side chain of Asp or its negative charge are not well tolerated in the context of neighboring structural elements.
The m212 allele affects the α-subunit part of the insert within FnIII2. m212 is a distinctive allele in that it has a strong temperature-sensitive Daf-c phenotype, yet is suppressed by daf-12(m20). m212 changes Cys 883 to Tyr. C883 is one of the three Cys residues (C882, C883, and C886 in DAF-2) that in hInsR form intermolecular disulfide bonds with the opposing α-chain (Cheatham and Kahn 1992; Schaffer and Ljungqvist 1992; Lu and Guidotti 1996; Sparrow et al. 1997; Wu and Guidotti 2002). The loss of this disulfide bridge may affect the quaternary structure of the receptor, perhaps accounting for the unusual properties of this allele.
Mutations in the tyrosine kinase domain:
There are five daf-2 alleles with missense mutations in the kinase domain: sa219, sa223, e1391, e1370, and m633 (Figure 1B). sa223, an unusual nonconditional allele (see discussion), changes the fully conserved Arg 1430 to Gln. The equivalent residue in hInsR is part of the loop that determines kinase substrate selectivity (Hubbard et al. 1994). In e1370, the fully conserved Pro 1465 is replaced with a Ser. The backbone carbonyl oxygen of Pro 1465 is expected to form a hydrogen bond with the side chain of the Arg that is mutated in sa223 and this mutation is therefore also predicted to have an effect on substrate binding. e1391, a severe class 2 allele, has a mutation only four residues away from sa223 (Kimura et al. 1997).
DISCUSSION
We have conducted a detailed analysis and survey of 24 daf-2 mutations. This includes phenotypic characterization of nonconditional daf-2 alleles (including epistasis analysis), a study of allele differences in downstream effectors, sequencing of 11 new daf-2 alleles, and modeling of the DAF-2 receptor and the likely mutational effects on its structure.
m65 is a probable daf-2 null mutation:
Three nonconditional alleles of daf-2 contain nonsense mutations: m65 (UAG, amber), m631 (UGA, opal), and m646 (UAA, ochre). While m631 and m646 are hypomorphic, m65 is a potential null allele, despite being the most 3′ nonsense mutation. This may reflect the fact that m65 affects a critical Trp residue in the active site of the receptor tyrosine kinase such that, if translational readthrough did occur (see below), the resulting receptor would have no tyrosine kinase activity.
Another possibility is that the truncated DAF-2 protein resulting from m65 has a poisonous effect, such that this allele is antimorphic. However, if this were the case, one might expect m65 to behave in a dominant-negative fashion in effects either on dauer formation (Gems et al. 1998) or on life span (this study), and it does not do so. The absence of any dominant effects on life span is more significant, since life span is more sensitive to the effects of reduced insulin/IGF-1 signaling than dauer formation. That m65 is nullimorphic rather than antimorphic was also implied by an early test using the deficiency mDf11, which removes the daf-2 gene region. There, daf-2(e1370)/mDf11 and daf-2(e1370)/daf-2(m65) animals were shown to exhibit a similar level of dauer formation (Gems et al. 1998). However, the only way to be certain of the character of the daf-2(0) phenotype will be to delete this gene in its entirety.
A further, slight possibility is that in the m65 strain there is a second-site mutation in a gene closely linked to daf-2, which enhances early larval lethality in the daf-16; daf-2 mutant. We explored this possibility by attempting to rescue the effects of m65 using a pdpy-30∷daf-2 transgene, which has previously been shown to rescue daf-2(e1370) (Wolkow et al. 2000). However, the transgene did not rescue the m65 Daf-c phenotype, probably reflecting non-native daf-2 expression from this transgene (data not shown).
The daf-2(tm1236) in-frame deletion removes the kinase domain and is a less severe allele than m65. This implies that, despite the absence of the kinase domain, some residual receptor function exists in tm1236 mutants. One possibility is that the DAF-2 C-terminal extension, present in tm1236 but not in m65, binds to and activates downstream effectors at a low level even in the absence of DAF-2 kinase activity.
