Abstract
During a 1000-generation evolution experiment, two types of morphologically and kinetically distinct bacteria repeatedly diverged from a common ancestor in a fully sympatric seasonal environment containing glucose and acetate. To investigate the metabolic modifications associated with this adaptive diversification, we compared transcription profiles of the two derived types and the common ancestor. Both derived types share a suite of common metabolic changes that may represent adaptation to the environment preceding the diversification event. These include improved translation efficiency, glucose uptake capacity via the mal/lamB genes, upregulation of various transporters during stationary phase, and likely the disruption of the rbs operon. The diversification event is associated with the overexpression of genes involved in the TCA cycle, glyoxylate shunt, acetate consumption, and anaerobic respiration in one type and in acetate excretion in the other. These results reveal that competition for both carbon and oxygen have likely played an important role in the adaptation of Escherichia coli during this adaptive diversification event, where one derived type mainly consumes glucose at a fast rate when oxygen is not limiting, and the other derived type consumes glucose and acetate at a slower rate, even when oxygen is limiting.
LINKING genetic, biochemical, and physiological changes to evolutionary processes is difficult. One of the most important reasons for this is due to the nature of descent with modification: evolutionary processes occur over long periods of time. One must thus often infer history and ancestry from the standing biological diversity without the actual knowledge of the ancestral states that would allow tracking biological changes. For the last few decades, experiments with quickly reproducing organisms, such as bacteria, evolving under controlled conditions in the lab for hundreds or thousands of generations, have been a powerful alternative to classic comparative studies for better understanding of various evolutionary processes (see Elena and Lenski 2003 and Kassen and Rainey 2004 for reviews). Another difficulty when linking cellular and evolutionary processes is the lack of precise genetic, biochemical, and physiological knowledge in most of the biological systems used as models in evolution studies. Classic genetic and physiological microbial models, such as yeast and bacteria, offer a powerful alternative. By coupling evolution experiments and genetic investigations in these microorganisms, several studies have identified cellular functions involved in evolutionary processes (e.g., Treves et al. 1998; Notley-McRobb and Ferenci 1999; Cooper et al. 2001; Spencer et al. 2007). However, in most of the cases, these studies focus on specific candidate genes, and general investigations of metabolic changes are rare (but see Kurlandzka et al. 1991; Cooper et al. 2003; Pelosi et al. 2006). Moreover, most such studies concentrate on understanding directional adaptation to optimize performance in a given environment.
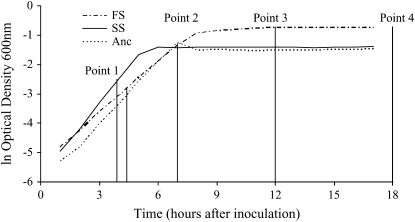
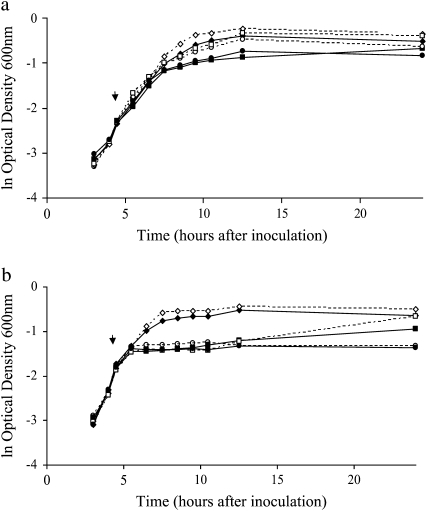
We investigate the evolutionary changes in metabolic functions that occur when a population diversifies. Diversification that leads to the coexistence of several types of bacteria from a single ancestral strain is a frequent outcome of evolution experiments (see, for example, Rosenzweig et al. 1994; Rainey and Travisano 1998; Rozen and Lenski 2000; Friesen et al. 2004). In fact, many of the reported diversification events are consistent with the process of adaptive diversification (Dieckmann et al. 2004) that occurs under sympatric (well-mixed) conditions due to competition for limiting resources. In this study, we take advantage of the short generation time and extensive genetic knowledge of the bacterium Escherichia coli to investigate metabolic changes that occurred during such an adaptive diversification event. According to theory, populations undergoing adaptive diversification experience two different evolutionary phases. They adapt to the environment following a directional selective regime, which ends up in disruptive selection generated by frequency-dependent competitive interactions, subsequently leading to the diversification of the population (Dieckmann and Doebeli 1999). In our experiments, an E. coli B clone (hereafter named A for ancestor) evolved during 1000 generations in a fully sympatric daily batch culture containing two carbon sources, glucose and acetate. In an environment containing glucose and acetate, metabolic trade-offs prevent the bacteria from concomitantly using the two carbon sources because genes implied in acetate consumptions are repressed by glucose (Saier et al. 1996). This results in a diauxic growth regime where bacteria first grow fast by consuming glucose and then more slowly after switching to acetate consumption when glucose is depleted. On the basis of the theory and patterns of diversification observed in other populations (Spencer et al. 2008), we expected that the bacteria would first undergo directional adaptation and then diversify when the selection regime turned disruptive due to ecological interactions. Specifically, we expected that one type, named SS (Spencer et al. 2007), would maximize its growth rate on glucose by first consuming glucose, switching to acetate consumption when glucose was exhausted, and finally growing on acetate alone and that a second type, named FS (Spencer et al. 2007), would minimize the lag time between glucose and acetate consumption by starting to consume both glucose and acetate and continuing to consume acetate when glucose was exhausted (Friesen et al. 2004). Under such a scenario, in each daily batch the second type would initially grow more slowly than the first one due to the metabolic trade-offs preventing efficient consumption of both glucose and acetate but then more quickly by reducing the lag time needed to switch from glucose to acetate-only consumption. Diversification repeatedly occurred in all replicates (10/10 replicates diversified into SS and FS types) of the evolution experiment, and the evolutionary dynamics conform to the theoretical expectations for adaptive diversification, showing a directional evolutionary phase followed by diversification into two types (Spencer et al. 2008). In a similar evolution experiment, the two derived types maintained a stable coexistence due to frequency-dependent interactions, where each of the two types increased its frequency in the population when it was initially rare (Friesen et al. 2004; Tyerman et al. 2005). In our study population, the two types differ by their growth kinetics in the evolution medium (Figure 1). As expected, the SS type seems to first consume glucose and produce acetate as a result of fermentation, while the FS type seems to start consuming both glucose and acetate (Spencer et al. 2007).
Figure 1.—
Growth kinetic curves of the three types of bacteria. Optical densities measured at 600 nm are plotted against time after inoculation. The four time points used to perform the RNA extraction are indicated.
Phenotypic assays (i.e., assessing colony morphology and growth profiles) and comparing transcription levels of candidate genes (Spencer et al. 2007) have limited power to unravel the general metabolic modifications that occurred during our evolution experiments. In this study, we tried to obtain a more general picture of the modified cellular functions by investigating how total gene expression changed during the adaptive diversification. To do so, we focused on a single microcosm and compared the global transcription profile of FS, SS, and A at four different time points during growth in a single batch (Figure 1). In particular, we investigated which modifications in cellular functions were common to both derived types, and hence presumably evolved prior to the diversification event, and which modifications distinguish the derived types and hence arose during diversification.
