Abstract
Some mutations arise in association with a potential sequence donor that consists of an imperfect direct or reverse repeat. Many such mutations are complex; that is, they consist of multiple close sequence changes. Current models posit that the primer terminus of a replicating DNA molecule dissociates, reanneals with an ectopic template, extends briefly, and then returns to the cognate template, bringing with it a locally different sequence; alternatively, a hairpin structure may form the mutational intermediate when processed by mismatch repair. This process resembles replication repair, in which primer extension is blocked by a lesion in the template; in this case, the ectopic template is the other daughter strand, and the result is error-free bypass of the lesion. We previously showed that mutations that impair replication repair can enhance templated mutagenesis. We show here that the intensity of templated mutation can be exquisitely sensitive to its local sequence, that the donor and recipient arms of an imperfect inverse repeat can exchange roles, and that double mutants carrying two alleles, each affecting both templated mutagenesis and replication repair, can have unexpected phenotypes. We also record an instance in which the mutation rates at two particular sites change concordantly with a distant sequence change, but in a manner that appears unrelated to templated mutagenesis.
TEMPLATED mutations are initiated when a DNA primer strand dissociates from its cognate template and anneals with a fragment of complementary sequence in an ectopic template. The relocated primer may be extended, acquiring a short sequence that is not fully complementary to the original template, and then reanneal with its cognate template. If the acquired noncomplementary bases escape correctly oriented proofreading and mismatch repair, mutations will result. Lynn Ripley (1982) described two models for templated mutagenesis based on imperfect palindromic repeats, in one of which the ectopic template was in the other parental strand and in the other of which the ectopic template was in the daughter strand itself. Examination of other mutations added imperfect direct repeats as mutagenic templates (Ripley et al. 1986; Shinedling et al. 1987). Templated mutations are usually invoked when they simultaneously acquire multiple changes for which a plausible donor template can be found. Despite the intrinsic fascination of templated complex mutations and their potential importance for certain evolutionary paths and some human genetic disorders, the enzymology that creates them has not been characterized.
Template switching also occurs in a mutation-avoiding mode called replication repair. In the canonical model for replication repair (Fujiwara and Tatsumi 1976; Higgins et al. 1976), a primer strand whose extension has been blocked by DNA damage switches to the other daughter strand as a template, extends briefly, and then switches back to its cognate template, thus accurately bypassing the damaged site.
In 1987, a mechanism for surviving DNA damage was described in bacteriophage T4, which was distinct from and additive to the classical T4 DNA recombination-repair and excision-repair systems (Wachsman and Drake 1987). This system was defined by mutant alleles of two T4 genes, 32 (encoding gp32, the “SSB” protein that binds to single-stranded DNA) and 41 (encoding gp41, the main replicative DNA helicase). Subsequent enzymological analyses using an eight-protein T4 DNA replication system in vitro showed that strand switching could indeed be promoted by DNA damage in the template strand and that the resulting replication repair was severely compromised when the mutant gp32 and gp41 proteins replaced their wild-type counterparts (Kadyrov and Drake 2003). Then, somewhat surprisingly, an alternative T4 system of replication repair was discovered in vitro in which gp32 and gp41 were replaced by the classical T4 recombinase UvsX and a different T4 DNA helicase, Dda (Kadyrov and Drake 2004). To date, these are the only genetically and enzymologically well-defined replication-repair systems.
Because both replication repair and the genesis of many complex mutations depend upon template switching, we tested whether the repair-defective alleles of genes 32, 41, and uvsX perturbed templated mutagenesis (Schultz et al. 2006). All three tested alleles displayed a general, nonspecific mutator activity that included templated mutagenesis, for which the mutator factor was particularly strong with the mutant alleles 32mms or 41uvs79. Thus, these defects in replication repair enhance templated mutagenesis.
These studies were facilitated by the availability of a hotspot of templated mutation in the T4 rI gene, although other more sporadic templated mutations were also observed. Here, we used this system to further explore templated mutagenesis in T4. We conducted a test to inquire whether the donor site for the hotspot was also a recipient site for the reciprocal templated mutation, and we examined rates of templated mutagenesis in two combinations of the three previously tested mutator mutations. Several surprising results ensued.
MATERIALS AND METHODS
Most of our bacteriophage T4 and Escherichia coli strains and methods have been described (Schultz et al. 2006). The T4 double-mutant 32mms 41uvs79 was constructed in an earlier millennium (Wachsman and Drake 1987). The T4 double-mutant gp32mms uvsXam (where uvsXam = uvsXam64am67) was constructed by recombination using a 10:1 ratio of uvsXam to 32mms. Progeny were screened first on E. coli Tab32, which does not support the growth of 32mms, and the uvsXam allele was then scored by DNA sequencing, which was also used to confirm the further genotypes of both double mutants.
The pseudo-wild-type (PsWT) replacement AGC → TCA at 145–147 and the strains carrying TAG → CGC at 171–173 or TAGT → CTGA at 171–174 were constructed as follows. A PCR product was obtained containing the wild-type rI gene using the downstream primer 5′-AATCAAATCTGGCAACT-3′ and the upstream primer 5′-TTATGACAGCTCGATT-3′. The PCR consisted of a 1-min preheating step followed by 30 cycles of 1 min at 94°, 1 min at 46°, and 1 min at 72° followed by an extension time of 10 min at 72° using Taq DNA polymerase (Invitrogen, Carlsbad, CA). This product was cloned into the plasmid vector pCR2.1-TOPO using the TOPO TA cloning kit (Invitrogen). The plasmid with the rI insert was transformed into One Shot TOP10 competent E. coli cells (Invitrogen). Plasmids were purified by miniprep (QIAGEN, Valencia, CA). Using the GeneTailor site-directed mutagenesis system (Invitrogen), the plasmids were methylated and then amplified in a reaction with the primers shown in Table 1 containing the mutant targets. The product of this reaction is linear double-stranded DNA containing the desired alteration. This DNA was then transformed into E. coli DH5α-TI cells (Invitrogen), which circularize the linear DNA. To rescue the introduced alleles from a plasmid into T4, log-phase DH5α-TI cells carrying the desired donor plasmid were infected with T4 at a phage/cell ratio of ∼10 and lysis was completed with chloroform at 40 min. The lysate was then plated on E. coli BB cells. For an expected r phenotype, the infecting T4 was r+ and r plaques were isolated. For an expected r+ phenotype, the infecting T4 carried an rI amber mutation (G → T at position 241) and r+ plaques were isolated. The desired genotypes were confirmed by sequencing.
TABLE 1.
Primers
| Construct | Expected phenotype | Direction | Primers |
|---|---|---|---|
| AGC → TCA at 145–147 | r+ | 5′ → 3′ | 127-ATGCTTTTTGAAAATAAATCAGTAGAATCGTC-158 |
| 3′ → 5′ | 116-CCGTGCAAATATACGAAAAACTTTTATTT-144 | ||
| TAG → CGC at 171–173 | r+ | 5′ → 3′ | 151-GAATCGTCTGAACAATTCTACGCTTTTATGAGAAC-185 |
| 3′ → 5′ | 141-ATTTTCGCATCTTAGCAGACTTGTTAAGAT-170 | ||
| ATAG → CTGA at 171–174 | r | 5′ → 3′ | 151-GAATCGTCTGAACAATTCTACTGATTTATGAGAAC-185 |
| 3′ → 5′ | 141-ATTTTCGCATCTTAGCAGACTTGTTAAGAT-170 |
The introduced sequence change is shown both in the first column and in the top of each pair of primers, the latter underlined.