Incomplete suppression of severe daf-2 alleles by daf-18 and daf-16:
Using severe daf-2 alleles, including the likely null allele m65, we tested whether daf-2 mutant phenotypes are fully dependent on daf-16 and daf-18, the first time that epistatic interactions between these genes have been tested using all null alleles. The increased longevity of daf-2 mutants was fully dependent on daf-16 but not on daf-18. The latter observation could mean that in the absence of DAF-2, PIP3 is produced at a very low level, such that removal of its sink, PTEN, can only weakly restore PIP3 levels. Alternatively, it could reflect a PIP3-independent mechanism of life extension by daf-2.
The embryonic arrest and the various morphological and behavioral abnormalities in daf-2; daf-18 and daf-16; daf-2 mutants imply action of insulin/IGF-1 signaling in early development that warrants further investigation. These defects could reflect an early requirement for DAF-2 that is detectable only in daf-18 or daf-16 mutant backgrounds. By this view, severe loss of daf-2 results in Emb and Daf-c, and only the latter may be suppressed by daf-18 and daf-16. Moreover, daf-2; daf-18 animals show some degree of dauer larva differentiation at 15°, suggesting that the daf-18 suppressor mutation is slightly temperature sensitive.
Possible readthrough in opal and ochre but not amber nonsense mutations:
Nonsense mutations are predicted to cause premature truncation of protein, which often results in a complete loss of gene function. However, some hypomorphic nonsense mutants have been reported. This can occur because the mutation is near the 3′-end of the gene, such that the truncated protein retains some activity. However, in some instances, nonsense mutations near the 5′-end of the gene are hypomorphic. This may reflect the occurrence of some translational readthrough of the nonsense codon, producing small amounts of full-length protein. This happens in opal (UGA) mutants since C. elegans, like many animals, has tRNA[Ser]Sec, which can insert selenocysteine at UGA codons (Lee et al. 1990). Moreover, in Escherichia coli, third-position wobble in codon–anticodon recognition results in substitution of Trp at opal stop codons; perhaps this happens in C. elegans, too.
Such readthrough does not seem to happen in amber (UAG) mutants. For example, here daf-2(m631) (opal) is hypomorphic and daf-2(m65) (amber) is potentially null. Likewise, daf-1(m40) (opal) is hypomorphic, while four daf-1 amber mutants all appear to be nulls (Gunther et al. 2000). Similarly, lin-1(e1275) is a hypomorphic opal (Beitel et al. 1995), and the hypomorphic lon-1(sp3) opal produces a full-length protein product (Morita et al. 2002). By contrast, hypomorphic amber alleles are usually found near the 3′-end of genes, e.g., in lin-1(e1777) (Beitel et al. 1995; Tuck and Greenwald 1995). Taken together, these findings suggest that, in C. elegans, translational readthrough occurs in opal but not in amber mutants. That the ochre mutation daf-2(m646) is also weaker than daf-2(m65) suggests that readthrough of ochre alleles may also occur, although we know of no mechanism by which this might happen.
Molecular basis of differences among nonconditional daf-2 mutants:
daf-2 mutations may be broadly grouped into class 1 (suppressed by daf-12, often weaker) and class 2 (not suppressed by daf-12, often stronger and more pleiotropic) (Gems et al. 1998). Comparison of the effects of mutations in the daf-2 gene on protein structure and phenotype allow some broad conclusions to be drawn about the molecular basis of these allele differences and suggest several new hypotheses.
First, apart from e1369 (which seems to lie beyond the daf-2 open reading frame), all class 1 mutations map to the CR, L2, and FnIII domains of the extracellular part of the receptor. Structural modeling implies that these mutations do not directly affect ligand contact residues, but instead cause subtle changes in the conformation of the extracellular domain. Studies in mammalian cells have shown that this can lead to mis-processing of the proreceptor and to a reduced number of functional receptors at the cell surface. Interestingly, mutations affecting the same residues as the class 1 alleles m577 and m596 have been studied in the human insulin receptor and have this effect (see RILM at http://www.biochem.ucl.ac.uk/rilm/) (Garza-Garcia et al. 2007). Possibly, reduced DAF-2 protein levels typify class 1 daf-2 mutants.
