Abstract
The Eurasian common shrew (Sorex araneus L.) is characterized by spectacular chromosomal variation, both autosomal variation of the Robertsonian type and an XX/XY1Y2 system of sex determination. It is an important mammalian model of chromosomal and genome evolution as it is one of the few species with a complete genome sequence. Here we generate a high-precision cytological recombination map for the species, the third such map produced in mammals, following those for humans and house mice. We prepared synaptonemal complex (SC) spreads of meiotic chromosomes from 638 spermatocytes of 22 males of nine different Robertsonian karyotypes, identifying each autosome arm by differential DAPI staining. Altogether we mapped 13,983 recombination sites along 7095 individual autosomes, using immunolocalization of MLH1, a mismatch repair protein marking recombination sites. We estimated the total recombination length of the shrew genome as 1145 cM. The majority of bivalents showed a high recombination frequency near the telomeres and a low frequency near the centromeres. The distances between MLH1 foci were consistent with crossover interference both within chromosome arms and across the centromere in metacentric bivalents. The pattern of recombination along a chromosome arm was a function of its length, interference, and centromere and telomere effects. The specific DNA sequence must also be important because chromosome arms of the same length differed substantially in their recombination pattern. These features of recombination show great similarity with humans and mice and suggest generality among mammals. However, contrary to a widespread perception, the metacentric bivalent tu usually lacked an MLH1 focus on one of its chromosome arms, arguing against a minimum requirement of one chiasma per chromosome arm for correct segregation. With regard to autosomal chromosomal variation, the chromosomes showing Robertsonian polymorphism display MLH1 foci that become increasingly distal when comparing acrocentric homozygotes, heterozygotes, and metacentric homozygotes. Within the sex trivalent XY1Y2, the autosomal part of the complex behaves similarly to other autosomes.
MEIOTIC recombination involves breakage and rejoining of DNA between homologous chromosomes. It plays a central role in the evolution of eukaryotes generating individual genetic variation, decreasing mutational load, and ensuring the genetic unity of species (Otto and Lenormand 2002). Recombination is crucially important for the orderly segregation of meiotic chromosomes and production of balanced gametes (Roeder 1997).
Meiotic recombination has been studied extensively both genetically and cytologically. Genetic linkage studies provide precise estimates of recombination between even closely linked genes, but they require large data sets involving well-controlled crosses or well-characterized pedigree records. The frequency of chiasmata along bivalents at diakinesis–metaphase I provides an estimate of the global rate of recombination. However, for recombination mapping, basic cytological studies are limited by difficulties in identification of individual bivalents, in measuring the position of chiasmata accurately and in combining data from cells that show different degrees of condensation of the chromosomes.
Recently, new methods of cytological recombination mapping have been developed, on the basis of the localization of recombination sites along the synaptonemal complex (SC) using fluorescently labeled antibodies to MLH1, a mismatch repair protein of mature recombination nodules (Sherman and Stack 1995; Baker et al. 1996; Barlow and Hultén 1998; Anderson et al. 1999; Froenicke et al. 2002; Koehler et al. 2002; Lynn et al. 2002). So far, these methods have been used to analyze the frequency and distribution of recombination events for only two species of mammal, humans and house mice.
Here we present detailed MLH1 recombination maps for all chromosomes of a third mammal, a small insectivore, the Eurasian common shrew (Sorex araneus L.: Soricidae, Eulipotyphla) and compare our results with those obtained for mice and humans. The common shrew is a good model for such studies for several reasons:
The efficiency of MLH1 mapping depends crucially on reliable chromosome identification for each bivalent (indeed, for metacentrics, identification of the individual chromosome arms). A characteristic feature of the common shrew is the ease in which DAPI patterns along chromosomes can be revealed in SC spreads (Belonogova et al. 2006).
The common shrew and related species show impressive diversification involving chromosomal rearrangements. Robertsonian fusions and whole-arm reciprocal translocations (WARTs) appear to have been involved in speciation in the S. araneus group (Searle and Wójcik 1998; Zima et al. 1998; Basset et al. 2006) and the influence of the rearrangements on recombination may have been crucial to this, following the model of Rieseberg (2001). The common shrew itself shows some of the most remarkable chromosomal variation in mammals. To date, 68 chromosome races have been described (Wójcik et al. 2003) and the actual number of distinct races probably goes far beyond 100. While the number of autosomal arms is constant within the common shrew (FNa = 40), the diploid chromosome number (2n) varies from 20 to 33. The source of this chromosomal variation is Robertsonian fusions, almost certainly with further modification by WARTs (Searle and Wójcik 1998). This high degree of chromosomal variation within the common shrew provides plenty of opportunity to study how Robertsonian fusions and WARTs affect recombination.
The common shrew has an XX/XY1Y2 sex chromosome system. The “X” in S. araneus represents a tandem fusion between the true mammalian X and an autosome (Sharman 1956; Fredga 1970; Pack et al. 1993). Since the XY pair in mammals differs in its meiotic behavior from the autosomal bivalents (Ashley 2002), it is of value to determine the meiotic behavior of the autosomal arm of the sex trivalent in male common shrews.
The basic karyotype of the common shrew is rather similar to the human karyotype. Comparative chromosome mapping indicates that the introduction of only 18 breaks in the human karyotype generates segments that can be fused to give the karyotype of the common shrew (Ye et al. 2006).
The common shrew is one of only a small number of species for which a complete genome sequence is available (http://www.broad.mit.edu/mammals/). A genetic map is clearly of value for future comparison with the physical map.
MATERIALS AND METHODS
Animals:
Twenty-two adult male common shrews were used in this study (Table 1). They were trapped at the beginning of the breeding season in 2006 in the hybrid zone between the Oxford and Wirral chromosome races (Zima et al. 1996), which is a continuation of the Oxford–Hermitage hybrid zone (Searle 1986a). The animals were karyotyped by the analysis of bone marrow chromosome spreads prepared according to Searle (1986a) and G-band stained (Seabright 1971). Chromosome nomenclature followed Searle et al. (1991) and Searle (1993), with chromosome arms represented by italicized letters of the alphabet and bi-armed chromosomes by a sequence of two letters (the first is the long arm, the second the short arm). Simple Robertsonian heterozygotes for a metacentric and twin acrocentric chromosomes, for example ko and k, o, are described as k/o. All animals used in this study were homozygous for the chromosomes af, bc, gm, hi, pr, and tu, and had the sex trivalent de(X)/s(Y1), dv(Y2). The individual karyotypes with respect to the chromosomes j, l, k, n, o, and q are listed in Table 1. Metacentric chromosomes kq and no were characteristic of the Oxford race and ko of the Wirral race; jl was found in both races. Homozygotes for the acrocentric chromosomes k, n, o, and q were often observed in the center of the hybrid zone and can be considered to represent a zone-specific “acrocentric race” (Searle 1986a).
TABLE 1.
Karyotypes of common shrews used in this study
| Karyotypea | 2n | Race | No. of specimens | No. of cells analyzed |
|---|---|---|---|---|
| jl, kq, no | 21 | Oxford | 2 | 23 |
| jl, kq, n, o | 23 | Oxford | 1 | 35 |
| jl, k/q, n/o | 23 | Oxford | 3 | 106 |
| jl, k/q, n, o | 24 | Oxford | 2 | 54 |
| jl, k, n/o, q | 24 | Oxford | 3 | 91 |
| j/l, k, n/o, q | 25 | Oxford | 1 | 27 |
| jl, ko, n, q | 23 | Wirral | 2 | 43 |
| jl, k/o, n, q | 24 | Wirral | 2 | 53 |
| jl, k, n, o, q | 25 | Acrocentric | 6 | 206 |
| Total | 22 | 638 |
Only the variable chromosomes are given; see text.