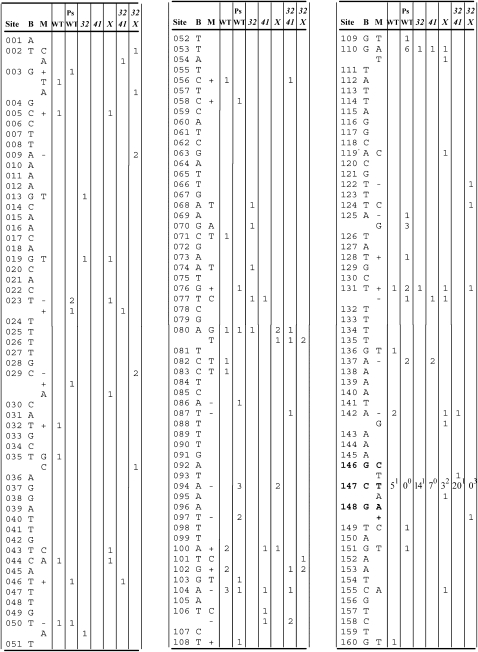
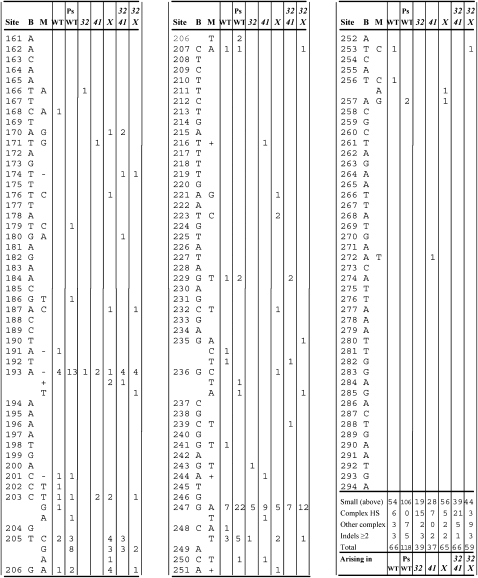
T4 r mutants produce large, sharp-edged plaques relative to the wild type (except for leaky mutants, which produce an intermediate phenotype). About three-fourths of the r mutants detected on E. coli BB cells or on K12 strains arise in the rI gene, which is a useful mutation reporter because it is not involved in DNA metabolism, displays a mutant phenotype with many missense mutations, and, at 294 bp, is well suited to DNA sequencing and for detecting mutational warm spots as well as hotspots. To grow stocks to be screened for r mutants, T4 strains were first plated on BB cells and individual plaques were recovered. These ministocks were then plated at ∼500 plaques/plate and the plates were screened for r plaques, plating wild type, 32mms, 41uvs79, and 32mms 41uvs79 on BB cells and uvsXam and 32mms uvsXam on CR63 suI + cells. Subsequent DNA sequencing and calculation of mutation rates have been described (Schultz et al. 2006). Sequencing identifies which r mutants are rI, and the rI/r ratios for the seven genotypes wild type, PsWT, 32mms, 41uvs79, uvsXam, 32mms 41uvs79, and 32mms uvsX were 66/88, 118/146, 39/65, 37/47, 65/93, 66/83, and 59/71, respectively. All mutation rates are per genome replication under the geometrical model.
RESULTS AND DISCUSSION
A QP mutational hotspot:
The first use of the T4 rI gene as a mutation reporter (Bebenek et al. 2001) revealed a hotspot of complex mutations consisting of GCG → CTA replacements at positions 146–148 (Schultz et al. 2006). These mutations were associated with a quasi-palindrome (QP; an imperfect inverse repeat, usually with a central spacer between the arms of the palindrome). Extending the canonical models of Ripley (1982), QP-mediated mutations might arise in three different ways (Figure 1). The top two pathways in Figure 1 involve primer melting from the cognate template strand, annealing with an ectopic template strand (the other parental strand in the top pathway, an earlier region of the same primer strand in the middle pathway); primer extension; and, finally, a return to the cognate template strand, thus introducing a sequence mismatch. The resulting mismatch must fall sufficiently far from the primer terminus to escape proofreading.
Figure 1.—
Models for mutations templated by quasi-palindromes. In the top two pathways (intermolecular and intramolecular template switching), donor sequences are blue, GCG target sequences are boldface black letters, and resulting mutations are red letters. In the bottom pathways (mismatch repair), donor and target sequences are reciprocally related and are both blue letters, while mutant sequences are red letters.
The bottom pathway of Figure 1 involves primer-strand melting and self-annealing of the full arms of the QP followed by DNA mismatch repair, which excises the mismatch within the stem and fills the gap using the other half of the stem as a template. When the primer strand reverts to full annealing with the cognate template strand, either of two possible mismatches are formed. Whether this pathway occurs in T4 is unclear. Mismatches produced by T4 polymerase insertion errors and escaping proofreading are subject to subsequent DNA mismatch repair in most (but not all) cellular organisms, but phage T4 seems to lack such mismatch repair as judged mainly by failure to observe the corresponding mutator mutations that would result from its inactivation (Drake and Ripley 1994). However, mismatches in T4 DNA that are formed by recombination are subject to a different kind of mismatch repair, provided that the mismatch involves an indel and thus forms a looped structure very close to the recombinational junction, in which case the looped-out bases are preferentially removed (Shcherbakov and Plugina 1991 and references therein; Shcherbakov et al. 1995). Thus, the substrate in the bottom pathway of Figure 1 might be processed by this kind of mismatch repair or might not because it has a loop–loop configuration rather than a loop on only one strand. However, when the “repaired” (homogenized) QP reannealed with its cognate template (not shown in Figure 1), one or the other mismatch would be recreated, but might be efficiently rerepaired to the wild type.
The rI complex hotspot and its wild-type and mutated DNA and amino-acid sequences are shown in Figure 2. (In all experiments to date, the additional central CGT … ACA has not contributed to QP mutations.) The apparent donor sequence whose complement CTA appears at 146–148 is TAG at 171–173. Not all complex hotspot mutations at 146–148 are GCG → CTA. Instead, some are GCG → ACTA (8 of 57 to date). As speculated previously (Schultz et al. 2006), the ACTA replacements could result if a primer strand that had elongated on the other parental strand then reannealed to the cognate template with its terminal TTTT out of register with the template AAAA, leaving one T unpaired. T4 A·T-runs are strongly prone to such single-base slippage mutations (Streisinger and Owen 1985).
Figure 2.—
The rI QP hotspot region, its potential mutations, and their coded amino acids. (A) The middle sequence is that of the wild type with the two arms of the QP underlined and separated by a CGTCTGAACA spacer. The noncomplementary three bases in each arm (at 146–148 and 171–173) are in larger boldface letters. (B) The coding consequences of the mutations with replacements in boldface letters. (C) The sequences are the pseudo-wild type. The engineered sequence in the left site now directs the mutation that would be introduced in the right site. (D) Coding at the left site remains unchanged while the templated mutation at the right site introduces a chain-termination (CT) mutation.