Second, class 2 and nonconditional alleles either map to the tyrosine kinase domain or cluster around the predicted ligand-binding cleft formed by parts of the L1 and CR domains (Figure 2B). It has been suggested that the properties of class 1 alleles (e.g., e1368) might reflect effects on ligand binding (Hsin and Kenyon 1999). However, the distribution of lesions in DAF-2 (Figure 2B) argues against this interpretation. Third, none of the 24 mutations lie in the N-terminal and C-terminal extension regions of DAF-2, hinting (although certainly not proving) that these play a relatively minor role in DAF-2 function. Fourth, each of the atypical class 1 alleles (i.e., those that are more severe in terms of Daf-c than many class 2 alleles) is idiosyncratic in terms of its molecular lesion. For example, the m41 lesion lies in a structurally obscure region of DAF-2 present only in nematodes; m212 may affect function at the level of quaternary structure; and the most atypical class 1 allele, e1369, appears to lie outside of the daf-2 open reading frame.
The results described here provide a more nuanced description of daf-2 allelic variation than the previously defined class 1/class 2 allele distinction, as follows. First, overall severity defines a basic allelic series, to which many alleles conform. For example, differences in effects of m577 (class 1) and e1370 (class 2) on signaling effectors reveal only that e1370 is more a severe allele. Second, the class 1/class 2 distinction is necessary, given the unusual alleles that are severely Daf-c yet not suppressed by daf-12: atypical class 1 alleles such as e1369, m41, and m212.
Potential complex effects on signaling in some class 2 daf-2 alleles:
Our analysis also suggests a new hypothesis relating to class 2 daf-2 mutants. Signaling into the cell from insulin/IGF-1 receptors involves several pathways (Nanji et al. 2005). Potentially, some aspects of daf-2 allele differences reflect differential effects of mutation on signaling via the PIP3 and other pathways, e.g., let-60/Ras (Nanji et al. 2005). For example, daf-2(sa223) is an unusual allele, which results in nonconditional pre-dauer (L2d) arrest that can be maternally rescued (Malone and Thomas 1994; Gems et al. 1998). daf-2(sa223) homozygotes derived from daf-2(sa223)/+ mothers form dauer larvae at 25°, but few or none at lower temperatures. sa223 results in R1430Q, and the equivalent mutation, R1174Q, has been found in hInsR in several patients with type A insulin resistance (Moller et al. 1994; Moritz et al. 1994) (in each case, the individual was heterozygous for the mutation). The effects of this mutation in the homozygous state were characterized by expression in CHO cells and found to cause impairment of insulin-stimulated receptor tyrosine phosphorylation, glycogen synthesis, and mitogenesis (Krook et al. 1996). Interestingly, however, insulin-stimulated IRS-1 phosphorylation and recruitment of PI 3-kinase was not fully blocked and at high concentrations of insulin it approached wild-type levels (Krook et al. 1996, 1997). However, there was an absence of insulin-stimulated tyrosine phosphorylation of Src homologous and collagen-like (Shc), GTP loading of Ras, and MAP kinase activation. Shc is an adaptor protein that binds to various activated receptors, usually at the PTB domain. daf-2(e1391) is equivalent to the human IR mutation P1178L (Kim et al. 1992; Krook et al. 1994; Kimura et al. 1997), which has effects on receptor function similar to the sa223-equivalent R1174Q (Krook et al. 1996, 1997).