Regarding the relationship of the races studied here, the common shrew is subdivided into several “karyotypic groups” of chromosomally related races (Searle and Wójcik 1998) and the Oxford, Wirral, and acrocentric races all belong to the “West European karyotypic group” (Searle 1984, 1986a; Searle and Wilkinson 1987; Searle and Wójcik 1998). It is reasonable to assume that these races are also closely related genically, although there are no direct data on this. The races are believed to have a common ancestry in a glacial refugium in southeastern Europe at the last glacial maximum, 20,000 years ago (Searle 1984; Bilton et al. 1998).
Here we study individuals homozygous for only metacentric chromosomes, individuals homozygous for a variety of metacentric and acrocentric chromosomes, and individuals that are simple heterozygotes for either one or two arm combinations. This range of karyotypes is commonly found elsewhere in the common shrew, often in hybrid zones between chromosomal races or other areas of Robertsonian polymorphism (Searle and Wójcik 1998). The particular situation that we have studied in Britain where two races characterized by different metacentrics are separated by a third acrocentric race has also been described in Sweden and Poland (Searle and Wójcik 1998). Therefore, we are studying a range of karyotypes that is typical for the common shrew and of wide comparative value; simple Robertsonian heterozygotes and their associated homozygotes have been found in many other mammals (Searle 1993).
Immunostaining, identification, and measurement of meiotic chromosomes:
Spermatocyte spreads were prepared from both left and right testes using the technique of Peters et al. (1997). The immunostaining protocol was similar to that of Anderson et al. (1999). The slides were incubated for 2 hr at 37° with a rabbit polyclonal antibody against rat lateral element protein SCP3 (a gift from C. Heyting) diluted to a concentration of 1:1000, a mouse monoclonal antibody to human mismatch repair protein MLH1 (Pharmingen, San Diego) at 1:50 dilution, and a human ANA-C antibody against centromeric proteins (Sigma-Aldrich, St. Louis) at a 1:100 dilution in 3% bovine serum albumin (BSA) in phosphate buffered saline (PBS). Slides were washed in 1× PBS and incubated for 40 min at 37° with donkey anti-rabbit Cy3-conjugated antibodies (Jackson, West Grove, PA) at 1:200 dilution, goat anti-mouse FITC-conjugated antibodies (Jackson) at 1:400 dilution and goat anti-human FITC-conjugated antibodies (Vector Laboratories, Burlingame, CA) at 1:100 dilution. Slides were washed with PBS, rinsed briefly with distilled water, dried, and mounted in Vectashield with DAPI (Vector Laboratories) to stain DNA and reduce fluorescence fading.
The preparations were visualized with an Axioplan 2 imaging microscope (Carl Zeiss) equipped with a CCD camera (CV M300, JAI), CHROMA filter sets, and ISIS4 image-processing package (MetaSystems GmbH). Brightness and contrast of all images were enhanced using PaintShopPro 7.0.
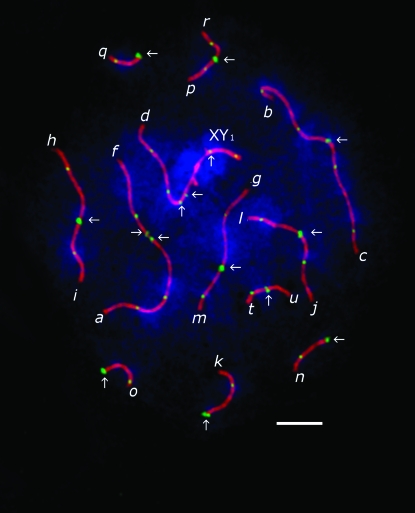
Only cells containing complete sets of chromosomes were analyzed. The number of cells studied for each karyotype is listed in Table 1. Each chromosome arm was identified by its specific DAPI pattern according to Belonogova et al. (2006). The centromere position for each SC was identified by an ANA-C focus. Although we used the same fluorochrome for detection of the ANA-C and MLH1 antibodies, ANA-C foci differed from MLH1 foci by their brighter and more diffuse staining (Figure 1). MLH1 signals were only scored if they were localized on an SC. The length of the SC of each chromosome arm was measured in micrometers using MicroMeasure 3.3 (Reeves 2001) and the positions of MLH1 foci in relation to the centromere were recorded.
Figure 1.—
SC spread from a shrew spermatocyte at pachytene, stained with DAPI (blue) and immunolabeled with antibodies to SCP3 (red), MLH1 (green), and centromere proteins (green). Bar, 5 μm. Chromosome arms (indicated by letters next to their telomeres) were identified by DAPI banding. Centromeres (indicated by arrows) differ from MLH1 foci by their brighter and more diffuse staining. Note that the centromeres on the af bivalent and on the d arm of the sex trivalent are misaligned and therefore generate weaker signals than aligned centromeres.
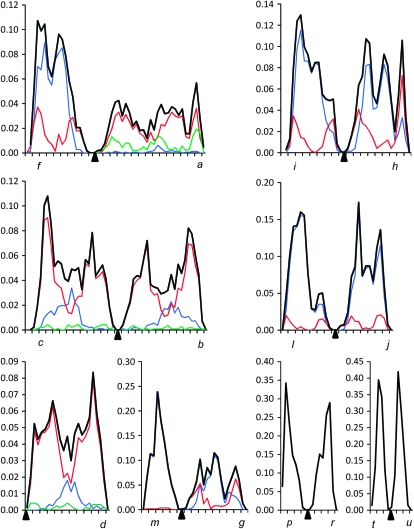
To generate recombination maps, we calculated the absolute position of each MLH1 focus multiplying the relative position of each focus by the average absolute length for the appropriate chromosome arm. These data were pooled for each arm and graphed to represent a recombination map (Figures 2 and 3).
Figure 2.—
Distribution of MLH1 foci along the arms of the Robertsonian-invariant chromosomes, i.e., those showing no Robertsonian polymorphism. One individual analyzed was a j/l heterozygote but it provided insufficient data for analysis and here we show the data from jl metacentric homozygotes. The x-axis shows the position of MLH1 foci; the marks on this axis are separated by 1 μm. Letters indicate the telomeric ends of the arms; arrows show where the centromeres are located. The y-axis indicates the frequency of MLH1 foci in each 0.5-μm interval (note the different scaling for each chromosome). The blue line shows the frequency for the arms containing a single MLH1 focus, the red line shows two MLH1 foci, and the green line shows three MLH1 foci. The black line shows the overall frequencies of MLH1 foci. Due to a very low frequency of arms containing more than one MLH1 focus per arm for the bivalents pr and tu, only the overall frequencies are shown for these bivalents.
Figure 3.—
Distribution of MLH1 foci along the arms of the Robertsonian-variable chromosomes, i.e., those showing Robertsonian polymorphism. The x-axis shows the position of MLH1 foci; the marks on this axis are separated by 1 μm. Letters indicate the telomeric ends of the arms; arrows show where the centromeres are located. The y-axis indicates the frequency of MLH1 foci in each 0.5-μm interval. The lines represent the overall frequency of MLH1 foci in each interval. The blue line shows the distribution in acrocentric homozygotes, the green line shows that in metacentric homozygotes, and the red line shows that in heterozygotes.