MATERIALS AND METHODS
Biological system:
A 150-day (∼1000 generations) evolution experiment was performed in a 24-hr batch culture environment (Tyerman et al. 2008; Spencer et al. 2008). Every 24 hr, 100 μl of culture was inoculated into 18-mm test tubes containing 10 ml of fresh medium composed of Davis minimum medium supplemented with 0.0002% thiamine HCl, 0.1% MgSO4, 250 mg · liter−1 of glucose, and 1323 mg · liter−1 of sodium acetate trihydrate (DMGA). Test tubes were incubated at 37° at 250 rpm. This experiment repeatedly led to the diversification and coexistence of two types of bacteria, SS and FS, from one ancestral E. coli B strain (the REL606 strain hereafter called A for ancestor). For each derived type, three clones were isolated from population 20 after 150 days of evolution. For each type, the three clones were pooled and samples of the resulting populations were frozen at −80°. Changes in gene expression associated with this adaptive diversification were investigated by comparing gene expression profiles between the pooled FS and A and between the pooled SS and A.
Transcript profiling using microarrays:
Transcription profiling was performed by using microarrays printed with 5978 70mer probes representing the genome of three strains of E. coli (K12, O157:H7 EDL933, and Sakai; Array-Ready Oligoset for the E. coli genome v1.0, QIAGEN, Valencia, CA). The slides were printed by the Gene Array Facility of the Prostate Centre at Vancouver General Hospital. The array platform was submitted at the NCBI's Gene Expression Omnibus (GEO) data bank (http://www.ncbi.nlm.nih.gov/geo/) under the accession no. GPL5767.
SS, FS, and A were incubated in DMGA at 37° and 250 rpm for ∼18 hr. We then inoculated 500 μl of these overnight cultures into 50 ml of DMGA in 500-ml flasks and grew them at 37° and 250 rpm. The growth kinetics were assessed by measuring the optical density at 600 nm (OD600) regularly during the incubation (Figure 1). Total RNA from the three types of bacteria was extracted using QIAGEN RNeasy mini kit as per the manufacturer's protocol at four points of the growth curve: point 1 after 3 hr and 30 min (SS) or 4 hr (FS and A) (two different extraction times where used to minimize the OD600 variability between SS replicates while capturing the fast increase in OD600 in FS and A); point 2 after 7 hr; point 3 after 12 hr; and point 4 after 18 hr of incubation (Figure 1). DNA was removed using Ambion's DNA-free kit. RNA quality was assessed using 2% agarose gels and an OD260/OD280 ratio. Indirect labeling methods were used to label 5 μg of RNA during reverse transcription. The reverse transcription was performed at 42° during 50 min using 600 units of superscript RT II (Invitrogen, San Diego); 3 μg of random hexamers; 0.6 mm dATP, dCTP, and dGTP; 0.2 mm dTTP; and 0.4 mm amino-allyl dUTP (Ambion). RNA was then degraded by incubation at 65° for 15 min after adding 10 μl of 0.5 m EDTA and 10 μl of 1 m NaOH. cDNA was cleaned using the QIAquick purification kit (QIAGEN) and labeled with either Cy3 (A) or Cy5 (FS and SS) following the manufacturer's instructions (Amersham Pharmacia). Labeled cDNA was purified using QIAquick purification kit (QIAGEN). For each of the four points of the growth curve, 80 pmol of the labeled cDNA was combined in two different ways: SS cDNA with A cDNA and FS cDNA with A cDNA. The combined cDNA were speed vacuum concentrated, resuspended in 4 μl 10 mm EDTA, and denaturated at 95° for 1 min before adding 76 μl of Ambion hybridization buffer 2. After 2 min of denaturation at 95°, the hybridization solution was placed on a microarray slide and covered by a cover glass. The slides were assembled with hybridization chambers (Corning) and hybridized for 20 hr in a 42° water bath. The slides were washed three times in 0.5× SSC, 0.5% SDS for 5 min and then three times in 0.5× SSC and were dried by centrifugation. The hybridized slides were scanned with a ScanArray Express scanner (Perkin-Elmer, Norwalk, CT). ImageGene v5.0 was used to quantify the spot intensities. Each transcript comparison (FS/A and SS/A at time points 1–4) had five replicates, leading to a total of 40 microarray slides that were submitted to the NCBI's Gene Expression Omnibus data bank (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession no. GSE8965.
Statistical preprocessing steps were conducted with ArrayPipe (version 1.7), a web-based software designed for processing of microarray data (http://www.pathogenomics.ca/arraypipe; Hokamp et al. 2004). The following preprocessing steps were applied: (1) flagging of markers and control spots, (2) subgrid-wise background correction using the median of the lower 10% foreground intensity as an estimate for the background noise, (3) data shifting, (4) limma's LOESS normalization by print tip, (5) merging of duplicate spots, and (6) merging of replicate arrays to yield overall fold changes and intensity ratios for each group. Merging of replicate arrays was performed in several ways:
For each time point, the results of the five slides comparing FS/A and SS/A (allowing the direct comparison between each of the derived types and the ancestor FS/A or SS/A and the indirect comparison between the two derived types FS/SS for each time point).
For each time point, the results of the 10 slides comparing FS/A and SS/A (considering FS and SS as a single derived type hereafter named D, allowing the comparison between the derived types and the ancestor D/A for each time point).
The results of the 20 slides comparing FS/A or SS/A from all four time points combined (allowing the overall comparison between each of the derived types and the ancestor FS/A or SS/A and the indirect comparison between the two derived types FS/SS).
The results of the 40 slides from all four time points combined (allowing the overall comparison between the derived types and the ancestor D/A).
Identification and analysis of differentially expressed genes:
Differentially expressed genes were identified using the optimal discovery procedure (Storey et al. 2007) as implemented in the software EDGE v1.1.208 (Leek et al. 2006). Tests were performed to identify differentially expressed genes between FS/A, SS/A, D/A, and FS/SS at each of the four time points and after pooling the four time points (20 tests). Each test was performed using 200 permutations. Genes were considered as differentially expressed when false discovery rates (FDR; Storey and Tibshirani 2003) gave q-values ≤0.2 and when absolute fold changes were ≥1.5. The P-values give the probability of an event when a single test is performed. In the case of multiple testing, FDR allows estimation of the probability q of one gene to be false positive, given the distribution of P-values.
To visualize the general trends of changes in gene expression in our experiment, the normalized intensity ratio of all differentially expressed genes for the comparisons FS/A and SS/A at each of the four points were clustered using the QT CLUST algorithm (Heyer et al. 1999) in the software TM4 (Saeed et al. 2003), using the Pearson correlation metric distance with a maximum cluster diameter of 0.75 and a minimum cluster population of 50.