Testing bidirectionality of transfer:
Results obtained in E. coli suggest a bias in the preferred direction of mutagenic templating between the two potential donor/recipient elements of a QP (Trinh and Sinden 1991; Rosche et al. 1997; Viswanathan et al. 2000; Yoshiyama et al. 2001). When the direction of DNA synthesis is fixed, as it is in E. coli, such polarity might reflect an underlying difference in the vulnerabilities of the leading and lagging strands or perhaps an association with the direction of transcription (Yoshiyama and Maki 2003). In T4, however, the direction of DNA synthesis at any point is variable, first, because multiple origins are used early in replication and, second, because random origins are created by recombination for most DNA replication (Kreuzer and Morrical 1994). While ∼80 GCG → CTA mutations have been observed to date at 146–148, the inverse mutation, TAG → CGC at 171–173, has never been observed. While the GCG → CTA mutation produces the protein change ser val → thr ile, the TAG → CGC mutation would produce only ser → ala (Figure 2B), which might not produce a detectable r phenotype. Thus, either of two hypotheses could explain the observed pattern of sequence transfer: (1) it is strongly nonreciprocal or (2) the TAG → CGC mutation goes undetected.
To explore this matter, the sequence at 145–148 was altered from the wild-type AGCG to the pseudo-wild-type (PsWT) TCAG (Figure 2C). As a result, the sequence at 145–148 encodes the same amino acids as does the wild type but would introduce a chain-termination mutation at 172–174 when its complement was inserted there (Figure 2D). The AGCG → TCAG replacement also reduces the length of the QP from 12 base complementarities between arms to 11 and increases the central region of noncomplementarity within the arms from 3 to 4 bp. Genotypes were constructed and tested to confirm our prior expectations about phenotypes: the replacement of TAG at 171–173 with the CGC expected to be introduced from 146 to 148 did indeed produce a wild phenotype, and the replacement of TAGT at 171–174 with the CTGA expected to be introduced from the PsWT TCAG at 145–148 did indeed present an r phenotype. Mutational spectra were then obtained using the wild-type and PsWT rI mutation-reporter genes. Unexpectedly, no complex mutations were detected in the PsWT spectrum at either 145–148 or 171–174 (Table 2). (Full descriptions of the two spectra are provided in Figures 4 and 5 and Table 4.) Because the wild-type and PsWT rI mutation rates are similar, the wild-type spectrum predicts about (118/66)(6) = 10.7 mutations at 145–148 in the PsWT spectrum, whereas the observed number was 0. If the reciprocal complex mutations arose at 171–174 at the same rate in the wild type but were invisible, then another 10.7 of detected complex mutations would be expected at 171–174 in the PsWT spectrum. These differences have P = 0.001 in a χ2 test in which genotypes were weighted in proportion to their mutation rates. We will return to this result later.
TABLE 2.
QP-associated complex mutations in the wild-type and PsWT spectra
| Sequence at 145–148 | Total rI mutations | μ(rI) | Complex mutations at 145–148 | μ(complex) at 145–148 | Complex mutations at 171–174 | μ(complex) at 171–174 |
|---|---|---|---|---|---|---|
| Wild type (AGCG) | 66 | 2.8 × 10−5 | 6 | 2.6 × 10−6 | 0 | ≤0.43 × 10−6 |
| PsWT(TCAG) | 118 | 4.0 × 10−5 | 0 | ≤0.34 × 10−6 | 0 | ≤0.34 × 10−6 |
μ, mutation rate per genome replication. “≤” values are calculated on the basis of one mutation.
Figure 4.—
T4 rI mutational spectra. Sites start with the first base of the initiation codon and end with the last base of the termination codon. B, base; M, mutation; “–,” single-base deletion; “+,” single-base duplication. Indels in a run are usually entered at the first (sometimes the last) base of the run. WT, wild type, PsWT, the silent modification at 145–148, 32, 32mms; 41, 41uvs79; X, uvsXam. CTA replacements at 146–148 are indicated at 147, where superscripts are the additional number of ACTA replacements (see text). Other mutations are described in Table 4 and Figure 5.
Figure 5.—
Other complex mutations. The numbers in the first column indicate the rI position from the start of the underlined left repeat to the end of the underlined right repeat. The second column lists the genetic backgrounds in which the mutations arose, where 32 41 = 32mms 41uvs79 and 32 X = 32mms uvsXam. The mutant sequences are in red letters above the corresponding wild-type sequence and the donor sequences are in blue letters.
TABLE 4.
Tandem duplications and deletions
| Background genotype | Tandem duplications | Deletions |
|---|---|---|
| Wild type | +31 (8–38), R = 0 | −3 (104–106), R = 0 |
| −3 (104–106), R = 0 | ||
| PsWT | +2 (55–58), R = TC | −9 (−7–8), R = GGCCTT |
| +2 (99–101), R = T | ||
| +2 (257–258), R = 0 | ||
| +130 (116–250), R = GGCAC | ||
| 32mms | +6 (169–175), R = T | −46 (−12–39), R = TAGGA |
| −258 (137–396), R = AA | ||
| 41uvs79 | −6 (104–110), R = G | |
| −98 (19–120), R = GCAC | ||
| uvsXam | +2 (55–58), T = TC | |
| +2 (55–58), T = TC | ||
| 32mms 41uvs79 | — | −2 (184–185), R = 0 |
| 32mms uvsXam | −153 (−65–96), R = TTGATAAA | |
| −153 (−65–96), R = TTGATAAA | ||
| −11 (104–115), R = A |
Numbers in parentheses indicate both the added (or deleted) bases and both repeats (R), where R = 0 means “no repeat.” The mutation adds (or deletes) one copy of the repeat.
Potential secondary structure in the rI gene:
Erlan Ramanculov kindly provided us with an estimate of secondary structure potential in the rI gene (Figure 3). The amount of internal complementarity in rI is somewhat unusual for a protein-coding gene, and the QP associated with the GCG → CTA hotspot turns out to be particularly complicated because each arm of the QP is also largely complementary to yet a different patch of sequence, which appears in Figure 3 as the left half of region III. However, this alternative region cannot act as a donor for the CTA replacement because the corresponding region is TGC rather than TAG. On the other hand, when the 145–148 TCAG PsWT sequence is in place (Figure 3, arrows in region III), that arm of the QP has considerable complementarity with the sequence shown in boldface type on the left side of region IV. If this region were acting as a donor, it would produce the mutation 149T → A with a corresponding val → gln replacement. This mutation has never been observed among ∼700 rI mutants, so either it is rarely produced or it lacks a mutant phenotype.