The absolute distances between neighboring foci were measured from the images. The relative distances between the foci across the centromere were calculated as fractions of chromosome length; relative distances within each arm were calculated as fractions of the arm length. The data on MLH1 foci for each arm were fitted to gamma distributions by a maximum-likelihood method using STATISTICA 6 (StatSoft, 2001) and the shape parameter (υ) was used as a measure of the strength of interference (De Boer et al. 2006). The STATISTICA package was also used for ANOVA, correlations, and other statistical analysis.
RESULTS AND DISCUSSION
Characteristics of the SCs:
We found a close correspondence between the average relative length of SCs compared with mitotic chromosomes (Spearman rank correlation, rs = 0.95, P < 0.001). However, several SCs were noticeably shorter (bc, d) or longer (hi, jl, k, n, tu) than would be expected on the basis of their relative mitotic lengths (Table 2). We also found substantial differences between SCs and mitotic chromosomes in the arm ratio of metacentrics. For example, arms p and t were the long arms in mitotic chromosomes pr and tu and the short arms in the SC. This confirms that it is dangerous to extrapolate from mitotic chromosome length for the identification of SCs and emphasizes the importance of using DAPI banding for this purpose.
TABLE 2.
Comparison between the lengths of synaptonemal complexes and mitotic metaphase chromosomes in the common shrew
| Absolute length of SC in μm
|
Relative length of SCa
|
Relative length of mitotic chromosomesb | % difference from expected relative length ratioc | |||
|---|---|---|---|---|---|---|
| Chromosome | Mean | SD | Mean | SD | ||
| af | 24.7 | 3.8 | 17.3 | 1.0 | 17.3 | 0.0 |
| bc | 24.9 | 4.0 | 17.5 | 1.0 | 20.5 | −14.9 |
| d | 11.1 | 1.9 | 7.8 | 0.7 | 9.4 | −17.2 |
| gm | 14.1 | 2.0 | 9.9 | 0.6 | 9.2 | 7.6 |
| hi | 17.0 | 2.7 | 11.9 | 0.8 | 10.8 | 10.1 |
| jl | 14.6 | 2.2 | 10.2 | 0.7 | 8.9 | 15.1 |
| kd | 7.7 | 1.2 | 5.4 | 0.5 | 4.5 | 20.0 |
| nd | 5.6 | 0.8 | 4.0 | 0.4 | 3.4 | 16.8 |
| od | 5.2 | 0.8 | 3.6 | 0.4 | 3.6 | 0.3 |
| pr | 7.7 | 1.1 | 5.4 | 0.4 | 5.8 | −6.9 |
| qd | 4.3 | 0.7 | 3.0 | 0.3 | 3.0 | −0.3 |
| tu | 5.8 | 0.9 | 4.1 | 0.4 | 3.4 | 19.4 |
Percentage of total autosomal SC length.
Percentage of total autosomal mitotic length (calculated from the data of Král and Radjabli 1974).
Calculated as [(relative SC length/relative mitotic chromosome length) − 1] × 100.
The data for the arms of the chromosomes showing Robertsonian variation were averaged for acrocentric homozygotes, metacentric homozygotes of various arm combinations, and heterozygotes.
The mean (± SD) total length of the autosomal SCs (including the autosomal arm d of the sex trivalent) was 142.8 ± 18.8 μm. An ANOVA revealed significant effects of individual (F1,19 = 14.1, P < 0.001) and race (F1,2 = 9.0, P < 0.001) on the variation for this trait. Studies on humans have also revealed significant individual variation (Lynn et al. 2002; Sun et al. 2004, 2005, 2006a,b). The causes of this variation are unclear but Lynn et al. (2002) suggested that allelic variation in loci encoding the proteins involved in chromosome pairing and recombination (such as SPO11, MRE11, RAD51, and DMC1) might mediate differences in SC length.
The number of MLH1 foci:
The mean (± SD) number of MLH1 foci over all autosomes (including the autosomal arm d of the sex trivalent) was 21.9 ± 2.0, with a range of 15–30 foci per cell. This is in accordance with the chiasma count per late diplotene/early diakinesis cell, estimated earlier for common shrews from the Oxford–Hermitage hybrid zone (21.8 ± 1.7, with a range between 18 and 28; Searle 1986b). We found no significant differences in number of MLH1 foci per cell between the Oxford (22.2 ± 2.0), Wirral (21.6 ± 1.9), and acrocentric (21.7 ± 1.9) shrews (F2,635 = 2.4, P = 0.09).
To estimate the recombination length of each chromosome in centimorgans, the average number of MLH1 foci for the chromosome was multiplied by 50 MU (one recombination event = 50 cM). The genetic length of each arm, calculated in this way, is shown in Table 3. We estimated the total autosomal map length for the male common shrew as 1095 cM. To estimate the total map length for the male genome we added 50 cM for the obligate crossover in the XY1 pairing region, giving 1145 cM. As the shrew genome is 2850 Mb (Vinogradov 1998), 1 cM of the shrew genetic map is equal to ∼2.5 Mb of sequence. This is rather close to the estimates obtained by MLH1 mapping for male mice (2.75; Anderson et al. 1999) but twice that for human males (1.20; Sun et al. 2006a).
TABLE 3.