To identify the cellular functions that are most often associated with change in gene expression, genes were classified in two kinds of functional categories: (1) in broad functional categories according to GenProtEC (http://genprotec.mbl.edu/) and (2) in more precise gene ontology (GO) cellular process categories defined in Riley et al. (2006). Overrepresentation of the categories was tested using one-tail Fisher's exact tests in R (http://www.r-project.org/) by comparing for each category the following: (1) the total number of differentially expressed genes to the number of genes on the array; (2) the number of genes found differentially expressed for each of the 20 statistical comparisons (FS/A, SS/A, D/A, and FS/SS at each of the four points and after pooling the four points) to the total number of differentially expressed genes; and (3) the number of differentially expressed genes in each cluster of genes and the total number of differentially expressed genes. Tests were performed only when more than one gene was found differentially expressed in the focal comparison. As Fisher's exact tests were performed multiple times for both GO and functional categories, FDRs were estimated using the QVALUE software (Storey and Tibshirani 2003) and categories were considered as overrepresented for q-values ≤0.2.
The statistical tests performed to identify differential expression allow the identification of individual genes based only on the expression of each particular gene. However, due to the small sample size associated with microarray analyses, these statistical tests often offer limited statistical power. Comparing patterns of expression for several genes involved in the same pathways offers an alternative to individual gene tests and may indicate relevant biological information, especially if several genes display an identical trend (even without statistical significance when referring to each individual gene), indicating an overall up- or downregulation of a pathway. To identify such trends, we used the software Pathway Tools v11.0 (Paley and Karp 2006) to visualize the difference in gene expression for our eight direct comparisons (FS/A and SS/A at the four points of comparison) on a map representing the various cellular pathways. We considered as a trend for up- or downregulation if, in pathways containing more than three genes, at least three-fourths of the genes linking two metabolic compounds on the metabolic map displayed an absolute fold change ≥1.5 in the same direction.
Gene operons and regulons were considered of special interest if several genes of the same regulon/operon were identified as differentially expressed in more than one statistical comparison.
Glucose, acetate, and glycerol concentrations:
Three independent cultures of each of FS, SS, and A were sampled at several points during growth in 10 ml DMGA to determine glucose, acetate, and glycerol concentrations. For each sample, we removed ∼1 ml of culture, pelleted the cells, and stored the supernatant at −80°. After all samples were obtained, we estimated the glucose concentrations using a glucose (HK) assay kit (Sigma, St. Louis), acetate concentrations using a UV-method acetic acid kit (Boehringer Mannheim/R-biopharm), and glycerol concentrations using a UV-method glycerin/glycerol kit (Boehringer Mannheim/R-biopharm). Manufacturer-supplied standards and non-inoculated media samples were used as controls.
Dissolved oxygen measurement:
Two overnight cultures of each FS and SS were inoculated (1/100 dilution) into 10 ml DMGA in test tubes and 50 ml DMGA in 500-ml flasks. A Microx-TX fiber optic oxygenmeter (PreSens) was used to measure oxygen concentration relative to sterile medium at several points during 24 hr of growth.
Growth curve assays:
Growth curve assays were performed to investigate the effect of glucose, glycerol, acetate, and aeration to the kinetic growth of FS and SS strains. Previously frozen FS and SS were incubated in DMGA at 37° and 250 rpm during ∼18 hr and 100 μl of these overnight cultures were inoculated in 10 ml of DMGA in test tubes and were grown at 37° and 250 rpm. After 4 hr and 30 min of incubation, 250 mg · liter−1 of glucose, 250 mg · liter−1 of acetate, extra aeration (by transfer to a sterile 150-ml bottle, vigorous shaking, and transfer back to the test tube), 250 mg · liter−1 of glucose and extra aeration, 250 mg · liter−1 of acetate and extra aeration, or nothing was added to the test tubes. Kinetic growth was assessed by measuring OD600 at regular intervals during the incubation.
Additional remark:
All the assays were performed growing FS and SS in isolation, because we were unable to distinguish them when grown together. However, it should be kept in mind that the two types diverged from their common ancestor in sympatry and that it is possible that, when grown together, the growth of one type is affected by compounds excreted by the other type. Such effects would not necessarily be reflected in our study.
RESULTS
Numerous genes are overexpressed in the two derived types at time points 1, 3, and 4 and overexpressed in FS at time point 2:
We compared global expression profiles of an ancestral E. coli strain (A) and two strains (FS and SS) derived in sympatry from the ancestor during 1000 generations of experimental evolution in serial batch culture. Microarrays were used to identify changes in gene expression at four time points during growth in a single batch in the evolution medium (Figure 1). A total of 644 genes were identified as differentially expressed (several genes were significant in more than one comparison). Table 1 indicates the number of differentially expressed genes for each of the 20 different tests performed (detailed gene lists are presented in supplemental Table 1 at http://www.genetics.org/supplemental/). In the 15 tests comparing the two derived types to the ancestor, many more genes were upregulated in the derived types compared to the ancestor (663 significant comparisons display a fold change ≥1.5 while 234 display a fold change ≤ −1.5, two-tailed Fisher's exact test, P < 0.0001). Interestingly, this is opposite to what has been found in another evolution experiment in which E. coli B evolved for 20,000 generations in batch culture containing glucose as the only carbon source and in which most of the difference in gene expression between the evolved and ancestral strains involved a downregulation in the evolved strains (Cooper et al. 2003; Pelosi et al. 2006). In the five tests comparing the two derived types SS and FS, more genes are upregulated in FS compared to SS (324 genes upregulated in FS and 34 in SS, two-tailed Fisher's exact test, P < 0.0001). As shown in Table 1, most of the differences (in terms of the number of genes differentially expressed) occur between the derived types (FS and SS pooled and considered as a single type D) and A at time points 1, 3, and 4, and between FS on the one hand and SS and A on the other at time point 2. The differences between the evolved strains and the ancestor probably reflect adaptation to the experimental environment preceding the divergence of the two strains. They indicate that the mutations accumulating during that stage of evolution had an effect not only during the log phase, corresponding to active cell divisions (time point 1 for FS, SS, and A and time point 2 for FS), but also during the stationary phase, when cells are no longer dividing and must survive in an environment containing very few nutrients (time points 3 and 4). The differences between FS and SS likely reflect mutations responsible for the divergence between the derived strains.
TABLE 1.
Genes differentially expressed for each of the 20 tests performed
| Type | TP1 | TP2 | TP3 | TP4 | Global |
|---|---|---|---|---|---|
| FS/A | 18 | 275 | 9 | 48 | 30 |
| SS/A | 44 | 4 | 12 | 46 | 19 |
| D/A | 147 | 21 | 117 | 88 | 19 |
| FS/SS | 6 | 344 | 2 | 0 | 6 |
One particular gene may be considered as differentially expressed in more than one comparison. Rows indicate the comparisons between two types of bacteria (FS/A, SS/A, D/A: FS and SS are considered as a single derived type; FS/SS: the indirect comparison between FS and SS). Each column corresponds to one point of comparison (TP1, -2, -3, -4, and global; all the points are pooled and considered as a single point). The various analyses were performed using different sample sizes (five microarray slides are indicated by roman, 10 slides by italic, 20 slides by underlining, and 40 slides by boldface type) and may thus result in different statistical power. TP, time point.