A third allele, daf-2(m579), results in R437C, equivalent to hInsR R252C, which is found in the homozygous state in subjects with type A insulin resistance (Hamer et al. 2002). It is therefore a less severe defect than the human equivalents of sa223 and e1391, which cause severe insulin resistance in the heterozygous state (Kim et al. 1992; Krook et al. 1994; Moller et al. 1994). However, it too results in severe reduction of signaling through Shc, but has little effect on activation of PI 3-kinase (Hamer et al. 2002). In this case, the authors suggest that the PI 3-kinase pathway activation may be due to demonstrably reduced endocytosis of the ligand-bound receptor, resulting in increased perdurance of the activated receptor signal. Enhancement of receptor signaling has also been observed in the EGF receptor upon inhibition of clathrin-mediated endocytosis (Vieira et al. 1996). Consistent with this, mutation of unc-101, which encodes a homolog of the AP47 clathrin-associated protein, suppresses hypomorphic alleles of the let-23 EGF receptor gene (Lee et al. 1994). Such a combination of overall reduction of insulin/IGF-1 receptor function, combined with selective rescue of PI 3-kinase signaling, may be a common characteristic among class 2 daf-2 mutants.
Conclusions:
Our study suggests the following overview of daf-2 allele variation: daf-2 mutations form a very approximate allelic series in which weaker alleles are often class 1, and more severe alleles are often class 2. A number of class 1 alleles are atypical and do not conform to this series, being unusually severe in terms of Daf-c. Mutations in class 1 alleles occur only in some regions of the receptor ectodomain while class 2 alleles are clustered in the ligand-binding cleft and the kinase domain. The mutations in atypical class 1 alleles each have idiosyncratic effects, whose outcome remains to be established. In class 2 alleles, signaling pathways other than PIP3-dependent signaling may be disproportionately impaired, as suggested by the properties of similar mutations in humans. The most severe loss of function in daf-2 results in recessive early embryonic lethality and effects on life span that are fully dependent on daf-16 but not on daf-18.
Acknowledgments
We thank Sindhuja Kadambi and Diana McCulloch for help with sequencing; Adam Antebi and Naoto Ueno for useful discussion; Cynthia Kenyon, Don Riddle, and Jim Thomas for providing daf-2 alleles; Kotaro Kimura and Gary Ruvkun for sharing information; and Ken Kemphues for a helpful critique of the manuscript. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources. This work was supported by the Biotechnology and Biological Sciences Research Council (UK), the European Union (Framework V), and the Wellcome Trust.
References
- Antebi, A., W. Yeh, D. Tait, E. Hedgecock and D. Riddle, 2000. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 14 1512–1527. [PMC free article] [PubMed] [Google Scholar]
- Avery, L., and S. Wasserman, 1992. Ordering gene function: the interpretation of epistasis in regulatory hierarchies. Trends Genet. 8 312–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyadevara, S., R. Alla, J. J. Thaden and R. J. Shmookler Reis, 2007. Remarkable longevity and stress resistance of nematode PI3K-null mutants. Aging Cell 7 13–22. [DOI] [PubMed] [Google Scholar]
- Beitel, G., S. Tuck, I. Greenwald and H. Horvitz, 1995. The Caenorhabditis elegans gene lin-1 encodes an ETS-domain protein and defines a branch of the vulval induction pathway. Genes Dev. 9 3149–3162. [DOI] [PubMed] [Google Scholar]
- Bluher, M., B. Kahn and C. Kahn, 2003. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 299 572–574. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassada, R. C., and R. L. Russell, 1975. The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev. Biol. 46 326–342. [DOI] [PubMed] [Google Scholar]
- Cheatham, B., and C. R. Kahn, 1992. Cysteine 647 in the insulin receptor is required for normal covalent interaction between alpha- and beta-subunits and signal transduction. J. Biol. Chem. 267 7108–7115. [PubMed] [Google Scholar]