Number of MLH1 foci per chromosome arm
| No. of MLH1 foci
|
Genetic length (cM) | No. of cells containing
|
||||||
|---|---|---|---|---|---|---|---|---|
| Arm | Karyotypea | Mean | SD | 0 foci | 1 focus | 2 foci | 3 foci | |
| a | af | 2.09 | 0.50 | 104.5 | 3 | 46 | 481 | 108 |
| b | bc | 1.72 | 0.49 | 86.0 | 1 | 188 | 438 | 11 |
| c | bc | 1.69 | 0.52 | 84.5 | 4 | 202 | 418 | 14 |
| d | de/s, dv | 1.83 | 0.44 | 91.5 | 0 | 128 | 493 | 17 |
| f | af | 1.14 | 0.38 | 57.0 | 6 | 534 | 98 | 0 |
| g | gm | 1.22 | 0.45 | 61.0 | 7 | 483 | 147 | 1 |
| h | hi | 1.19 | 0.43 | 59.5 | 11 | 495 | 132 | 0 |
| i | hi | 1.09 | 0.35 | 54.5 | 12 | 556 | 70 | 0 |
| j | jl | 1.06 | 0.28 | 53.0 | 6 | 561 | 44 | 0 |
| j | j/l | 1.04 | 0.19 | 52.0 | 0 | 26 | 1 | 0 |
| k | ko | 1.05 | 0.21 | 52.5 | 0 | 41 | 2 | 0 |
| k | k/o | 0.94 | 0.30 | 47.0 | 4 | 48 | 1 | 0 |
| k | kq | 0.98 | 0.23 | 49.0 | 2 | 55 | 1 | 0 |
| k | k/q | 1.06 | 0.32 | 53.0 | 4 | 143 | 13 | 0 |
| k | k | 1.06 | 0.29 | 53.0 | 4 | 299 | 20 | 1 |
| l | jl | 1.03 | 0.25 | 51.5 | 10 | 572 | 29 | 0 |
| l | j/l | 0.96 | 0.19 | 48.0 | 1 | 26 | 0 | 0 |
| m | gm | 0.96 | 0.23 | 48.0 | 31 | 602 | 5 | 0 |
| n | no | 0.96 | 0.21 | 48.0 | 1 | 22 | 0 | 0 |
| n | n/o | 1.00 | 0.09 | 50.0 | 1 | 222 | 1 | 0 |
| n | n | 1.00 | 0.14 | 50.0 | 4 | 383 | 4 | 0 |
| o | ko | 0.98 | 0.15 | 49.0 | 1 | 42 | 0 | 0 |
| o | k/o | 0.91 | 0.30 | 45.5 | 5 | 48 | 0 | 0 |
| o | no | 1.00 | 0.00 | 50.0 | 0 | 23 | 0 | 0 |
| o | n/o | 0.97 | 0.17 | 48.5 | 7 | 217 | 0 | 0 |
| o | o | 0.98 | 0.15 | 49.0 | 6 | 288 | 1 | 0 |
| p | pr | 0.94 | 0.26 | 47.0 | 42 | 593 | 3 | 0 |
| q | kq | 0.98 | 0.13 | 49.0 | 1 | 57 | 0 | 0 |
| q | k/q | 0.96 | 0.21 | 48.0 | 7 | 153 | 0 | 0 |
| q | q | 0.97 | 0.17 | 48.5 | 12 | 407 | 1 | 0 |
| r | pr | 0.90 | 0.30 | 45.0 | 64 | 573 | 1 | 0 |
| t | tu | 0.53 | 0.50 | 26.5 | 297 | 341 | 0 | 0 |
| u | tu | 0.54 | 0.50 | 27.0 | 296 | 342 | 0 | 0 |
A karyotype ko will produce a metacentric bivalent, k/o will produce a trivalent (metacentric ko and acrocentrics k and o), and k will produce an acrocentric bivalent. The same is true for other chromosomes. de/s, dv is the sex trivalent.
Altogether, 7095 autosomal SCs were analyzed (including SCs for the autosomal arm d of the sex trivalent). Of these, 51 (0.7%) lacked an MLH1 focus; these were found in 48 of the 638 cells scored (7.5%). These frequencies are within the limits of variation found in studies on mice and humans. Koehler et al. (2002) showed that 0.1% of autosomal SCs in males of four inbred mouse strains had 0 foci, while in the study of Anderson et al. (1999) involving C57BL males, the frequency of such bivalents was 4%. Sun et al. (2006b) found substantial variation in the frequency of autosomal SCs lacking MLH1 among 10 normal men, although the overall mean frequency of achiasmate autosomal bivalents was rather low (0.3%). The frequency of cells containing one or more such bivalents varied among individuals between 1 and 11%. The studies on humans and mice demonstrate that small bivalents suffer a much higher risk of recombination failure than large bivalents (Anderson et al. 1999; Koehler et al. 2002; Sun et al. 2006b). In our study, achiasmate bivalents were predominantly small metacentrics and acrocentrics; 24 were chromosome tu, 11 q, 6 o, 4 k, 3 n, 2 gm, and 1 pr. The cells containing achiasmate bivalents did not appear to be exceptional (e.g., at late pachytene or poorly stained) and did not differ significantly from the whole sample in their total SC length (147.5 ± 20.6 μm). Searle (1986b) analyzed the frequency of autosomal univalents at diakinesis–metaphase I in common shrews from the Oxford–Hermitage hybrid zone. He found a similar frequency of achiasmate bivalents as detected in this study (0.4–1.0%). Thus, although achiasmate bivalents might be expected to trigger a pachytene checkpoint leading to apoptosis (Roeder and Bailis 2000), there is no evidence for this in the common shrew.
Rather often we observed metacentric bivalents with no MLH1 foci at one arm and one or more foci in the other arm (overall frequency among metacentric bivalents, 15.4%). This was fairly common in pr and a typical feature of chromosome tu (Table 3), supporting previous chiasma data (Searle 1986b). Only 10.8% of tu bivalents had MLH1 foci on both arms. Chromosome tu is the smallest and the most ancient among the autosomal metacentrics of the common shrew (Searle and Wójcik 1998). Interestingly, it evolved by a centric shift or pericentric inversion from an acrocentric chromosome while all other autosomal metacentrics are derived from Robertsonian fusions (Wójcik and Searle 1988). Presumably the acrocentric ancestral to tu was characterized by a single chiasma. The conversion of that acrocentric into a metacentric did not lead to a requirement for separate chiasmata in each arm. These results are of interest because it is a common perception that there is a minimum requirement of one chiasma per chromosome arm for correct segregation (Dumas and Britton-Davidian 2002 and references therein).
The number of MLH1 foci that we observed per arm of chromosomes involved in Robertsonian polymorphism did not depend on the karyotype (homozygous metacentric, homozygous acrocentric, and heterozygous) (Table 3). The effect of karyotype was nonsignificant for all arms tested: k (F2,635 = 1.2, P = 0.31), n (F2,635 = 0.9, P = 0.42), o (F2,635 = 1.8, P = 0.16), and q (F2,635 = 0.7, P = 0.49). These results contrast to the chiasma data obtained by Bidau et al. (2001) and Dumas and Britton-Davidian (2002) for the house mouse, which showed a significant decrease in recombination frequency in the arms of metacentric chromosomes compared to twin acrocentrics. In the mouse the decrease in recombination frequency in the arms of metacentric chromosomes compared to twin acrocentrics was determined mainly by a reduction in double chiasmata (Bidau et al. 2001; Dumas and Britton-Davidian 2002). In the common shrew all the arms of the variable chromosomes were rather small and very rarely accommodated more than one MLH1 focus even in the acrocentric state.
We also did not find significant differences in the number of MLH1 foci for any particular chromosome arm in a metacentric, comparing between different arm combinations. Thus, arm k had a similar number of foci in kq and ko bivalents (t1,99 = 1.4, P = 0.16), and arm o had a similar number of foci in ko and no bivalents (t1,64 = 0.7, P = 0.48).
Correlation between SC length and number of MLH1 foci:
As for previous studies in mice and humans (Anderson et al. 1999; Sun et al. 2004, 2006a), we also found in common shrews a very strong positive correlation between the average length of SCs of individual chromosome arms and the number of MLH1 foci on those arms (Spearman rank correlation, rs = 0.95, P < 0.001). On the other hand, the correlation between the total length of SCs and number of MLH1 foci per cell was much weaker (rs = 0.17, P < 0.001) as found for male mice (Froenicke et al. 2002), but in contrast to the strong correlation found in human males (Lynn et al. 2002).
Crossover interference:
The occurrence of a crossover usually decreases the probability that another will occur close by. This phenomenon is called positive interference (see Jones and Franklin 2006 for a review). The strength of interference can be estimated from the distances between adjacent recombination sites. Table 4 shows the average distances between neighboring MLH1 foci within chromosome arms and across centromeres in metacentric bivalents. The absolute distance varied from 1.0 to 15.6 μm within an arm and from 1.2 to 27.1 μm across the centromere, with the transcentromere distances significantly larger (P < 0.001). These results indicate that there may be crossover interference across centromeres as well as within chromosome arms as suggested for a variety of organisms including humans (Colombo and Jones 1997; Broman and Weber 2000; Drouaud et al. 2006). There is the following expectation with transcentromere interference, which we tested for metacentrics in the common shrew: The most proximal MLH1 focus on one chromosome arm should influence the most proximal focus on the other arm, such that their distances from the centromere will be inversely correlated. We did indeed find inverse correlations in the shrew metacentrics af, bc, gm, hi, jl, and pr (r = −0.14, −0.28, −0.12, −0.23, −0.10, −0.10, respectively; P < 0.05). In other words, transcentromere interference explains 1–8% of total variance in distances between the centromeres and the most proximal foci in these bivalents. For the remaining metacentrics the correlations were not significant, but in all cases this is reasonably ascribed to small sample sizes.