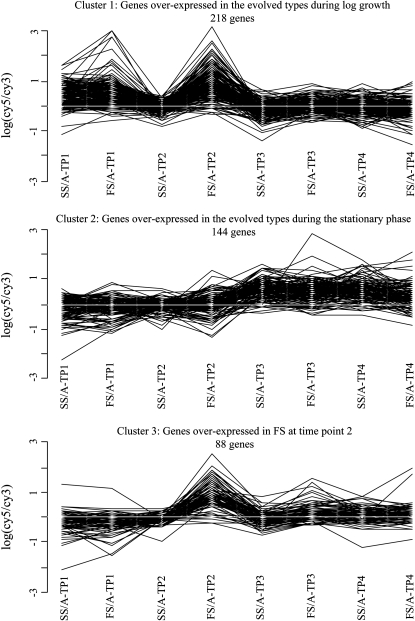
Differentially expressed genes can be grouped in three main clusters:
To identify genes with parallel changes in gene expression at the four time points, we performed a cluster analysis on the whole set of differentially expressed genes to group together genes presenting the same pattern of differential expression. The analysis identified three main clusters of genes (Figure 2). The first cluster groups 218 genes more highly expressed in both FS and SS at time point (TP) 1 (D/A-TP1) and in FS at time point 2 (FS/A-TP2 and FS/SS-TP2). This cluster identifies genes likely to be linked to the adaptation of the derived types to log growth in the evolution environment and indicates that the responsible mutations probably accumulated before the diversification event. The second cluster identifies 144 genes that are more expressed in both derived types (FS and SS) at the points 3 and 4 (D/A-TP3, and D/A-TP4), indicating that at these two points, many of the differentially expressed genes are identical. At these two time points, the bacteria are in stationary phase and are not rapidly dividing anymore due to low resource concentrations. These genes may thus be involved in adaptation to survival in starvation conditions that the bacteria have faced at the end of each 24-hr growth batch during the evolution experiment. The third cluster is composed of 88 genes that have higher expression in FS at time point 2 (FS/A-TP2, FS/SS-TP2) and are therefore most likely involved in allowing FS to grow while SS is already in stationary phase.
Figure 2.—
The three clusters of differentially expressed genes. Each line corresponds to one gene and indicates the changes in gene expressions between the eight different comparisons. Positive values indicate that the gene has higher expression in the derived strain (FS or SS), and negative values indicate that the gene has higher expression in the ancestor (A). Numbers of genes in each cluster are indicated; 194 of the differentially expressed genes are not clustered.
Cellular functions associated with the differentially expressed genes:
To identify the physiological functions associated with the differential expression profiles and to gain a clearer picture of the molecular mechanisms involved in the adaptive diversification event, three approaches were used. First, genes were classified in broad functional categories and then in more precise GO cellular process categories. We performed a total of 563 tests to identify the overrepresented categories, 48 of which were significant, identifying 9 and 17 overrepresented functional and GO categories, respectively (Table 2). Second, observed fold changes were mapped onto E. coli metabolic pathways to identify trends of up- or downregulation in specific pathways. This showed that genes involved in the TCA cycle, glyoxylate shunt, and acetate excretion/consumption are differentially expressed in FS and SS at time point 1 (Figure 3). Third, we looked for genes involved in the same operon or regulon that were identified as differentially expressed in numerous statistical comparisons. This allowed the identification of three groups of genes. Results are detailed separately for the comparisons between the evolved strains and A (comparisons D/A, cluster 1 and 2) and between the FS and SS types (comparisons FS/SS, cluster 3).
TABLE 2.
Overrepresented functional and gene ontology (GO) categories in the differentially expressed genes
| Functional category | Focal point | Focal point: no. of significant genes | Overall: no. of significant genes | No. of genes on the arrays | P-value | q-Value |
|---|---|---|---|---|---|---|
| Cell structure, membrane | D/A-TP3 | 43 | 131 | 784 | <0.001 | 0.03 |
| Cluster 2 | 43 | <0.001 | 0.003 | |||
| Cell structure, ribosome | FS/A-TP2 | 27 | 38 | 69 | 0.004 | 0.08 |
| D/A-TP1 | 23 | <0.001 | <0.001 | |||
| Cluster 1 | 33 | <0.001 | <0.001 | |||
| Overall | NA | <0.001 | <0.001 | |||
| Information transfer, protein related | D/A-TP1 | 35 | 65 | 295 | <0.001 | <0.001 |
| Cluster 1 | 45 | <0.001 | <0.001 | |||
| Overall | NA | <0.001 | 0.02 | |||
| Location, cytoplasm | D/A-TP1 | 50 | 140 | 849 | 0.007 | 0.1 |
| Cluster 1 | 68 | 0.003 | 0.07 | |||
| Location, inner membrane | D/A-TP3 | 32 | 99 | 531 | 0.005 | 0.09 |
| Overall | NA | 0.006 | 0.1 | |||
| Location, periplasmic space | D/A-TP1 | 9 | 14 | 128 | 0.003 | 0.07 |
| Metabolism, carbon compound utilization | FS/A-TP1 | 7 | 50 | 363 | 0.003 | 0.07 |
| Metabolism, energy metabolism (carbon) | Cluster 3 | 13 | 39 | 229 | 0.003 | 0.07 |
| Transport | D/A-TP3 | 32 | 104 | 616 | 0.006 | 0.1 |
| Cluster 2 | 33 | 0.002 | 0.07 | |||
| GO category | Focal point | Focal point: no. of significant genes | Overall no. of significant genes | No. of genes on the arrays | P-value | q-Value |
| Acetyl-CoA biosynthesis from pyruvate | Overall | NA | 2 | 2 | 0.03 | 0.18 |
| Amino acid activation | FS/A | 2 | 4 | 31 | 0.008 | 0.12 |
| Amino sugar biosynthesis | Cluster3 | 2 | 2 | 15 | 0.02 | 0.15 |
| Anaerobic respiration | Cluster3 | 7 | 22 | 139 | 0.03 | 0.18 |
| ATP-synthesis-coupled proton transport | Overall | NA | 9 | 12 | <0.001 | <0.001 |
| Carbohydrate catabolism | FS/A-TP1 | 8 | 36 | 235 | <0.001 | <0.001 |
| FS/A-TP3 | 3 | 0.03 | 0.19 | |||
| SS/A-TP1 | 8 | 0.004 | 0.07 | |||
| DNA catabolism | SS/A-TP4 | 2 | 3 | 49 | 0.01 | 0.12 |
| Fermentation | Cluster3 | 3 | 4 | 33 | 0.01 | 0.12 |
| Flagella biogenesis | Cluster3 | 3 | 5 | 35 | 0.02 | 0.17 |
| Glycerol metabolism | FS/A-TP1 | 2 | 5 | 16 | 0.02 | 0.14 |
| FS/A-TP4 | 3 | 0.003 | 0.06 | |||
| FS/A | 2 | 0.01 | 0.15 | |||
| D/A-TP2 | 2 | 0.01 | 0.12 | |||
| D/A | 2 | 0.005 | 0.07 | |||
| FS/SS-TP1 | 2 | 0.002 | 0.04 | |||
| FS/SS | 2 | 0.001 | 0.04 | |||
| Leucine biosynthesis | SS/A-TP4 | 2 | 2 | 16 | 0.004 | 0.06 |
| Polysaccharide biosynthesis | D/A-TP4 | 2 | 2 | 8 | 0.01 | 0.12 |
| Cluster2 | 2 | 0.02 | 0.14 | |||
| Protein biosynthesis | Overall | NA | 44 | 78 | <0.001 | <0.001 |
| FS/A-TP2 | 30 | 0.002 | 0.04 | |||
| D/A-TP1 | 28 | <0.001 | <0.001 | |||
| Cluster1 | 37 | <0.001 | <0.001 | |||
| Protein modification | Cluster2 | 3 | 5 | 64 | 0.02 | 0.14 |
| Purine nucleotide biosynthesis | Overall | NA | 5 | 10 | 0.01 | 0.14 |
| Translational attenuation | Overall | NA | 5 | 6 | <0.001 | 0.02 |
| Tryptophan biosynthesis | FS/A-TP4 | 2 | 4 | 11 | 0.03 | 0.19 |
| SS/A-TP4 | 2 | 0.02 | 0.15 |