Figure 3.—
Potential secondary structure in the rI gene, courtesy of Erlan Ramanculov. The sequence begins in the gene upstream of rI whose TAG terminus is separated from rI by 9 intervening bases. The rI gene starts with the boldface ATG at the top of region I and ends with the boldface TGA at the base of region V. Vertical lines indicate no base, implying that the opposite base(s) would be looped out. Potential base pairs in secondary structures, which may include G·T, are joined by dots. The palindrome associated with the GCG → CTA hotspot is shown in red letters with bases 146–148 in boldface red letters and 171–173 in boldface blue letters. The engineered PsWT change at 145–148 is indicated by arrows and might be capable of an alternative pairing with the boldface sequence on the left side of region IV. The hotspot bases 205 and 247 are in boldface red letters near the base of region IV and between IV and V, respectively.
The presence of a complement to part of the PsWT sequence might suggest that interactions between the two sequences affect the ability of the PsWT sequence to donate to its intended 171–174 target, but this possibility remains speculative.
Modulation of site-specific mutation rates by a distant sequence change:
Base-pair substitution (BPS) mutation rates vary hugely but mostly inexplicably across virtually all mutational spectra (Benzer 1961; Salts and Ronen 1971; Ronen and Rahat 1976; Ronen et al. 1976). The experimental modification of site-specific mutation rates by changes in neighboring bases has been studied both in vitro (Kunkel and Soni 1988; Bebenek et al. 1993) and in vivo in T4, where Koch (1971) was the first to demonstrate changes in site-specific mutation rates when their neighboring bases were altered. At least in the cases of T4 (which lacks conventional DNA mismatch repair) and of DNA synthesis in vitro by various polymerases, the rate of formation and/or disposition of a mismatch during synthesis can reasonably be expected to vary as a function of its close neighbors, but the number of template and primer bases contacted by the polymerase and/or proofreading exonuclease is small, so the effects of more distant bases might be expected to extinguish quickly. It was therefore surprising to observe that a particular BPS increased mutation rates at sites separated from it by 11 and 12 intervening bases (Conkling et al. 1980; Sugino and Drake 1984 as recently confirmed by unpublished results of G. T. Carver and J. W. Drake).
Relative rates of mutation in the wild-type and PsWT spectra were mostly indistinguishable across classes of mutations, with three exceptions. One was the absence (described above) of either class of hotspot mutations in the PsWT spectrum. Two other differences were changes in site-specific BPS rates (Table 3). At 205, the number of T → G transversions was predicted from the wild-type spectrum to be ≤1 in the PsWT spectrum, whereas 8 were observed. At 247, the number of G → A transitions predicted by the wild-type spectrum to appear in the PsWT spectrum was (118/66)(7) = 12.5, whereas 22 were observed. (Both of these mutations appeared in most of the other five spectra, sporadically in the case of T205G and frequently in the case of G247A.) Their different representations between the wild-type and PsWT spectra have P = 0.021 and 0.136 for T205G and G247A, respectively, in χ2 tests in which genotypes were weighted in proportion to their mutation rates. The number of bases intervening between 148 and 205 is 56, and between 148 and 247 it is 92, both numbers far exceeding any simply conceived footprint of the T4 DNA polymerase. Thus, either these two apparent actions at a distance are merely sampling deviations, or they imply as-yet-unconceived aspects of replication fidelity. These two sites are in boldface red type in Figure 3 near the bottom of regions IV and V, and we could find no credible nearby templating donor in either the QP or the direct-repeat configuration, nor an inviting impact upon secondary-structure potential in the rI gene. We searched the rest of the T4 genome for possible donor sites and found one QP of size 9 for the hotspot at 205 and two direct repeats of size 10 for the hotspot at 247, all three far from the rI gene, but there is no precedent involving a complex mutation with such a high rate of templated mutagenesis from a distant donor.
TABLE 3.
BPS hotspots in the wild-type and PsWT spectra
| Sequence at 145–148 | Total rI mutations | μ(rI) | No. of 205T → G | μ(T → G) at 205 | No. of 247G → A | μ(G → A) at 247 |
|---|---|---|---|---|---|---|
| Wild type (AGCG) | 66 | 2.8 × 10−5 | 0 | ≤0.43 × 10−6 | 7 | 3.0 × 10−6 |
| PsWT(TCAG) | 118 | 4.0 × 10−5 | 8 | 2.7 × 10−6 | 22 | 7.5 × 10−6 |
μ, mutation rate per genome replication. The “≤” value is calculated on the basis of one mutation.
Determinants of replication repair and templated mutagenesis:
Replication repair is a model that invokes template switching to achieve error-free bypass of DNA lesions, for example, by the transient use of the other daughter strand as a template (Fujiwara and Tatsumi 1976; Higgins et al. 1976). Experimentally, replication repair has been demonstrated only in phage T4, where it is well defined by an epistasis group whose alleles increase sensitivity to the killing effects of diverse agents that damage DNA (Wachsman and Drake 1987) and by reconstruction in vitro using wild-type and mutant proteins of T4 DNA replication (Kadyrov and Drake 2003, 2004). Unexpectedly, both the genetic and the biochemical evidence suggests that two different epistasis groups are involved in two different but perhaps parallel biochemical pathways of replication repair.
In the first pathway, gp32 (the SSB protein, encoded by gene 32) and gp41 (the main helicase of DNA replication, encoded by gene 41) support replication repair in vivo, and certain nonlethal mutations in either gene block this pathway both in vivo and in vitro. Gp32 binds cooperatively to single-stranded DNA and can thereby enhance its activity as a template and promote either melting or annealing depending on the circumstances, while gp41 is a 5′ → 3′ DNA helicase that unwinds double-stranded DNA and thus also promotes DNA synthesis. Both proteins interact with other proteins of DNA replication and may thereby influence diverse aspects of both normal and perturbed DNA replication (Nossal 1994).
In the second pathway, a knockout mutation of the uvsX gene (encoding UvsX, a RecA-related recombinase) blocks a pathway classically called “recombination” repair because UvsX is also required for most conventional genetic recombination in T4. UvsX can also mediate replication repair in vitro. While the mutant alleles of genes 32 and 41 depress overall DNA synthesis only modestly, the uvsXam mutation depresses it strongly because the majority of replication is initiated by random recombination, leaving only origin-initiated early DNA synthesis (Kreuzer and Morrical 1994). A uvsX mutation reduces conventional genetic recombination by only about threefold, indicating the operation of other recombination pathways. Another member of the uvsX epistasis group, uvsW, encodes a DNA helicase (Webb et al. 2007). For replication repair in vitro, the UvsX protein also requires a helicase, Dda, yet a third in the T4 repertory. It is not known whether UvsW can replace Dda in vitro; a dda deletion does not alter the resistance to ultraviolet irradiation of a uvsW deletion in vivo (G. T. Carver and J. W. Drake, unpublished results).
Because both replication repair and templated mutation proceed via ectopic primer–template pairing, we previously investigated the impact of genetic defects in both pathways of replication repair upon templated mutation. The mutations 32mms, 41uvs79, and uvsXam each exhibited a modest general mutator activity using the T4 rI mutation reporter, but both 32mms and 41uvs79 exhibited strong mutator activities toward templated complex mutations, particularly at the 146–148 site (Schultz et al. 2006); the mutator effect of uvsXam was about the same for templated complex mutations as for BPSs and small indels. For ease of comparison and accessibility, we have drawn together all the relevant rI mutational spectra into Figures 4 and 5 and Table 4, where we quote our previously published wild-type, 32mms, 41uvs79, and uvsXam spectra to which we add the PsWT spectrum discussed above plus two new spectra described and discussed below. These spectra are summarized as to kind and rate of mutation in Tables 5 and 6. The mutation rates for 32mms have been revised slightly downward from the previous report as a result of additional sampling.