TABLE 4.
The average absolute and relative distances between neighboring MLH1 foci including the υ-value (shape parameter) of the gamma distribution for the within-arm data
| Distance across centromere
|
Distance within arms
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Absolute (μm)
|
Relativea
|
Absolute (μm)
|
Relativea
|
||||||||
| Chromosome | N | Mean | SD | Mean | SD | N | Mean | SD | Mean | SD | υ |
| af | 625 | 10.5 | 3.4 | 0.42 | 0.11 | 788 | 6.4 | 2.2 | 0.42 | 0.13 | 11.1 |
| bc | 634 | 10.0 | 3.3 | 0.40 | 0.12 | 904 | 6.1 | 1.9 | 0.48 | 0.13 | 13.6 |
| d | 525 | 5.4 | 1.6 | 0.48 | 0.13 | 13.8 | |||||
| gm | 601 | 7.3 | 2.2 | 0.52 | 0.13 | 152 | 5.2 | 1.8 | 0.54 | 0.13 | 15.3 |
| hi | 621 | 8.7 | 2.6 | 0.51 | 0.13 | 199 | 5.0 | 1.5 | 0.55 | 0.13 | 14.5 |
| jl | 621 | 9.8 | 2.3 | 0.66 | 0.12 | 71 | 4.3 | 1.1 | 0.55 | 0.13 | 15.6 |
| j/l | 26 | 8.8 | 2.4 | 0.60 | 0.13 | ||||||
| ko | 42 | 8.8 | 1.7 | 0.65 | 0.11 | ||||||
| k/o | 44 | 8.3 | 2.2 | 0.64 | 0.10 | ||||||
| kq | 55 | 8.7 | 2.2 | 0.63 | 0.12 | ||||||
| k/q | 153 | 7.3 | 1.7 | 0.60 | 0.12 | ||||||
| no | 22 | 8.1 | 1.4 | 0.67 | 0.12 | ||||||
| n/o | 216 | 7.2 | 1.5 | 0.64 | 0.11 | ||||||
| pr | 533 | 5.3 | 1.2 | 0.69 | 0.12 | ||||||
| tu | 69 | 3.6 | 0.8 | 0.57 | 0.10 | ||||||
Relative interfocus distance across the centromere was calculated in terms of fractions of the chromosome length; within a chromosome arm it was calculated as a fraction of the arm length.
Transcentromere absolute distance is greater on average for the larger bivalents (Table 4). This is associated with another phenomenon, the prominent misalignment of centromeres, which is most frequently seen on large configurations such as af, bc, and the sex trivalent (Figure 1). Recombination events are thought to occur at sites of initiation of synapsis (Bishop and Zickler 2004; Henderson and Keeney 2004) and, therefore, DNA homology is required at this stage. However, synapsis may proceed in between recombination sites without a homology check (Ashley 1988; Zickler and Kleckner 1999). The larger the region that may synapse without a homology check, the greater the likelihood that prominent misalignment may occur, explaining why such misalignments tend to be observed on large configurations. As well as misalignment of centromeres, sometimes DAPI patterns between two MLH1 foci within a chromosome arm were misaligned.
The absolute distances between two foci separated by the centromere were usually larger in metacentric homozygotes for chromosomes showing Robertsonian polymorphism compared to the corresponding heterozygotes (jl, t1,645 = 2.1, P < 0.05; kq, t1,206 = 4.3, P < 0.01; no, t1,236 = 2.9, P < 0.01; although ko, t1,84 = 1.2, P = 0.12; see also Table 4). This can be interpreted as a weakening of transcentromere interference in Robertsonian heterozygotes relative to metacentric homozygotes. It is well known that discontinuity of the SC or the switching of pairing partners decreases the strength of interference (see Zickler and Kleckner 1999 for a review). This can also be interpreted in terms of the “stress relief” model of crossover interference suggested by Kleckner et al. (2004). According to this model, recombination is induced by mechanical stress. A crossover leads to local stress relief and therefore to inhibition of further crossovers nearby. Since the SC mediates the interference, its interruption at the centromeres of the twin-acrocentrics in a Robertsonian trivalent may diminish transcentromere interference in heterozygotes compared to metacentric homozygotes.
The mean intra-arm distances between adjacent MLH1 foci in the shrew varied from 4.3 to 6.4 μm, according to the chromosome arm, with greater distances for larger arms (Table 4). In the mouse, which has a similar overall MLH1 focus density as the shrew, similar mean distances between MLH1 foci (4.1–7.0 μm) and a similar between-arm variation were found for arms of comparable sizes (Anderson et al. 1999). Recently De Boer et al. (2006, 2007) estimated the strength of crossover interference in the mouse using parameters of the gamma distribution. The gamma distribution describes the probability of the distances that would occur if MLH1 focus precursors were randomly placed along the bivalent and only every nth precursor would result in a focus. The higher the n value, the stronger the interference. The n value can be estimated via the shape parameter (υ) of a gamma distribution. We determined the υ-value for which the observed frequency distribution of interfocus distances fitted best to a gamma distribution. The υ-values estimated for the chromosome arms of the male common shrew varied between 11.1 and 15.6 (Table 4) and were similar to those obtained for mouse bivalents of comparable sizes (υ = 13.7–14.4, De Boer et al. 2006; υ = 8.9–11.7, De Boer et al. 2007). These estimates suggest that the strength of crossover interference is rather conservative among mammals when arms of similar SC size and recombination density are compared.
Recombination maps of Robertsonian invariant chromosomes:
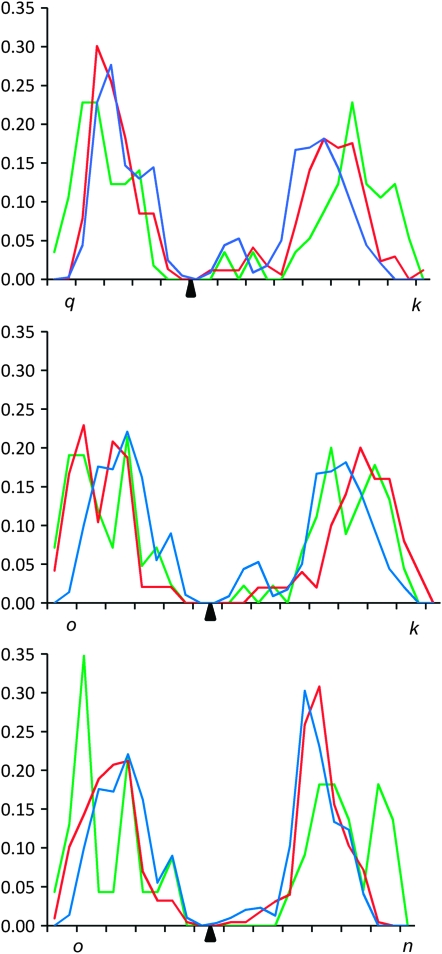
Individual distributions of MLH1 foci for each of the chromosomes not involved in Robertsonian variation differed little among representatives of the two parental races and acrocentric individuals from the center of the hybrid zone. Of all distributions cross-tested between the races (24 tests) only one test comparing values for each 0.5-μm interval gave a P-value <0.01 (between the acrocentric shrews and the Oxford shrews for bivalent tu: 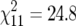 , P < 0.01). For this reason we pooled the data from all karyotypes and plotted recombination maps for each arm of the invariant bivalents (Figure 2).