For each category identified as overexpressed, the number of significant genes in the focal comparison and in the overall data set, the number of genes on the arrays, the P-values of the one-tail Fisher's exact tests, and the q-values of the false discovery rate are indicated. For example, the first line of the table indicates that 784 genes represented on the array encode for proteins associated with the structure of the membrane, that 131 of these genes are identified as differentially expressed in at least 1 of the 20 statistical comparisons performed, and that 43 genes are identified as differentially expressed between the evolved and the ancestor at the time point 3, which is more than would be expected if the significant genes were randomly distributed in the different functional categories, with a P-value <0.001 and a q-value of 0.03. NA indicates that the overall number of genes identified as differentially expressed is greater than would be randomly expected (for example, the 38 genes differentially expressed in the functional category “Cell structure, ribosome” is more than would be randomly expected).
Figure 3.—
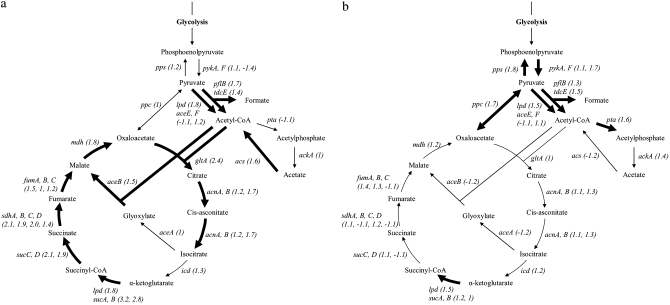
TCA, glyoxylate shunt, and acetate consumption/excretion metabolic pathways showing different trends in FS/A and SS/A at the first point of comparison. Enzymes are indicated by italics and fold changes are in parentheses. A thick arrow indicates a fold change ≥1.5. (a) Comparison between FS and A. (b) Comparison between SS and A.
Modification of cellular functions prior to diversification (comparisons of D/A at the various time points, clusters 1 and 2):
Numerous functions seem to differ between the two evolved strains and the ancestral strain (see Table 2), indicating that numerous metabolic functions were modified during the adaptation process that probably occurred before the diversification event.
During the stationary phase (time points 3 and 4), the products of the differentially expressed genes associated with the transport function (comparisons D/A-TP3, cluster 2; Table 2) were more differentially expressed than would be expected randomly. This result suggests that adaptation to survival during the stationary phase is associated with the ability to take up rare nutrients efficiently via overexpression of numerous transporters.
During log growth (time point 1 for FS, SS, and A and time point 2 for FS), one very clear result was that numerous genes associated with protein biosynthesis functions are more expressed in D than in A. This is indicated by the overrepresentation of the following categories: cell structure, ribosome (comparisons FS/A-TP2, D/A-TP1, cluster 1; Table 2); information transfer, protein related (comparisons D/A-TP1, cluster 1; Table 2); and protein biosynthesis (comparisons FS/A-TP2, D/A-TP1, cluster 1; Table 2). This result may indicate the evolution of increased translation efficiency prior to the diversification event. Another possible explanation would be that, as a result of adaptation, the derived types are able to grow faster than A in the environment in which they evolved for 1000 generations without any genetic modification directly associated with the translation efficiency. As during the first time point A seems to grow at least as fast as FS (see Figure 1), this explanation seems unlikely. Translation efficiency has been identified as an important target for selection under continuous growth conditions (Mikkola and Kurland 1992; Herring et al. 2006). This is the first time, however, that it is reported as a potential important adaptive factor in a seasonal (batch culture) environment.
Three groups of genes belonging to the same operon or regulon are found differentially expressed in several of the statistical comparisons between D and A. First, several of the genes encoded in the rbs operon were often downregulated in the comparisons between the two evolved types and the ancestor (at least one gene downregulated in the comparisons SS/A-TP1, -2, -3, -4; FS/A-TP1, -2, -3, -4; D/A-TP1, -2, -3, -4; and D/A, SS/A, FS/A; see supplemental Table 1). This operon contains genes allowing E. coli to use D-ribose as a carbon source. During another evolution experiment containing glucose as the only carbon source, all 12 replicate populations lost D-ribose catabolic function. This loss of function was due to the deletion of part or all of the rbs operon and conferred a fitness advantage in the evolution environment (Cooper et al. 2001). Thus, it is probable that, before the diversification event, deletion of part of the rbs operon also occurred in our experiment. Indeed, PCR amplification of the rbs operon suggested a 9-kb deletion in both FS and SS (data not shown).
Second, numerous genes involved in maltose uptake (mal/lamB genes) are found overexpressed in the evolved strains (at least one gene upregulated in the comparisons SS/A, SS/A-TP1, FS/A-TP1, -2, D/A, and FS/SS-TP2; see supplemental Table 1). These genes are grouped mainly in cluster 1 (Figure 2) and are thus more expressed during the log growth of the two evolved types in the evolution environment. This group of genes has already been identified as involved in adaptation to a better glucose uptake rate in other evolution experiments. For example, in all replicate populations of a 280-generation experiment performed under continuous growth (chemostat culture) on glucose, the mal/lamB genes were found overexpressed, leading to a significant increase in glucose uptake (Notley-McRobb and Ferenci 1999; see Kurlandzka et al. 1991 for other examples). This improvement is probably linked to the LamB protein, known as being the most important contributor to glucose uptake under nutrient limitation (Death et al. 1993). In another evolution experiment performed in a glucose-limited seasonal environment (batch culture), the investigated populations presented a lower expression level of mal/lamB genes after 20,000 generations, which was very often associated with mutations in the malT gene responsible for a fitness increase in the evolution environment (Pelosi et al. 2006). Third, genes of the glycerol-3-phosphate regulon (glp genes) are often found upregulated in D compared to A (see supplemental Table1). The glp genes are found differentially expressed not only when comparing D to A but also when comparing FS to SS. Results concerning this regulon are therefore detailed in the following section.