TABLE 5.
Kinds and rates of rI mutations
| Wild type
|
PsWT
|
32mms
|
41uvs79
|
uvsXam
|
32 + 41
|
32 + X
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | μ | N | μ | N | μ | N | μ | N | μ | N | μ | N | μ | |
| All mutations | 66 | 2.82 | 118 | 4.01 | 39 | 38.14 | 37 | 24.22 | 65 | 23.79 | 66 | 38.72 | 59 | 20.47 |
| Transitions | 21 | 0.90 | 49 | 1.66 | 10 | 9.78 | 15 | 9.82 | 29 | 10.61 | 15 | 8.80 | 23 | 7.98 |
| A·T → G·C | 5 | 0.21 | 11 | 0.37 | 2 | 1.96 | 2 | 1.31 | 14 | 5.12 | 6 | 3.52 | 5 | 1.73 |
| G·C → A·T | 16 | 0.68 | 38 | 1.29 | 8 | 7.82 | 13 | 8.51 | 15 | 5.49 | 9 | 5.28 | 18 | 6.25 |
| Transversions | 13 | 0.56 | 19 | 0.65 | 7 | 6.85 | 3 | 1.96 | 15 | 5.49 | 9 | 5.28 | 7 | 2.43 |
| A·T → T·A | 0 | ≤0.04 | 0 | ≤0.03 | 4 | 3.91 | 1 | 0.66 | 3 | 1.10 | 2 | 1.17 | 3 | 1.04 |
| A·T → C·G | 1 | 0.04 | 8 | 0.27 | 0 | ≤0.98 | 1 | 0.66 | 5 | 1.83 | 3 | 1.76 | 3 | 1.04 |
| G·C → C·G | 2 | 0.09 | 0 | ≤0.03 | 0 | ≤0.98 | 0 | ≤0.66 | 1 | 0.37 | 0 | ≤0.59 | 0 | ≤0.35 |
| G·C → T·A | 10 | 0.43 | 11 | 0.37 | 3 | 2.93 | 1 | 0.66 | 6 | 2.20 | 4 | 2.35 | 1 | 0.35 |
| BPSs | 34 | 1.45 | 68 | 2.31 | 17 | 16.6 | 18 | 11.8 | 44 | 16.10 | 24 | 14.08 | 30 | 10.4 |
| Small indels | 20 | 0.86 | 38 | 1.29 | 2 | 1.96 | 10 | 6.55 | 12 | 4.39 | 15 | 8.80 | 14 | 4.86 |
| −A·T | 11 | 0.47 | 27 | 0.92 | 1 | 0.98 | 7 | 4.58 | 6 | 2.20 | 10 | 5.87 | 8 | 2.78 |
| −G·C | 1 | 0.04 | 1 | 0.03 | 0 | ≤0.98 | 0 | ≤0.66 | 0 | ≤0.37 | 0 | 0.00 | 2 | 0.69 |
| +A·T | 4 | 0.17 | 6 | 0.20 | 1 | 0.98 | 3 | 1.96 | 5 | 1.34 | 3 | 1.76 | 1 | 0.35 |
| +G·C | 4 | 0.17 | 4 | 0.14 | 0 | ≤0.98 | 0 | 0.00 | 1 | 0.37 | 2 | 1.17 | 3 | 1.04 |
| Larger indels | 3 | 0.13 | 5 | 0.17 | 3 | 2.93 | 2 | 1.31 | 2 | 0.73 | 1 | 0.59 | 3 | 1.04 |
| Tandem duplications | 1 | 0.04 | 4 | 0.14 | 1 | 0.98 | 0 | ≤0.66 | 2 | 0.73 | 0 | ≤0.59 | 0 | ≤0.35 |
| Deletions | 2 | 0.09 | 1 | 0.03 | 2 | 1.96 | 2 | 1.31 | 0 | ≤0.37 | 1 | 0.59 | 3 | 1.04 |
| Complex mutations | 9 | 0.38 | 7 | 0.24 | 17 | 16.62 | 7 | 4.58 | 7 | 2.56 | 26 | 15.25 | 12 | 4.16 |
| Hotspot | 6 | 0.26 | 0 | ≤0.03 | 15 | 14.67 | 7 | 4.58 | 5 | 1.83 | 21 | 12.32 | 3 | 1.04 |
| Other | 3 | 0.13 | 7 | 0.24 | 2 | 1.96 | 0 | ≤0.66 | 2 | 0.73 | 5 | 2.93 | 9 | 3.12 |
32 + 41 = 32mms 41uvs79. 32 + X = 32mms uvsXam. N, no. of mutations. μ, mutation rate × 106/genome replication on the basis of 7, 7, 17, 7, 7, 7, 8, and 11 stocks, respectively, for the seven genotypes. When no mutation was observed, the value of μ is less than or equal to the rate for one mutation.
TABLE 6.
Mutation rates relative to wild-type T4
| Mutation rate relative to wild-type T4
|
|||||||
|---|---|---|---|---|---|---|---|
| Wild type | PsWT | 32mms | 41uvs79 | uvsXam | 32mms41uvs79 | 32mmsuvsXam | |
| All mutations | 1.0 | 1.4 | 13.5 | 8.6 | 8.4 | 13.7 | 7.3 |
| Transitions | 1.0 | 1.9 | 10.9 | 10.9 | 11.8 | 9.8 | 8.9 |
| A·T → G·C | 1.0 | 1.8 | 9.2 | 6.1 | 24.0 | 16.5 | 8.1 |
| G·C → A·T | 1.0 | 1.9 | 11.4 | 12.4 | 8.0 | 7.7 | 9.1 |
| Transversions | 1.0 | 1.2 | 12.3 | 3.5 | 9.9 | 9.5 | 4.4 |
| A·T → T·A | ≤1.0 | (ud) | ≥91 | ≥15 | ≥26 | ≥27 | ≥24 |
| A·T → C·G | 1.0 | ≈6 | ≤23 | ≈15 | ≈43 | ≈41 | ≈24 |
| G·C → C·G | 1.0 | ≤0.4 | ≤11 | ≤8 | 4 | ≤7 | ≤4 |
| G·C → T·A | 1.0 | 0.9 | 6.9 | 1.5 | ≈5 | 5.5 | ≈0.8 |
| BPSs | 1.0 | 1.6 | 11.4 | 8.1 | 11.1 | 9.7 | 7.2 |
| Small indels | 1.0 | 1.5 | 2.3 | 7.7 | 5.1 | 10.3 | 5.7 |
| −A·T | 1.0 | 2.0 | ≈2 | 9.7 | 4.7 | 12.5 | 5.9 |
| −G·C | 1.0 | ≈0.8 | ≤23 | ≤15 | ≤9 | ≤14 | ≈16 |
| +A·T | 1.0 | 1.2 | ≈6 | 11.5 | 10.7 | 10.3 | ≈2 |
| +G·C | 1.0 | 0.8 | ≤6 | ≤0.4 | ≈2 | 6.9 | 6.1 |
| Larger indels | 1.0 | 1.3 | 22.9 | 10.2 | 5.7 | ≈5 | 8.1 |
| Tandem duplications | 1.0 | ≈3 | ≈23 | ≤15 | ≈17 | ≤14 | ≤8 |
| Deletions | 1.0 | ≈0.4 | ≈23 | 15.3 | ≤4.3 | 6.9 | 12.2 |
| Complex mutations | 1.0 | 0.6 | 43.2 | 11.9 | 6.7 | 39.6 | 10.8 |
| Hotspot | 1.0 | ≤0.3 | 57.2 | 17.9 | 7.1 | 48.0 | 4.1 |
| Other | 1.0 | 1.9 | 15.2 | ≤5 | 5.7 | 22.9 | 24.3 |
No A·T → T·A transversions were observed in the wild type, and the entries for the other genotypes in that row are based on one mutation in wild-type T4. The value for PsWT, 0/0, is (ud), undefined.