- Choe, S. E., M. Boutros, A. M. Michelson, G. M. Church and M. S. Halfon, 2005. Preferred analysis methods for Affymetrix GeneChips revealed by a wholly defined control dataset. Genome Biol. 6 R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy, D., D. Gems, L. G. Harshman, S. Oldham, E. Hafen et al., 2001. Extension of lifespan by loss of chico, a Drosophila insulin receptor substrate protein. Science 292 104–106. [DOI] [PubMed] [Google Scholar]
- Friedman, D. B., and T. E. Johnson, 1988. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics 118 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi, S., J. Santelli, D. G. Mitchell, J. W. Stiles and D. Raosanadi, 1980. A simple method for maintaining large, aging populations of Caenorhabditis elegans. Mech. Ageing Dev. 12 137–150. [DOI] [PubMed] [Google Scholar]
- Garigan, D., A. Hsu, A. Fraser, R. Kamath, J. Ahringer et al., 2002. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics 161 1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Garcia, A., D. S. Patel, D. Gems and P. C. Driscoll, 2007. RILM: a web-based resource to aid comparative and functional analysis of the insulin and IGF-1 receptor family. Hum. Mutat. 28 660–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems, D., and D. L. Riddle, 2000. Defining wild-type life span in Caenorhabditis elegans. J. Gerontol. A Biol. Sci. Med. Sci. 55 B215–B219. [DOI] [PubMed] [Google Scholar]
- Gems, D., A. J. Sutton, M. L. Sundermeyer, P. L. Larson, P. S. Albert et al., 1998. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics 150 129–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch, B., C. Weitzel, C. Kober-Eisermann, V. Rottiers and A. Antebi, 2001. A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev. Cell 1 841–851. [DOI] [PubMed] [Google Scholar]
- Gil, E., E. Malone Link, L. Liu, C. Johnson and J. Lees, 1999. Regulation of the insulin-like developmental pathway of Caenorhabditis elegans by a homolog of the PTEN tumor suppressor gene. Proc. Natl. Acad. Sci. USA 96 2925–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb, S., and G. Ruvkun, 1994. daf-2, daf-16 and daf-23: genetically interacting genes controlling dauer formation in Caenorhabditis elegans. Genetics 137 107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther, C., L. Georgi and D. Riddle, 2000. A Caenorhabditis elegans type I TGF beta receptor can function in the absence of type II kinase to promote larval development. Development 127 3337–3347. [DOI] [PubMed] [Google Scholar]
- Hamer, I., M. Foti, R. Emkey, M. Cordier-Bussat, J. Philippe et al., 2002. An arginine to cysteine(252) mutation in insulin receptors from a patient with severe insulin resistance inhibits receptor internalisation but preserves signalling events. Diabetologia 45 657–667. [DOI] [PubMed] [Google Scholar]
- Henderson, S. T., and T. E. Johnson, 2001. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr. Biol. 11 1975–1980. [DOI] [PubMed] [Google Scholar]
- Hertweck, M., C. Gobel and R. Baumeister, 2004. C. elegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and life span. Dev. Cell 6 577–588. [DOI] [PubMed] [Google Scholar]
- Holzenberger, M., J. Dupont, B. Ducos, P. Leneuve, A. Geloen et al., 2003. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature 421 182–187. [DOI] [PubMed] [Google Scholar]
- Hopper, N. A., 2006. The adaptor protein soc-1/Gab1 modifies growth factor receptor output in Caenorhabditis elegans. Genetics 173 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin, H., and C. Kenyon, 1999. Signals from the reproductive system regulate the lifespan of C. elegans. Nature 399 362–366. [DOI] [PubMed] [Google Scholar]
- Hsu, A., C. Murphy and C. Kenyon, 2003. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300 1142–1145. [DOI] [PubMed] [Google Scholar]
- Huang, L. S., and P. W. Sternberg, 1995. Genetic dissection of developmental pathways, pp. 98–122 in Caenorhabditis elegans: Modern Biological Analysis of an Organism, edited by H. F. Epstein and D. C. Shakes. Academic Press, San Diego.