, P < 0.01). For this reason we pooled the data from all karyotypes and plotted recombination maps for each arm of the invariant bivalents (Figure 2).
Bivalents in the common shrew reproduce a general distribution of recombination events that is similar across a wide variety of vertebrates such as fish (Moens 2006), birds (Pigozzi and Solari 1999; Pigozzi 2001; Calderon and Pigozzi 2006), mice (Anderson et al. 1999; Froenicke et al. 2002), and humans (Barlow and Hultén 1998; Lynn et al. 2002; Tease et al. 2002; Sun et al. 2004, 2006a): a pronounced recombination peak near to the telomere, a deficiency near to the centromere, and a bi- or a multimodal distribution along the chromosome arms. (Note that where we use the term “telomere” in this article, we always mean the structure at the distal end of a chromosome or a chromosome arm; we disregard telomeres that are situated next to the centromere in single-armed chromosomes.)
Suppression of recombination in the pericentromeric area of the mouse chromosomes has been interpreted as an effect of the blocks of centromeric heterochromatin that reside there (Froenicke et al. 2002). In the common shrew there is only a very small amount of centromeric C-heterochromatin (Schmid et al. 1982; Belonogova et al. 2006) and it appears less likely that this causes the recombination suppression.
Regarding recombination near the telomere, this was not precisely in the terminal segment. Instead the peak was usually located at 2–3 μm from the telomeres. An excess of recombination near the telomeres is apparently due to early involvement of this region in chromosome pairing (Scherthan et al. 1996; Zickler and Kleckner 1999). Recombination too close to the centromere or telomere may be suppressed as the result of natural selection. It has been shown in humans that chiasmata in such locations often lead to nondisjunction (Hassold and Hunt 2001).
The distribution of MLH1 foci along all the large arms of metacentric chromosomes, except a and h, was usually bimodal with a major peak near the distal end and the other peak in the middle of the arm or nearer the centromere. Arms a and h displayed clear trimodal distributions. The arms of the chromosomes pr and tu were too small to accommodate more than one crossover. Each of them contained one peak.
It is interesting to compare the distributions of MLH1 foci between chromosome arms of approximately equal size in metacentric bivalents. The distributions were rather similar in the arms p and r ( , P = 0.33) and drastically different between the arms b and c (
, P = 0.33) and drastically different between the arms b and c ( , P < 0.001), h and i (
, P < 0.001), h and i ( , P < 0.001), and j and l (
, P < 0.001), and j and l (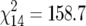 , P < 0.001).
, P < 0.001).
These comparisons show that the size of the chromosome arm is an important but not the only factor controlling the distribution of crossovers. Local hot and cold regions of recombination can significantly modulate the overall crossover distribution along a chromosome. Analysis of factors controlling local recombination rate in human, rat, and mouse have demonstrated the importance of GC/AT ratio, CpG density, and the occurrence of repetitive elements as well as chromosome size and proximity to the telomere and centromere, and the relative contribution of these factors varied between species (Jensen-Seaman et al. 2004).
Recombination maps of Robertsonian variable chromosomes:
For the chromosomes that do show Robertsonian polymorphism we found a substantial difference in MLH1 distribution for the same arms between homozygotes for metacentric chromosomes, homozygotes for acrocentrics, and heterozygotes (Figure 3).
In all homozygotes for acrocentrics we observed unimodal distributions with a peak of MLH1 foci in the middle of the arm. In heterozygotes there was still generally a single peak but the distribution was shifted towards a more distal localization. For arm o of k/o heterozygotes the pattern became bimodal with a second peak occurring near to the telomere. For metacentric homozygotes the distalization and presence of a second peak were even more pronounced in all arms, a result consistent with limited studies comparing chiasma position in trivalents and homologous metacentric bivalents in shrews (Giagia-Athanasopoulou and Searle 2003). The data are also in accordance with those of Dumas and Britton-Davidian (2002) for the house mouse. They too found a reduction in the number of proximal chiasmata and a more distal chiasma distribution in metacentric homozygotes compared to acrocentric homozygotes and proposed that the suppressive effect of the centromere on recombination was more pronounced in metacentric chromosomes.
From an evolutionary perspective, it is of significance that there is a greater tendency for reduced recombination in proximal chromosomal regions in metacentric homozygotes compared with simple Robertsonian heterozygotes. This makes it difficult to argue that recombination characteristics of heterozygotes are causing a reduction in gene flow across the hybrid zones between the Oxford, acrocentric, and Wirral races. It also suggests that speciation through recombination suppression (Rieseberg 2001) is not possible in hybrid zones of the common shrew characterized by simple heterozygotes. It would be of interest to determine the recombination characteristics of complex heterozygotes in the common shrew, i.e., hybrids between races that differ by metacentrics with a common chromosome arm and that form chain configurations of four or more elements at meiosis I (Searle 1993). The greater potency of complex heterozygotes than simple heterozygotes in promoting reproductive isolation is shown by Basset et al. (2006) for the speciation event that separated S. araneus and S. antinorii, and recombination suppression in complex heterozygotes has been suggested as a factor in a speciation event in the house mouse (Piálek et al. 2001).
There are other considerations relating to the metacentric or acrocentric “condition” in shrews and mice. In a chromosome race defined by a metacentric there may be the possibility of groups of epistatically interacting genes close to the metacentric centromere being able to segregate as a unit because of the lack of recombination. This could lead to different alternative adaptive multilocus genotypes within a population or geographic region (“coadaptive gene complexes,” e.g., Burton et al. 1999; De Jong and Nielsen 2002), and resultant polymorphism or regional genetic subdivision. In an acrocentric chromosome race, such coadapted gene complexes are less likely to evolve.
In both shrews and mice, some races are characterized by a large number of acrocentrics and an associated high diploid number, and other races by a large number of metacentrics and associated low diploid number. In terms of number of segregating units, this is clearly higher in the acrocentric than in the metacentric races, so the former should show greater genic variation (Qumsiyeh 1994). Clearly, recombination also affects genic variability, and so the reduced centromeric recombination in the metacentric races relative to the acrocentric races, will increase the differential in genic variability. Qumsiyeh (1994) argues that chromosomal forms with different levels of genic variability may be adapted to different conditions, e.g., adaptation to “pliable habitats” in the case of the acrocentric races and adaptation to “constant or specialized habitats” in the case of the metacentric races. There have, however, been no rigorous tests of this idea.