In addition to these determinants of templated mutations, we also note a role for the T4 DNA polymerase. T4 DNA replication can be conducted either by the cognate gp43 polymerase–exonuclease or by the corresponding protein encoded by the related phage RB69, in which case the overall rate of mutagenesis is unchanged, although site-specific mutation rates show some differences (Dressman et al. 1997). With replication driven by the RB69 enzyme, GCG → CTA replacements at 146–148 were 16/79 (20%) of all rI mutations at a rate of (16/79)(4.28 × 10−6) = 8.9 × 10−7, whereas when replication was driven by the T4 enzyme, the hotspot mutations were 6/66 (9%) of all rI mutations at a rate of (6/66)(2.82 × 10−6) = 2.6 × 10−7. This difference has P = 0.141 in a χ2 test in which the genotypes were weighted in proportion to their mutation rates. In addition, various mutator RB69 polymerases show increased frequencies of the hotspot mutations (A. Bebenek, G. T. Carver and J. W. Drake, unpublished results).
Rates of templated mutagenesis in the 32mms 41uvs79 double mutant:
These two mutations occupy the same epistasis group, each single mutant exhibiting sensitivities to irradiation with ultraviolet light (defined by terminal survival slopes on semilog plots) of 1.4 relative to the wild type, while the double mutant has a relative sensitivity of 1.5 (Wachsman and Drake 1987). If the same pattern of epistasis held for the impact of these mutations on templated mutagenesis, the mutator activity of the double mutant might be similar to that of 32mms, which has the stronger mutator activity of these two mutations. On the other hand, if local melting of the primer terminus promoted templated mutagenesis, then such melting might be assisted by wild-type gp32 in the presence of mutant gp41 and/or by wild-type gp41 in the presence of mutant gp32, in which case the double mutant might show less melting than either single mutant and also less templated mutagenesis. (The same reasoning applies to any reaction to which both proteins contribute.) As noted above, the mutational spectra of these mutants are shown in Figures 4 and 5 and Table 4, and the kinds and rates of mutations are summarized in Tables 5 and 6. At the 146–148 hotspot, relative rates for 32mms, 41uvs79, and the double mutant are 57, 18, and 48 on the basis of 15, 7, and 21 mutations, respectively (Table 6). This trend is approximately maintained for the other complex mutations, where relative rates are 15, ≤5, and 23 on the basis of 2, 0, and 5 mutations, respectively. The combined relative rates are 43, 12, and 40. This pattern is much closer to simple epistasis where, however, the impact of the single mutations on templated mutagenesis is larger in 32mms than in 41uvs79, in contrast to their impacts on survival after ultraviolet irradiation. The pattern is inconsistent with any substantial reduction in templated mutagenesis in the double mutant, faulting the hypothesis of impact via local melting of the primer terminus. An alternative interpretation of this result is that the melting that initiates replication repair and templated mutagenesis is promoted neither by gp32 nor by gp41, spontaneous melting perhaps sufficing. Gp32 binds to single-stranded DNA and helps, directly or indirectly, to load several replication proteins and to stabilize the replication fork. Although gp32 does not bind gp41, it does interact with gp59 which in turn loads gp41 (Ma et al. 2004; Delagoutte and Von Hippel 2005; Xi et al. 2005; Zhang et al. 2005). Thus, when gp32 is partly dysfunctional, as in gp32mms, the primer terminus would more often be exposed, and/or the single-stranded portion of template would more often not be coated with gp32, increasing primer-terminus melting and its mutagenic consequences; the allelic state of gene 41 would not matter very much, as observed (Tables 5 and 6). However, another T4-encoded helicase, Dda, which operates in the 3′ → 5′ direction and can displace gp32 from single-stranded DNA (Byrd and Raney 2006), might also play a role in this initial step.
Rates of templated mutagenesis in the 32mms uvsXam double mutant:
These two mutations occupy different epistasis groups with respect to rates of inactivation by ultraviolet irradiation: 32mms has a relative sensitivity of 1.4 and uvsX− (the tested allele having been uvsX1) a relative sensitivity of 1.6, whereas the double mutant has a relative sensitivity of 2.1 (Wachsman and Drake 1987). It is unclear why the survival of DNA damage in T4 has two pathways of apparent replication repair exhibiting multiplicative increases in sensitivity when both are inactivated. We therefore asked whether the effects of the single mutants on templated mutagenesis were also synergistic in the double mutant (Figures 4 and 5 and Tables 4–6). They were not. The relative rates of the hotspot complex mutation for 32mms, uvsXam, and 32mms uvsXam were 57, 7, and 4, respectively, the two smaller values being indistinguishable because of sample sizes. (The relative rates for the other complex mutations are based on sample sizes too small to justify comparison.) In this double mutant, the relative rate at the complex hotspot was smaller than that of its 32mms component and resembled that of its uvsXam component alone. This observation might be explained if wild-type UvsX initiates or promotes the excess of templated mutagenesis in the 32mms mutant and this effect is blocked by wild-type gp32 but not by gp32mms. Interactions between gp32 and UvsX are well documented and can be either cooperative or competitive, depending on the parameter and experimental conditions (Villemain et al. 2000; Liu et al. 2006). For instance, when gp32 is defective, UvsX might promote the annealing of a melted primer terminus to the other parental strand, a mutagenic pathway, rather than to the other daughter strand, an antimutagenic pathway. When both proteins are defective, as in the double-mutant 32mms uvsXam, then UvsX would not promote the mutagenic pathway. Thus, UvsX may control access to each pathway.
There is also the possibility that the effect of mutations in either pathway of replication repair is not only to decrease strand switching substantially but also to allow the residuum to occur promiscuously (in the words of one insightful reviewer), thus simultaneously both reducing survival after DNA damage and increasing the rate of templated mutation.