- Hubbard, S., L. Wei, L. Ellis and W. Hendrickson, 1994. Crystal structure of the tyrosine kinase domain of the human insulin receptor. Nature 372 746–754. [DOI] [PubMed] [Google Scholar]
- Kenyon, C., J. Chang, E. Gensch, A. Rudener and R. Tabtiang, 1993. A C. elegans mutant that lives twice as long as wild type. Nature 366 461–464. [DOI] [PubMed] [Google Scholar]
- Kim, H., H. Kadowaki, H. Sakura, M. Odawara, K. Momomura et al., 1992. Detection of mutations in the insulin receptor gene in patients with insulin resistance by analysis of single-stranded conformational polymorphisms. Diabetologia 35 261–266. [DOI] [PubMed] [Google Scholar]
- Kimura, K. D., H. A. Tissenbaum, Y. Liu and G. Ruvkun, 1997. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277 942–946. [DOI] [PubMed] [Google Scholar]
- Krook, A., S. Kumar, I. Laing, A. J. Boulton, J. A. Wass et al., 1994. Molecular scanning of the insulin receptor gene in syndromes of insulin resistance. Diabetes 43 357–368. [DOI] [PubMed] [Google Scholar]
- Krook, A., D. E. Moller, K. Dib and S. O'Rahilly, 1996. Two naturally occurring mutant insulin receptors phosphorylate insulin receptor substrate-1 (IRS-1) but fail to mediate the biological effects of insulin. Evidence that IRS-1 phosphorylation is not sufficient for normal insulin action. J. Biol. Chem. 271 7134–7140. [DOI] [PubMed] [Google Scholar]
- Krook, A., J. P. Whitehead, S. P. Dobson, M. R. Griffiths, M. Ouwens et al., 1997. Two naturally occurring insulin receptor tyrosine kinase domain mutants provide evidence that phosphoinositide 3-kinase activation alone is not sufficient for the mediation of insulin's metabolic and mitogenic effects. J. Biol. Chem. 272 30208–30214. [DOI] [PubMed] [Google Scholar]
- Labbe, J., J. Burgess, L. Rokeach and S. Hekimi, 2000. ROP-1, an RNA quality-control pathway component, affects Caenorhabditis elegans dauer formation. Proc. Natl. Acad. Sci. USA 97 13233–13238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen, P. L., P. S. Albert and D. L. Riddle, 1995. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics 139 1567–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B., M. Rajagopalan, Y. Kim, K. You, K. Jacobson et al., 1990. Selenocysteine tRNA[Ser]Sec gene is ubiquitous within the animal kingdom. Mol. Cell. Biol. 10 1940–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J., G. Jongeward and P. Sternberg, 1994. unc-101, a gene required for many aspects of Caenorhabditis elegans development and behavior, encodes a clathrin-associated protein. Genes Dev. 8 60–73. [DOI] [PubMed] [Google Scholar]
- Lee, R., J. Hench and G. Ruvkun, 2001. Regulation of C. elegans DAF-16 and its human orthologue FKHRL1 by the daf-2 insulin-like signaling pathway. Curr. Biol. 11 1950–1957. [DOI] [PubMed] [Google Scholar]
- Lin, K., J. B. Dorman, A. Rodan and C. Kenyon, 1997. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278 1319–1322. [DOI] [PubMed] [Google Scholar]
- Lin, K., H. Hsin, N. Libina and C. Kenyon, 2001. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat. Genet. 28 139–145. [DOI] [PubMed] [Google Scholar]
- Lu, K., and G. Guidotti, 1996. Identification of the cysteine residues involved in the class I disulfide bonds of the human insulin receptor: properties of insulin receptor monomers. Mol. Biol. Cell 7 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig, A., C. Kober-Eisermann, C. Weitzel, A. Bethke, K. Neubert et al., 2004. A novel nuclear receptor/coregulator complex controls C. elegans lipid metabolism, larval development, and aging. Genes Dev. 18 2120–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi, D., and R. Cordera, 2001. Cys 786 and Cys 776 in the posttranslational processing of the insulin and IGF-I receptors. Biochem. Biophys. Res. Commun. 280 836–841. [DOI] [PubMed] [Google Scholar]