Modification of cellular functions associated with the diversification event (comparisons FS/SS and cluster 3):
Concentrating on the differences between the two evolved types, no functional groups are found overrepresented at time points 3 and 4. At time point 1, only functions corresponding to glycerol metabolism (comparison FS/SS-TP1; Table 2) are more represented than randomly expected. This function (via the glp genes; see below) is found overrepresented not only between the two evolved strains, but also between the evolved strains and the ancestor (comparisons FS/A-TP1, FS/A-TP4, FS/A, D/A-TP2, and D/A; Table 2). The other differences between the two evolved strains are concentrated at time point 2 and involve mainly genes associated with energy metabolism, especially anaerobic respiration and fermentation (cluster 3; Table 2). Overrepresentation of these categories suggests that oxygen may, at some point, be a limiting resource in the evolution environment.
Observed fold changes were mapped onto the E. coli metabolic pathway to identify trends of up- or down-regulation in specific pathways. Only two pathways were identified, highlighting differences between FS and A and between SS and A at the first time point of growth kinetics. At this point, genes involved in the TCA cycle, the glyoxylate shunt, and acetate consumption are much more expressed in FS than in the ancestor (Figure 3a), indicative of a higher consumption of acetate and TCA cycle activity in FS. At the same time point, genes involved in acetate excretion are more expressed in SS than in A (Figure 3b). This result may indicate that, at this first point, SS consumes glucose and releases acetate in the environment while FS consumes both glucose and acetate. This difference is fully consistent with previous results for genetic differences in acetate metabolism between strains that diversified in a seasonal batch culture environment (Spencer et al. 2007). The upregulation in FS of the entire TCA cycle observed in this study is also consistent with acetate consumption in E. coli (Oh et al. 2002).
As briefly mentioned above, the genes of the glycerol-3-phosphate regulon (glp genes) are often upregulated in the two derived types compared to the ancestor and even more upregulated in FS compared to SS (at least one gene was identified as upregulated in the comparisons SS/A; SS/A-TP4; FS/A; FS/A-TP1, -2, -3, -4; D/A; D/A-TP1, -2, -3, -4; FS/SS; FS/SS-TP1, -2; see supplemental Table 1). These genes are associated with glycerol metabolism, one of the very few functional gene categories that are identified as different between FS and SS. The medium used for the evolution experiment does not contain any glycerol. However, glycerol is a frequent product of glucose fermentation, and in previous evolution experiments with glucose as the sole carbon source, glycerol has been found to be involved in the cross-feeding polymorphism allowing the coexistence of three bacterial types (Rosenzweig et al. 1994). Glycerol thus might play a role in the sympatric coexistence of the two types of bacteria arising in our evolution experiment.
Both carbon and oxygen are limiting resources in the evolution environment:
The comparison of the transcription profiles among FS, SS, and A suggests that (1) glucose and acetate are used in different ways by FS and SS; (2) glycerol, a carbon source not part of the abiotic evolution environment, might be an alternative carbon source excreted by the bacteria and involved in a cross-feeding polymorphism; and (3) oxygen may be a limiting resource in the evolution environment. To verify these hypotheses, we measured glucose, acetate, glycerol, and oxygen concentration at different points during growth in the evolution medium for FS and SS.
No appreciable amount of glycerol was detected at any point (the minimum detection level was 0.04 g · liter−1), challenging the idea, resulting from the overexpression of the glp genes in the derived types, that glycerol may be an important carbon source in our experiment. However, as suggested by Rosenzweig et al. (1994), glycerol might still be an important parameter if glycerol fluxes are important and the concentration very low. Alternatively, the excretion of a molecule related to glycerol but not detected by our method, such as glycerol-3-P, might play a role. The glp genes are also involved in the catabolism of this molecule during the metabolism of lipids and growth under anaerobic conditions. As genes involved in lipid metabolism are not overrepresented in the set of genes identified as upregulated, this metabolic pathway does not seem to be a likely explanation for the overexpression of the glp genes. Given our results, a role of the glp genes in anaerobic respiration would be a more likely explanation. This would require further investigation.
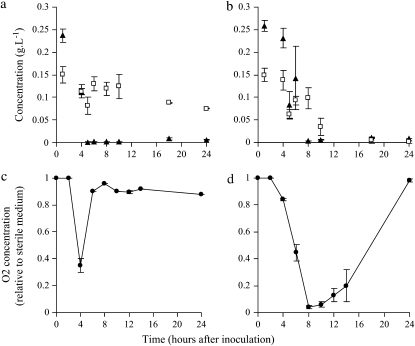
As predicted (Friesen et al. 2004), measured in a similar experiment (Spencer et al. 2007), and suggested by the microarray results, glucose concentration decreases more rapidly in SS than in FS cultures during growth in a single batch (Figure 4, a and b). Moreover, it seems that, in FS cultures, acetate concentration is completely consumed after 18 hr while in SS cultures there is still 0.08 g · liter −1 of acetate after 24 hr (Figure 4, a and b). This result suggests that the SS type is unable to efficiently switch to acetate consumption after exhausting the glucose.
Figure 4.—
Change in oxygen, glucose, and acetate concentrations during growth of SS and FS in the evolution environment. (a and b) Glucose (solid triangles) and acetate (open squares) concentration for SS and FS cultures. (c and d) Oxygen concentration during growth of SS and FS, respectively. Standard errors are indicated.
Oxygen saturation, relative to a sterile medium, was measured in SS and FS cultures (Figure 4, c and d). In full concordance with the microarray results, the dissolved oxygen level decreases during exponential growth in the evolution environment and decreases to a lower level in FS. In FS culture, oxygen level remains very low until both glucose and acetate are exhausted and then increases to the level observed in a sterile culture after 24 hr. In SS culture, oxygen level quickly increases once glucose is depleted and remains ∼0.9 until the end of the 24-hr growth period. The fact that at the end of the 24-hr growth period the oxygen level is 1 in FS and 0.9 in SS may indicate that SS are trying but are unable to efficiently consume acetate.