Varieties of templated mutations:
Most of the “other” complex mutations could be modeled as templated and are illustrated in Figure 5, including those quoted from Schultz et al. (2006). They are instructive. The top section of Figure 5 involves QPs in which a noncomplementary segment internal to one arm appears to act as the donor for a mutation appearing in the corresponding part of the other arm; the hotspot mutation illustrated in Figure 2 also falls into this category. The second and third entries of this section represent reciprocal transfers exactly of the kind that we sought to detect in the case of the complex hotspot. The fourth entry of this section depicts a partial transfer: inspection of the within-arm noncomplementarities reveals that they are discontinuous: only the two single bases at the ends and the central base are noncomplementary. In the mutation, only two of these three were transferred, suggesting either that the ectopically extending primer returned to its cognate template before extending to the final base or that the third mispair was transported but was removed by proofreading. The fifth example in this section (rI region 153–245) is interesting because the putatively transferred bases are flanked by only short regions of complementarity—1 base on one side and 3 on the other—indicating that the flanking complementarities can be quite short, at least anecdotally if not frequently. (The tandem duplications and deletions in Table 4 often displayed repeats of size 2, 1, or even 0.) In addition, the donor and recipient species are not of identical length, so that the 4 bases replacing “GGA” must have been accommodated by the polymerase and extended without proofreading when bridged by only 1 or 3 correct bases. The final example in this section is interesting in two ways. First, the flanking complementarities are minimal—1 base each; no other direct or indirect donor was found in the T4 genome with two flanking complementarities. Such short patching-in sequences might represent a rare kind of event. Second, the distance between arms was large, >85 kb, compared to the 25, 7, and 76 bases in the previous three QPs. However, because replicating T4 DNA is a pool of many permuted molecules, a wandering primer in principle could find a complement in another genome at any sequence distance from its own.
The second section is a single example in which the palindrome was perfect except for a spacer between its arms and in which the spacer appears to have templated its own replacement. This kind of mutation can arise only when the ectopic template is the other parental strand (Ripley 1982; Schultz et al. 2006).
The mutations in the third section are modeled as transfers between direct repeats. Their most striking trait is the very large distance (tens of kilobases) between the repeats in two of the three examples, perhaps for the same reason as mentioned above.
The mutations in the final section are complex, but we could find few or no compelling candidate donors in the T4 genome. The first entry, AGG → CGT, has no potential direct-repeat or QP donor with two or more flanking bases at each end, but does have 36 potential GCGTC direct-repeat donors and 56 potential GACGC QP donors. Perhaps these potential donors are individually poor because of their single flanking bases but donation occurs sporadically because of their high copy number. Alternatively, the mutation may simply represent two close but independent mispairing errors. The second entry, A → GC, might be templated by four copies of the direct repeat CGGCGG in the T4 genome with two flanking bases on each side or by 11 copies of the QP CCGCCG with two flanking bases on each side. Alternatively, this mutation may have been initiated by a slippage error within a G-rich sequence that also replaced an A with a C, the details remaining forever obscure.
Eight of the mutations in Figure 5 were detected only once. The first is structurally similar to the hotspot mutation, but the other six may be anecdotal and individually improbable, surfacing only sporadically from a sea of possibilities.
As noted above, our PsWT construct produced two surprises: there were no examples of either the complex hotspot at 146–148 or the reciprocal mutation. The mutations described in Figure 5 reveal that reciprocal transfer can occur, that an unequal number of bases between donor and recipient sites is not an absolute barrier to transfer, and that a very short complementary flanking sequence is also compatible with templating. We are left with the conclusion that any of these factors, or their combination, might have reduced the frequency of transfer in the PsWT strain sufficiently to render it undetectable among 118 isolates.
All of the spectra contained indels of two or more bases (Table 4), consisting altogether of eight tandem duplications and 11 deletions. Most (14/19) of these arose in association with short, direct sequence repeats with one of the repeat components and all of the intervening bases being duplicated or deleted. Such indels are widely interpreted as arising when a primer terminus melts and reanneals with a complementary sequence nearby (behind in the case of duplications, ahead in the case of deletions). Some of the repeats are of only one or two bases, and in five cases there is no repeat. If only one base can serve as a repeat, then perhaps “no” base can also, a situation that could arise when a mispair is followed by primer-terminus melting and reannealing so that the base in the template strand that is the recipient of the wandering primer terminus is complementary to the incorrectly inserted base. (The misinsertion itself might also promote primer-terminus melting.) Of the five cases for which R = 0, two could be mediated by the frequent mispair G·T and three would require pyrimidine–pyrimidine or purine–purine mispairs. Switching to an ectopic template is key to all these errors and thus might be promoted by our gene 32, 41, and uvsX mutations; such is the case (Tables 5 and 6), although usually not as strong a one as are the complex mutations.
Mutator activities promoting other kinds of mutations:
The three single mutants and the two doubles increased rates not only of apparently templated complex mutations but also of most other kinds of mutations (Tables 5 and 6; Schultz et al. 2006). BPSs were enhanced 7- to 11-fold (slightly more for transitions than for transversions) and single-base deletions and additions were enhanced 2- to 10-fold. We have no special insights into the mechanisms involved. Unimpressive speculations include a mispair or slippage that promotes primer melting, plus transiently lower proofreading efficiency upon reannealing with the cognate template, or either direct or indirect interactions between the three mutant proteins and the polymerase that result in increased insertion errors or decreased proofreading.
Acknowledgments
We thank Erlan Ramanculov for his exploration of potential secondary structure in the rI gene and Farid Kadyrov, Matt Longley, Jeff Stumpf, and Stephanie Nick McElhinny for critical readings of the manuscript. This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences.
References
- Bebenek, A., H. K. Dressman, G. T. Carver, S. Ng, V. Petrov et al., 2001. Interacting fidelity defects in the replicative DNA polymerase of bacteriophage RB69. J. Biol. Chem. 276 10387–10397. [DOI] [PubMed] [Google Scholar]
- Bebenek, K., J. Abbotts, S. H. Wilson and T. A. Kunkel, 1993. Error-prone polymerization by HIV-1 reverse transcriptase: contribution of template-primer misalignment, miscoding and termination probability to mutational hot spots. J. Biol. Chem. 268 10324–10334. [PubMed] [Google Scholar]
- Benzer, S., 1961. On the topography of the genetic fine structure. Proc. Natl. Acad. Sci. USA 47 403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd, A. K., and K. D. Raney, 2006. Displacement of a DNA binding protein by Dda helicase. Nucleic Acids Res. 34 3020–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conkling, M. A., R. E. Koch and J. W. Drake, 1980. Determination of mutation rates in bacteriophage T4 by unneighborly base pairs: genetic analysis. J. Mol. Biol. 143 303–315. [DOI] [PubMed] [Google Scholar]
- Delagoutte, E., and P. H. von Hippel, 2005. Mechanistic studies of the T4 DNA (gp41) replication helicase: functional interactions of the C-terminal tails of the helicase subunits with the T4 (gp59) helicase loader protein. J. Mol. Biol. 347 257–275. [DOI] [PubMed] [Google Scholar]
- Drake, J. W., and L. S. Ripley, 1994. Mutagenesis, pp. 98–124 in Molecular Biology of Bacteriophage T4, edited by J. D. Karam. American Society for Microbiology, Washington, DC.