- Malone, E. A., and J. H. Thomas, 1994. A screen for nonconditional dauer-constitutive mutations in Caenorhabditis elegans. Genetics 136 879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee, J. J., E. Schuster, E. Blanc, J. H. Thomas and D. Gems, 2004. Shared transcriptional signature in C. elegans dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J. Biol. Chem. 279 44533–44543. [DOI] [PubMed] [Google Scholar]
- Mihaylova, V., C. Borland, L. Manjarrez, M. Stern and H. Sun, 1999. The PTEN tumor suppressor homolog in Caenorhabditis elegans regulates longevity and dauer formation in an insulin receptor-like signaling pathway. Proc. Natl. Acad. Sci. USA 96 7427–7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller, D. E., O. Cohen, Y. Yamaguchi, R. Assiz, F. Grigorescu et al., 1994. Prevalence of mutations in the insulin receptor gene in subjects with features of the type A syndrome of insulin resistance. Diabetes 43 247–255. [DOI] [PubMed] [Google Scholar]
- Morita, K., A. J. Flemming, Y. Sugihara, M. Mochii, Y. Suzuki et al., 2002. A Caenorhabditis elegans TGF-beta, DBL-1, controls the expression of LON-1, a PR-related protein, that regulates polyploidization and body length. EMBO J. 21 1063–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz, W., E. Froesch and M. Boni-Schnetzler, 1994. Functional properties of a heterozygous mutation (Arg1174→Gln) in the tyrosine kinase domain of the insulin receptor from a type A insulin resistant patient. FEBS Lett. 351 276–280. [DOI] [PubMed] [Google Scholar]
- Morris, J. Z., H. A. Tissenbaum and G. Ruvkun, 1996. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature 382 536–538. [DOI] [PubMed] [Google Scholar]
- Motola, D. L., C. L. Cummins, V. Rottiers, K. K. Sharma, T. Li et al., 2006. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell 124 1209–1223. [DOI] [PubMed] [Google Scholar]
- Murphy, C. T., S. A. McCarroll, C. I. Bargmann, A. Fraser, R. S. Kamath et al., 2003. Genes that act downstream of DAF-16 to influence the lifespan of C. elegans. Nature 424 277–284. [DOI] [PubMed] [Google Scholar]
- Nanji, M., N. A. Hopper and D. Gems, 2005. LET-60 RAS modulates effects of insulin/IGF-1 signaling on development and aging in Caenorhabditis elegans. Aging Cell 4 235–245. [DOI] [PubMed] [Google Scholar]
- Ogg, S., and G. Ruvkun, 1998. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol. Cell 2 887–893. [DOI] [PubMed] [Google Scholar]
- Ogg, S., S. Paradis, S. Gottlieb, G. I. Patterson, L. Lee et al., 1997. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389 994–999. [DOI] [PubMed] [Google Scholar]
- Paradis, S., and G. Ruvkun, 1998. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. 12 2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis, S., M. Ailion, A. Toker, J. H. Thomas and G. Ruvkun, 1999. A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans. Genes Dev. 13 1438–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce, S. B., M. Costa, R. Wisotzkey, S. Devadhar, S. A. Homburger et al., 2001. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev. 15 672–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle, D. L., and P. S. Albert, 1997. Genetic and environmental regulation of dauer larva development, pp. 739–768 in C. elegans II, edited by D. L. Riddle, T. Blumenthal, B. J. Meyer and J. R. Priess. Cold Spring Harbor Laboratory Press, Plainview, NY. [PubMed]
- Rouault, J., P. E. Kuwabara, O. M. Sinilnikova, L. Duret, D. Thierry-Mieg et al., 1999. Regulation of dauer larva development in Caenorhabditis elegans by daf-18, a homologue of the tumour suppressor PTEN. Curr. Biol. 9 329–332. [DOI] [PubMed] [Google Scholar]