Recombination pattern of the sex trivalent:
The X chromosome of the common shrew represents a tandem fusion between the true mammalian X and an autosome (Sharman 1956; Fredga 1970; Pack et al. 1993). At male meiosis a trivalent comprising the X chromosome (de), the true Y (Y1 or the s chromosome), and the autosomal counterpart (Y2 or the dv chromosome) is formed (Wallace and Searle 1990; Borodin 1991; Pack et al. 1993). We usually detected a single MLH1 focus in the X–Y1 pairing region (Figure 1). The mean (± SD) size of the Y1 SC was 3.4 ± 0.7 μm. Although in some cells the Y1 was paired completely with the short arm of the X chromosome, the MLH1 focus was always located very close to the telomere. Its average distance from the telomere was 0.3 ± 0.2 μm with a range from 0 to 0.7 μm. This indicates that the pseudoautosomal region (Burgoyne 1982) is located in the most distal part of the true X and Y1 chromosomes. Thus, despite being fused with an autosome, the gonosomal part of the X remained autonomous in its meiotic behavior and showed characteristics displayed by a typical unfused mammalian X.
We did not observe MLH1 foci on the short arm v of the autosomal homolog of the sex trivalent. For the autosomal arm d of the sex trivalent we usually observed two MLH1 foci. Single and triple foci were rare (Table 3). For sex trivalents containing only one MLH1 focus on autosomal arm d, these foci were located mainly in the middle of the arm (Figure 2). The group with two foci showed a trimodal distribution. In general, arm d was rather similar to other autosomal arms of comparable size (b and c) in the frequency of MLH1 foci, their distribution, and interference parameters (Table 4). Previously we have demonstrated that arm d also acts like a typical autosome in chromosome pairing at male meiosis and is not involved in the process of X inactivation in female somatic cells (Pack et al. 1993).
Conclusion:
In summary, we have presented immunocytological recombination maps for every autosome in the male common shrew. Our maps demonstrate a global pattern rather similar to that found in human and mouse. We observed an excess of recombination in distal parts of bivalents and its suppression near the centromere. As for other vertebrates studied, male common shrews show a very strong correlation of SC length with the number of MLH1 foci, and, therefore, with genetic length of the chromosome. Interference estimates for the common shrew, i.e., average absolute and relative distances between neighboring crossovers, and the shape parameter of the gamma distribution, were comparable with corresponding estimates obtained for mouse chromosomes. All these comparisons suggest that a general pattern of recombination is conserved across mammals and perhaps all vertebrates.
As for other vertebrates, in the common shrew we observed that every chromosome arm has its own specific pattern of recombination, which is a complex function of its length, centromere and telomere effects, interference, and sequence characteristics. Chromosome arms of the same length may differ significantly in their recombination pattern. These recombination patterns remain rather conservative in representatives of different chromosome races of the common shrew that apparently have evolved independently for several thousand years (Searle and Wilkinson 1987). We found no difference in recombination patterns of the invariant chromosomes between the Oxford and Wirral races of the common shrew.
On the other hand, the recombination pattern of an individual chromosome arm varied substantially depending on whether it is an acrocentric chromosome or part of a metacentric chromosome. In contrast to Bidau et al. (2001) and Dumas and Britton-Davidian (2002) for the house mouse, in common shrews we did not find a significant decrease in numbers of MLH1 foci on the arms of metacentric chromosomes compared to acrocentrics. However, in agreement with them, we found a substantial redistribution of the crossovers to more distal positions in the metacentric chromosomes.
Acknowledgments
We thank C. Heyting, S. Garagna, A. Agoulnik, and N. Belyaev for the generous gift of antibodies and the Microscopic Center of the Siberian Branch of the Russian Academy of Sciences (RAS) for granting access to microscopic equipment. This work was supported by research grants from The International Association for the Promotion of Cooperation with Scientists from the New Independent States of the Former Soviet Union, the Russian Foundation for Basic Research, and the Programs of RAS “Biosphere Origin and Evolution” and “Biodiversity.”
References
- Anderson, L. K., A. Reeves, L. M. Webb and T. Ashley, 1999. Distribution of crossovers on mouse chromosomes using immunofluorescent localization of MLH1 protein. Genetics 151 1569–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley, T., 1988. G-band position effects on meiotic synapsis and crossing over. Genetics 118 307–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley, T., 2002. X-autosome translocations, meiotic synapsis, chromosome evolution and speciation. Cytogenet. Genome Res. 96 33–39. [DOI] [PubMed] [Google Scholar]
- Baker, S. M., A. W. Plug, T. A. Prolla, C. E. Bronner, A. C. Harris et al., 1996. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 13 336–342. [DOI] [PubMed] [Google Scholar]
- Barlow, A. L., and M. A. Hultén, 1998. Crossing over analysis at pachytene in man. Eur. J. Hum. Genet. 6 350–358. [DOI] [PubMed] [Google Scholar]
- Basset, P., G. Yannic, H. Brünner and J. Hausser, 2006. Restricted gene flow at specific parts of the shrew genome in chromosomal hybrid zones. Evolution 60 1718–1730. [PubMed] [Google Scholar]
- Belonogova, N. M., T. V. Karamysheva, L. S. Biltueva, E. A. Perepelov, J. M. Minina et al., 2006. Identification of all pachytene bivalents in the common shrew using DAPI-staining of synaptonemal complex spreads. Chromosome Res. 14 673–679. [DOI] [PubMed] [Google Scholar]
- Bidau, C. J., M. D. Giménez, C. L. Palmer and J. B. Searle, 2001. The effects of Robertsonian fusions on chiasma frequency and distribution in the house mouse (Mus musculus domesticus) from a hybrid zone in northern Scotland. Heredity 87 305–313. [DOI] [PubMed] [Google Scholar]
- Bilton, D. T., P. M. Mirol, S. Mascheretti, K. Fredga, J. Zima et al., 1998. Mediterranean Europe as an area of endemism for small mammals rather than a source for northwards postglacial colonization. Proc. R. Soc. Lond. B 265 1219–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, D. K., and D. Zickler, 2004. Early decision: meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117 9–15. [DOI] [PubMed] [Google Scholar]
- Borodin, P. M., 1991. Synaptonemal complexes of the common shrew, Sorex araneus L., in spermatocyte spreads. Cytogenet. Cell Genet. 56 61–62. [DOI] [PubMed] [Google Scholar]
- Broman, K. W., and J. L. Weber, 2000. Characterization of human crossover interference. Am. J. Hum. Genet. 66 1911–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne, P. S., 1982. Genetic homology and crossing over in the X and Y chromosomes of mammals. Hum. Genet. 61 85–90. [DOI] [PubMed] [Google Scholar]
- Burton, R. S., P. D. Rawson and S. Edmands, 1999. Genetic architecture of physiological phenotypes: Empirical evidence for coadapted gene complexes. Am. Zool. 39 451–462. [Google Scholar]