- Dressman, H., C.-C. Wang, J. D. Karam and J. W. Drake, 1997. Retention of replication fidelity by a DNA polymerase in a homeologous environment. Proc. Natl. Acad. Sci. USA 94 8042–8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara, Y., and M. Tatsumi, 1976. Replicative bypass repair of ultraviolet damage to DNA of mammalian cells: caffeine sensitive and caffeine resistant mechanisms. Mutat. Res. 377 91–110. [DOI] [PubMed] [Google Scholar]
- Higgins, N. P., K. Kato and B. Strauss, 1976. A model for replication repair in mammalian cells. J. Mol. Biol. 101 417–425. [DOI] [PubMed] [Google Scholar]
- Kadyrov, F. A., and J. W. Drake, 2003. Properties of bacteriophage T4 proteins deficient in replication repair. J. Biol. Chem. 278 25247–25255. [DOI] [PubMed] [Google Scholar]
- Kadyrov, F. A., and J. W. Drake, 2004. UvsX recombinase and Dda helicase rescue stalled bacteriophage T4 DNA replication forks in vitro. J. Biol. Chem. 279 35735–35740. [DOI] [PubMed] [Google Scholar]
- Koch, R. E., 1971. The influence of neighboring base pairs upon base-pair substitution mutation rates. Proc. Natl. Acad. Sci. USA 68 773–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer, K. N., and S. W. Morrical, 1994. Initiation of DNA replication, pp. 28–42 in The Molecular Biology of Bacteriophage T4, edited by J. D. Karam. American Society for Microbiology, Washington, DC.
- Kunkel, T. A., and A. Soni, 1988. Mutagenesis by transient misalignment. J. Biol. Chem. 263 14784–14789. [PubMed] [Google Scholar]
- Liu, J., N. Qian and S. W. Morrical, 2006. Dynamics of bacteriophage T4 presynaptic filament assembly from extrinsic fluorescence measurements of pg32-single-stranded DNA interactions. J. Biol. Chem. 281 26308–26319. [DOI] [PubMed] [Google Scholar]
- Ma, Y., T. Wang, J. L. Villemain, D. P. Giedroc and S. W. Morrical, 2004. Dual functions of single-stranded DNA-binding protein in helicase loading at the bacteriophage T4 DNA replication fork. J. Biol. Chem. 279 19035–19045. [DOI] [PubMed] [Google Scholar]
- Nossal, N. G., 1994. The DNA replication fork, pp. 43–53 in Molecular Biology of Bacteriophage T4, edited by J. D. Karam. American Society for Microbiology, Washington, DC.
- Ripley, L. S., 1982. Model for the participation of quasi-palindromic DNA sequences in frameshift mutation. Proc. Natl. Acad. Sci. USA 79 4128–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripley, L. S., A. Clark and J. G. deBoer, 1986. Spectrum of spontaneous frameshift mutations. Sequences of bacteriophage T4 rII gene frameshifts. J. Mol. Biol. 191 601–613. [DOI] [PubMed] [Google Scholar]
- Ronen, A., and A. Rahat, 1976. Mutagen specificity and position effects on mutation in T4rII nonsense sites. Mutat. Res. 34 21–34. [DOI] [PubMed] [Google Scholar]
- Ronen, A., A. Rahat and C. Halevy, 1976. Marker effects on reversion of T4rII mutants. Genetics 84 423–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche, W. A., T. Q. Trinh and R. R. Sinden, 1997. Leading strand specific spontaneous mutation corrects a quasipalindrome by an intermolecular strand switch mechanism. J. Mol. Biol. 269 176–187. [DOI] [PubMed] [Google Scholar]
- Salts, Y., and A. Ronen, 1971. Neighbor effects in the mutation of ochre triplets in the T4rII gene. Mutat. Res. 13 109–113. [DOI] [PubMed] [Google Scholar]
- Schultz, G. E., Jr., G. T. Carver and J. W. Drake, 2006. A role for replication repair in the genesis of templated mutations. J. Mol. Biol. 358 963–973. [DOI] [PubMed] [Google Scholar]
- Shcherbakov, V. P., and L. A. Plugina, 1991. Marker-dependent recombination in T4 bacteriophage. III. Structural prerequisites for marker discrimination. Genetics 128 673–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakov, V. P., L. A. Plugina and E. A. Kudryashova, 1995. Marker-dependent recombination in T4 bacteriophage. IV. Recombinational effects of antimutator T4 DNA polymerase. Genetics 140 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinedling, S., B. S. Singer, M. Gayle, D. Pribnow, E. Jarvis et al., 1987. Sequences and studies of bacteriophage T4 rII mutants. J. Mol. Biol. 195 471–480. [DOI] [PubMed] [Google Scholar]
- Streisinger, G., and J. Owen, 1985. Mechanisms of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics 109 633–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino, A., and J. W. Drake, 1984. Modulation of mutation rates in bacteriophage T4 by a base-pair change a dozen nucleotides removed. J. Mol. Biol. 176 239–249. [DOI] [PubMed] [Google Scholar]
- Trinh, T. Q., and R. R. Sinden, 1991. Preferential DNA secondary structure mutagenesis in the lagging strand of replication in E. coli. Nature 352 544–547. [DOI] [PubMed] [Google Scholar]
- Villemain, J. W., Y. Ma, D. P. Giedroc and S. W. Morrical, 2000. Mutations in the N-terminal cooperativity domain of gene 32 protein alter properties of the T4 DNA replication and recombination systems. J. Biol. Chem. 275 31496–31504. [DOI] [PubMed] [Google Scholar]
- Viswanathan, M., J. U. Lacirignola, R. L. Hurley and S. T. Lovett, 2000. A novel mutational hotspot in a natural quasipalindrome in Escherichia coli. J. Mol. Biol. 302 553–564. [DOI] [PubMed] [Google Scholar]
- Wachsman, J. T., and J. W. Drake, 1987. A new epistasis group for the repair of DNA damage in bacteriophage T4: replication repair. Genetics 115 405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, M. R., J. L. Plank, D. T. Long, T. Hsieh and K. N. Kreuzer, 2007. The phage T4 protein UvsW drives Holliday junction branch migration. J. Biol. Chem. 282 34401–34411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi, J., Z. Zhang, Z. Zhuang, J. Yang, M. M. Spiering et al., 2005. Interaction between the T4 helicase loading protein (gp59) and the DNA polymerase (gp43): unlocking of the gp59-gp43-DNA complex to initiate assembly of a fully functional replisome. Biochemistry 44 7747–7756. [DOI] [PubMed] [Google Scholar]
- Yoshiyama, K., and H. Maki, 2003. Spontaneous hotspot mutations resistant to mismatch correction in Escherichia coli: transcription-dependent mutagenesis involving template-switching mechanisms. J. Mol. Biol. 327 7–18. [DOI] [PubMed] [Google Scholar]
- Yoshiyama, K., K. Higuchi, H. Matsumura and H. Maki, 2001. Directionality of DNA replication fork movement strongly affects the generation of spontaneous mutations in Escherichia coli. J. Mol. Biol. 307 1195–1206. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., M. M. Spiering, M. A. Trakselis, F. T. Ishmael, J. Xi et al., 2005. Assembly of the bacteriophage T4 primosome: single-molecule and ensemble studies. Proc. Natl. Acad. Sci. USA 102 3254–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]