- Schaffer, L., and L. Ljungqvist, 1992. Identification of a disulfide bridge connecting the alpha-subunits of the extracellular domain of the insulin receptor. Biochem. Biophys. Res. Commun. 189 650–653. [DOI] [PubMed] [Google Scholar]
- Schuster, E. F., E. Blanc, L. Partridge and J. M. Thornton, 2007. Estimation and correction of non-specific binding in a large-scale spike-in experiment. Genome Biol. 8 R126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, B., M. Avidan and C. Crowder, 2002. Regulation of hypoxic death in C. elegans by the insulin/IGF receptor homolog DAF-2. Science 296 2388–2391. [DOI] [PubMed] [Google Scholar]
- Shim, J., S. Im and J. Lee, 2003. Tissue-specific expression, heat inducibility, and biological roles of two hsp16 genes in Caenorhabditis elegans. FEBS Lett. 537 139–145. [DOI] [PubMed] [Google Scholar]
- Sparrow, L. G., N. M. McKern, J. J. Gorman, P. M. Strike, C. P. Robinson et al., 1997. The disulfide bonds in the C-terminal domains of the human insulin receptor ectodomain. J. Biol. Chem. 272 29460–29467. [DOI] [PubMed] [Google Scholar]
- Sulston, J., and J. Hodgkin, 1988. Methods, pp. 587–606 in The Nematode Caenorhabditis elegans, edited by W. B. Wood. Cold Spring Harbor Laboratory Press, Plainview, NY.
- Tatar, M., A. Kopelman, D. Epstein, M.-P. Tu, C.-M. Yin et al., 2001. Mutations in the Drosophila insulin receptor homologue retard senescence and impair neuroendocrine function. Science 292 107–110. [DOI] [PubMed] [Google Scholar]
- Taylor, S., A. Cama, D. Accili, F. Barbetti, M. Quon et al., 1992. Mutations in the insulin receptor gene. Endocr. Rev. 13 566–595. [DOI] [PubMed] [Google Scholar]
- Tuck, S., and I. Greenwald, 1995. lin-25, a gene required for vulval induction in Caenorhabditis elegans. Genes Dev. 9 341–357. [DOI] [PubMed] [Google Scholar]
- Vieira, A., C. Lamaze and S. Schmid, 1996. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science 274 2086–2089. [DOI] [PubMed] [Google Scholar]
- Vowels, J. J., and J. H. Thomas, 1992. Genetic analysis of chemosensory control of dauer formation in Caenorhabditis elegans. Genetics 130 105–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, G. A., and G. J. Lithgow, 2003. Lifespan extension in C. elegans by a molecular chaperone dependent upon insulin-like signals. Aging Cell 2 131–139. [DOI] [PubMed] [Google Scholar]
- Walker, G., T. White, G. McColl, N. Jenkins, S. Babich et al., 2001. Heat shock protein accumulation is upregulated in a long-lived mutant of Caenorhabditis elegans. J. Gerontol. A Biol. Sci. Med. Sci. 56 B281–B287. [DOI] [PubMed] [Google Scholar]
- Whittaker, J., and L. Whittaker, 2005. Characterization of the functional insulin binding epitopes of the full-length insulin receptor. J. Biol. Chem. 280 20932–20936. [DOI] [PubMed] [Google Scholar]
- Wolkow, C., K. Kimura, M. Lee and G. Ruvkun, 2000. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science 290 147–150. [DOI] [PubMed] [Google Scholar]
- Wolkow, C., M. Munoz, D. Riddle and G. Ruvkun, 2002. Insulin receptor substrate and p55 orthologous adaptor proteins function in the Caenorhabditis elegans daf-2/insulin-like signaling pathway. J. Biol. Chem. 277 49591–49597. [DOI] [PubMed] [Google Scholar]
- Wu, J. J., and G. Guidotti, 2002. Construction and characterization of a monomeric insulin receptor. J. Biol. Chem. 277 27809–27817. [DOI] [PubMed] [Google Scholar]
- Yu, H., and P. Larsen, 2001. DAF-16-dependent and independent expression targets of DAF-2 insulin receptor-like pathway in Caenorhabditis elegans include FKBPs. J. Mol. Biol. 314 1017–1028. [DOI] [PubMed] [Google Scholar]