To investigate whether carbon and oxygen availability are limiting the growth, FS and SS types were inoculated into the evolution environment. After 4 hr and 30 min, either one of the two carbon sources (glucose and acetate) and/or extra aeration were added to the medium (Figure 5). The results show that (1) growth is often improved by extra aeration, indicating that oxygen availability may be a limiting resource in the evolution environment; (2) addition of glucose improves the growth in both types, indicating that glucose is also a limiting resource; (3) aeration only did not improve the growth of the SS type, indicating that, for SS type, carbon is the main limiting resource; (4) aeration only improved the growth of the FS type, indicating that in FS both oxygen and carbon are limiting; and (5) in both FS and SS, acetate addition did not immediately improve growth, indicating that acetate is not a strongly limiting resource. Oxygen is not considered a limiting resource in most evolution experiments. This may be because most E. coli evolution experiments are performed in flasks with low glucose concentration (Lenski et al. 1991; Rosenzweig et al. 1994; but see Friesen et al. 2004) while this one was performed in test tubes with high glucose concentration. Even with constant agitation, oxygenation is probably slower in test tubes than in flasks. Transcription profiling was performed in flasks and oxygen was still identified as a potentially limiting resource by the microarray experiment. We measured O2 saturation in both flasks and test tubes. Although oxygen level was higher in flasks than in test tubes, oxygenation in both FS and SS flask cultures decreased to levels that were substantially lower than in sterile medium (as low as 0.86 and 0.69 for SS and FS cultures, respectively; data not shown).
Figure 5.—
Growth curves of (a) FS and (b) SS in the evolution environment. Four hours and 30 min after inoculation (indicated by the arrow), aeration and carbon content of the cultures were manipulated. Dotted lines and open symbols indicate extra aeration, and continuous lines and solid symbols indicate no extra aeration. Circles indicate that no carbon source was added after 4 hr and 30 min, squares indicate acetate addition, and diamonds indicate glucose addition.
DISCUSSION
Models of adaptive diversification based on the theory of evolutionary branching predict that, under competition for limiting resources, a population would experience two different evolutionary regimes. In the first phase, evolution is directional. This directional phase ends in a regime of disruptive selection generated by frequency-dependent competitive interactions that subsequently lead to diversification of the population into two types, each exploiting the available limiting resources in different ways (Dieckmann and Doebeli 1999). These two evolutionary phases have been documented in a 1000-generation evolution experiment with E. coli (C. C. Spencer, J. Tyerman, M. Bertrand and M. Doebeli, unpublished results). In this study, we aimed to investigate the metabolic changes associated with this adaptive diversification by observing differences in the global transcription profile between two derived types and their common ancestor.
Focalizing on a single microcosm, in terms of both number of genes and diversity of metabolic functions, the changes in gene expression that occurred during the evolution experiment seem to involve mainly differences between the two derived types (pooled and considered as a single type D) and the ancestor. These differences are probably due to changes that have occurred prior to the diversification event and may thus reflect the directional adaptation to the environment predicted by adaptive diversification theory. During the directional evolutionary phase, the ancestral lineage probably improved translation efficiency, glucose uptake capacity via the mal/lamB genes, and survival rate during the stationary phase through the upregulation of various transporters and deleted part of the rbs operon. It is interesting to see that most of these metabolic functions have been implied in adaptation processes in previous E. coli experimental studies (see results). It is also striking that, after evolving in very similar environments (minimal medium supplemented with glucose, homogenized liquid culture, and identical temperature) from the same ancestral clone, the mal/lamB genes are in some cases strongly upregulated (Pelosi et al. 2006) and in others strongly downregulated (our study). This may illustrate that “fine tuning” the adaptation in very similar environments may result in dramatic differences in gene expression.
The other differences in gene expression observed during the evolution experiment involve mainly the two evolved types, FS and SS. They indicate an upregulation of the entire TCA cycle, of acetate consumption and anaerobic respiration genes in FS; and of acetate excretion in SS. These modifications are of special interest for the study of diversification because they may result from the disruptive selection forces responsible for the adaptive diversification. As theoretically predicted (Friesen et al. 2004) and experimentally assessed in a previous experiment (Spencer et al. 2007), our results indicate that the two derived types differ in how they consume the two available carbon sources: SS consumes glucose at a fast rate while FS consumes, more slowly, both glucose and acetate. This result is in full agreement with the notion that competition for carbon sources can be a major driver of diversification and result in the specialization of the derived types for differential usage of the available carbon sources. Despite the fact that SS is able to grow on acetate as the only carbon source (data not shown), it seems that, after depleting the glucose, SS is unable to switch to acetate consumption, as indicated by our acetate concentration measures (Figure 4). This inability must be linked to environmental conditions. Gene expression comparisons, oxygen measurements, and growth curve assays indicate that oxygen may be a limiting resource in the evolution environment. As oxygen limitation is known to repress the glyoxylate shunt, a bypass of the TCA cycle required to grow on acetate (White 2000), this factor may have played an important role in the evolution of the difference in acetate consumption by FS and SS.
During the time course of the evolution experiment, it seems that both carbon (especially glucose) and oxygen were limiting resources. However, limitation operates differently for these two resources: a fixed amount of carbon is provided every 24 hr (in a typical batch culture resource) while oxygen probably diffuses in the environment at a rate lower than the maximum consumption rate of the bacteria. In this situation, competition for both carbon and oxygen may have played a role in the diversification event. The relative importance of these two factors is unknown. We can imagine two scenarios, which would require further investigation implying direct competition between FS and SS to explain the diversification and frequency-dependent coexistence of FS and SS at the end of the evolution experiment. First, it is possible that, as previously suggested (Friesen et al. 2004), competition for carbon resources may have led to the specialization of one ecotype (SS) for fast glucose consumption (maximizing the growth rate on glucose) and another (FS) for an efficient carbon consumption (maximizing the yield on both glucose and acetate), oxygen limitation being a side factor involved only in the specialization for an efficient carbon consumption by FS. This would be the case if, when FS and SS are competing, glucose is completely depleted before oxygen becomes a limiting resource. Second, it is also possible that competition for oxygen may have been the primary factor leading to the diversification event and to the specialization of one ecotype (SS) for growth when oxygen is not limiting and of the other (FS) for growth when oxygen is limiting. This would be the case if oxygen becomes limiting when there is still glucose available in the environment and if, in this situation, FS grows faster than SS.
Our results are consistent with theoretical expectations (Dieckmann and Doebeli 1999) and previous experimental work (Spencer et al. 2008), indicating that adaptive diversification is the result of a directional selective pressure turning disruptive due to competition for limiting resource. During the course of the evolution experiment, which resulted in sympatric diversification into two coexisting bacterial ecotypes, the transcription of numerous genes was increased. Most of these modifications likely occurred before the diversification event during the directional adaptation to the evolution environment. Preceding the diversification event, the ancestral lineage probably improved translation efficiency, glucose uptake capacity via the mal/lamB genes, and survival rate during the stationary phase via the upregulation of various transporters and deleted part of the rbs operon. The diversification event resulted in the two ecotypes coexisting via frequency-dependent competition. Our results indicate that this diversification event likely results from competition for both carbon and oxygen resources. The resulting ecotypes use different strategies to exploit these resources, with one type (SS) being specialized for fast glucose consumption, probably when oxygen is not limiting, and the other type (FS) being adapted to efficient consumption of both glucose and acetate, even when oxygen is limiting.
Acknowledgments
We thank Minghui Yang for technical assistance; Manjeet Bains and Reza Falsafi for help with the microarray scanner; Linda Hanson and Tony Farrell for lending us the Microx-TX; Neeloffer Mookherjee, Joerg Overhage, Jennifer Gardy, and Tatiana Giraud for helpful discussions; and Jeffrey Lawrence and two anonymous reviewers for helpful comments. Michael Doebeli was supported by the James S. McDonnell Foundation (United States) and by the National Sciences and Engineering Research Council (Canada).
Microarray platform and data from this article have been deposited in the NCBI's Gene Expression Omnibus (GEO) data bank (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO accession no. GPL5767 and GEO series accession no. GSE8965.
References
- Cooper, T. F., D. E. Rozen and R. E. Lenski, 2003. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc. Natl. Acad. Sci. USA 100 1072–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, V. S., D. Schneider, M. Blot and R. E. Lenski, 2001. Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of Escherichia coli B. J. Bacteriol. 183 2834–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Death, A., L. Notley and T. Ferenci, 1993. Derepression of LamB protein facilitates outer-membrane permeation of carbohydrates into Escherichia coli under conditions of nutrient stress. J. Bacteriol. 175 1475–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann, U., and M. Doebeli, 1999. On the origin of species by sympatric speciation. Nature 400 354–357. [DOI] [PubMed] [Google Scholar]
- Dieckmann, U., H. Metz, M. Doebeli and D. Tautz (Editors), 2004. Adaptive Speciation. Cambridge University Press, Cambridge, UK/London/New York.
- Elena, S. F., and R. E. Lenski, 2003. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat. Rev. Genet. 4 457–469. [DOI] [PubMed] [Google Scholar]
- Friesen, M. L., G. Saxer, M. Travisano and M. Doebeli, 2004. Experimental evidence for sympatric ecological diversification due to frequency-dependent competition in Escherichia coli. Evolution 58 245–260. [PubMed] [Google Scholar]
- Herring, C. D., A. Raghunathan, C. Honisch, T. Patel, M. K. Applebee et al., 2006. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat. Genet. 38 1406–1412. [DOI] [PubMed] [Google Scholar]
- Heyer, L. J., S. Kruglyak and S. Yooseph, 1999. Exploring expression data: identification and analysis of co-expressed genes. Genome Res. 9 1106–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokamp, K., F. M. Roche, M. Acab, M. E. Rousseau, B. Kuo et al., 2004. ArrayPipe: a flexible processing pipeline for microarray data. Nucleic Acids Res. 32 W457–W459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassen, R., and P. B. Rainey, 2004. The ecology and genetics of microbial diversity. Annu. Rev. Microbiol. 58 207–231. [DOI] [PubMed] [Google Scholar]
- Kurlandzka, A. A., R. F. Rosenzweig and J. Adams, 1991. Identification of adaptive changes in an evolving population of Escherichia coli: the role of changes with regulatory and highly pleiotropic effects. Mol. Biol. Evol. 8 261–281. [DOI] [PubMed] [Google Scholar]
- Leek, J. T., E. C. Monsen, A. R. Dabney and J. D. Storey, 2006. EDGE: extraction and analysis of differential gene expression. Bioinformatics 22 507–508. [DOI] [PubMed] [Google Scholar]
- Lenski, R. E., M. R. Rose, S. C. Simpson and S. C. Tadler, 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138 1315–1341. [Google Scholar]
- Mikkola, R., and C. G. Kurland, 1992. Selection of laboratory wild-type phenotype from natural isolates in Escherichia coli in chemostats. Mol. Biol. Evol. 9 394–402. [DOI] [PubMed] [Google Scholar]
- Notley-McRobb, L., and T. Ferenci, 1999. The generation of multiple co-existing mal-regulatory mutations through polygenic evolution in glucose limited populations of Escherichia coli. Environ. Microbiol. 1 45–52. [DOI] [PubMed] [Google Scholar]
- Oh, M.-K., L. Rohlin, K. C. Kao and J. C. Liao, 2002. Global expression profiling of acetate-grown Escherichia coli. J. Biol. Chem. 277 13175–13183. [DOI] [PubMed] [Google Scholar]
- Paley, S. M., and P. D. Karp, 2006. The Pathway Tools cellular overview diagram and Omics Viewer. Nucleic Acids Res. 34 3771–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi, L., L. Kuhn, D. Guetta, J. Garin, J. Geiselmann et al., 2006. Parallel changes in global protein profiles during long-term experimental evolution in Escherichia coli. Genetics 173 1851–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey, P. B., and M. Travisano, 1998. Adaptive radiation in a heterogeneous environment. Nature 394 69–72. [DOI] [PubMed] [Google Scholar]
- Riley, M., T. Abe, M. B. Arnaud, M. K. B. Berlyn, F. R. Blattner et al., 2006. Escherichia coli K-12: a cooperatively developed annotation snapshot—2005. Nucleic Acids Res. 34 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig, R. F., R. R. Sharp, D. S. Treves and J. Adams, 1994. Microbial evolution in a simple unstructured environment: genetic differentiation in Escherichia coli. Genetics 137 903–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, D. E., and R. E. Lenski, 2000. Long-term experimental evolution in Escherichia coli. VIII. Dynamics of a balanced polymorphism. Am. Nat. 155 24–35. [DOI] [PubMed] [Google Scholar]
- Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang et al., 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34 374–378. [DOI] [PubMed] [Google Scholar]
- Saier, M. H., T. M. Ramseier and J. Reizer, 1996. Regulation of carbon utilization, pp. 1325–1329 in Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, edited by F. C. Neidhart. American Society for Microbiology, Washington, DC.
- Spencer, C. C., M. Bertrand, M. Travisano and M. Doebeli, 2007. Adaptive diversification in genes that regulate resource use in Escherichia coli. PLoS Genet. 3 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer, C. C., J. Tyerman, M. Bertrand and M. Doebeli, 2008. An experimental test of evolutionary branching. Proc. Natl. Acad. Sci. USA (in press).
- Storey, J. D., and R. Tibshirani, 2003. Statistical significance for genome-wide studies. Proc. Natl. Acad. Sci. USA 100 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey, J. D., J. Y. Dai and J. T. Leek, 2007. The optimal discovery procedure for large-scale significance testing, with applications to comparative microarray experiments. Biostatistics 8 414–432. [DOI] [PubMed] [Google Scholar]
- Treves, D. S., S. Manning and J. Adams, 1998. Repeated evolution of an acetate-crossfeeding polymorphism in long-term populations of Escherichia coli. Mol. Biol. Evol. 15 789–797. [DOI] [PubMed] [Google Scholar]
- Tyerman, J., N. Havard, G. Saxer, M. Travisano and M. Doebeli, 2005. Unparallel diversification in bacterial microcosms. Proc. Biol. Sci. 272 1393–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyerman, J., C. C. Spencer, M. Bertrand and M. Doebeli, 2008. Experimental demonstration of ecological character displacement in Escherichia coli. BMC Evol. Biol. (in press). [DOI] [PMC free article] [PubMed]
- White, D., 2000. The Physiology and Biochemistry of Prokaryotes, Ed. 2. Oxford University Press, Oxford.