- Calderon, P. L., and M. I. Pigozzi, 2006. MLH1-focus mapping in birds shows equal recombination between sexes and diversity of crossover patterns. Chromosome Res. 14 605–612. [DOI] [PubMed] [Google Scholar]
- Colombo, P. C., and G. H. Jones, 1997. Chiasma interference is blind to centromeres. Heredity 79 214–227. [DOI] [PubMed] [Google Scholar]
- De Boer, E., P. Stam, A. J. J. Dietrich, A. Pastink and C. Heyting, 2006. Two levels of interference in mouse meiotic recombination. Proc. Natl. Acad. Sci. USA 103 9607–9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer, E., A. J. J. Dietrich, C. Hoog, P. Stam and C. Heyting, 2007. Meiotic interference among MLH1 foci requires neither an intact axial element structure nor full synapsis. J. Cell Sci. 120 731–736. [DOI] [PubMed] [Google Scholar]
- De Jong, P. W., and J. K. Nielsen, 2002. Host plant use of Phyllotreta nemorum: Do coadapted gene complexes play a role? Entomol. Exp. Appl. 104 207–215. [Google Scholar]
- Drouaud, J., C. Camilleri, P. Y. Bourguignon, A. Canaguier, A. Berard et al., 2006. Variation in crossing-over rates across chromosome 4 of Arabidopsis thaliana reveals the presence of meiotic recombination “hot spots”. Genome Res. 16 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas, D., and J. Britton-Davidian, 2002. Chromosomal rearrangements and evolution of recombination: comparison of chiasma distribution patterns in standard and Robertsonian populations of the house mouse. Genetics 162 1355–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredga, K., 1970. Unusual sex chromosome inheritance in mammals. Phil. Trans. R. Soc. Lond. B 759 15–36. [DOI] [PubMed] [Google Scholar]
- Froenicke, L., L. K. Anderson, J. Wienberg and T. Ashley, 2002. Male mouse recombination maps for each autosome identified by chromosome painting. Am. J. Hum. Genet. 71 1353–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giagia-Athanasopoulou, E. B., and J. B. Searle, 2003. Chiasma localization in male common shrews Sorex araneus, comparing Robertsonian trivalents and bivalents. Mammalia 67 295–299. [Google Scholar]
- Hassold, T., and P. Hunt, 2001. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2 280–291. [DOI] [PubMed] [Google Scholar]
- Henderson, K. A., and S. Keeney, 2004. Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. Proc. Natl. Acad. Sci. USA 101 4519–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, G. H., and F. C. Franklin, 2006. Meiotic crossing-over: obligation and interference. Cell 126 246–248. [DOI] [PubMed] [Google Scholar]
- Jensen-Seaman, M. I., T. S. Furey, B. A. Payseur, Y. Lu, K. M. Roskin et al., 2004. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 14 528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner, N., D. Zickler, G. H. Jones, J. Henle, J. Dekker et al., 2004. A mechanical basis for chromosome function. Proc. Natl. Acad. Sci. USA 101 12592–12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler, K. E., J. P. Cherry, A. Lynn, P. A. Hunt and T. J. Hassold, 2002. Genetic control of mammalian meiotic recombination. I. Variation in exchange frequencies among males from inbred mouse strains. Genetics 162 297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Král, B., and S. I. Radjabli, 1974. Banding patterns and Robertsonian fusions in the western Siberian population of Sorex araneus (Insectivora, Soricidae). Folia Zool. 23 217–227. [Google Scholar]
- Lynn, A., K. E. Koehler, L. Judis, E. R. Chan, J. P. Cherry et al., 2002. Genetic length of mammalian genomes: inter-individual variation and dependence on synaptonemal complex length. Science 296 2222–2225. [DOI] [PubMed] [Google Scholar]
- Moens, P. B., 2006. Zebrafish: chiasmata and interference. Genome 49 205–208. [DOI] [PubMed] [Google Scholar]
- Otto, S. P., and T. Lenormand, 2002. Resolving the paradox of sex and recombination. Nat. Rev. Genet. 3 252–261. [DOI] [PubMed] [Google Scholar]
- Pack, S. D., P. M. Borodin, O. L. Serov and J. B. Searle, 1993. The X-autosome translocation in the common shrew (Sorex araneus L.): late replication in female somatic cells and pairing in male meiosis. Chromosoma 102 355–360. [DOI] [PubMed] [Google Scholar]
- Peters, A. H. F. M., A. W. Plug, M. J. Van Vugt and P. De Boer, 1997. A drying-down technique for spreading of mammalian spermatocytes from the male and female germline. Chromosome Res. 5 66–71. [DOI] [PubMed] [Google Scholar]
- Piálek, J., H. C. Hauffe, K. M. Rodríguez-Clark and J. B. Searle, 2001. Raciation and speciation in house mice from the Alps: the role of chromosomes. Mol. Ecol. 10 613–625. [DOI] [PubMed] [Google Scholar]
- Pigozzi, M. I., 2001. Distribution of MLH1 foci on the synaptonemal complexes of chicken oocytes. Cytogenet. Cell Genet. 95 129–133. [DOI] [PubMed] [Google Scholar]
- Pigozzi, M. I., and A. J. Solari, 1999. Recombination nodule mapping and chiasmata distribution in spermatocytes of the pigeon, Columba livia. Genome 42 308–314. [DOI] [PubMed] [Google Scholar]
- Qumsiyeh, M. B., 1994. Evolution of number and morphology of mammalian chromosomes. J. Hered. 85 455–465. [DOI] [PubMed] [Google Scholar]
- Reeves, A., 2001. Micromeasure: a new computer program for the collection and analysis of cytogenetic data. Genome 44 439–443. [PubMed] [Google Scholar]
- Rieseberg, L. H., 2001. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 16 351–358. [DOI] [PubMed] [Google Scholar]
- Roeder, G. S., 1997. Meiotic chromosomes: it takes two to tango. Genes Dev. 11 2600–2621. [DOI] [PubMed] [Google Scholar]
- Roeder, G. S., and J. M. Bailis, 2000. The pachytene checkpoint. Trends Genet. 16 395–403. [DOI] [PubMed] [Google Scholar]
- Scherthan, H., S. Weich, H. Schwegler, C. Heyting, M. Harle et al., 1996. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J. Cell Biol. 134 1109–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M., W. Schempp and J. Olert, 1982. Comparative analysis of karyotypes in European shrew species. II. Constitutive heterochromatin, replication patterns, and sister chromatid exchanges in Sorex araneus and S. gemellus. Cytogenet. Cell Genet. 34 124–135. [DOI] [PubMed] [Google Scholar]
- Seabright, M., 1971. A rapid banding technique for human chromosomes. Lancet ii 971–972. [DOI] [PubMed] [Google Scholar]
- Searle, J. B., 1984. Three new karyotypic races of the common shrew Sorex araneus (Mammalia: Insectivora) and a phylogeny. Syst. Zool. 33 184–194. [Google Scholar]
- Searle, J. B., 1986. a Factors responsible for a karyotypic polymorphism in the common shrew, Sorex araneus. Proc. R. Soc. Lond. B 229 277–298. [DOI] [PubMed] [Google Scholar]
- Searle, J. B., 1986. b Meiotic studies of Robertsonian heterozygotes from natural populations of the common shrew, Sorex araneus L. Cytogenet. Cell Genet. 41 154–162. [DOI] [PubMed] [Google Scholar]
- Searle, J. B., 1993. Chromosomal hybrid zones in eutherian mammals, pp. 309–353 in Hybrid Zones and the Evolutionary Process, edited by R. G. Harrison. Oxford University Press, New York.
- Searle, J. B., and P. J. Wilkinson, 1987. Karyotypic variation in the common shrew (Sorex araneus) in Britain—a “Celtic fringe”. Heredity 59 345–351. [Google Scholar]
- Searle, J. B., and J. M. Wójcik, 1998. Chromosomal evolution: the case of Sorex araneus, pp. 219–268 in Evolution of Shrews, edited by J. M. Wójcik and M. Wolsan. Mammal Research Institute, Polish Academy of Sciences, Bialowieza, Poland.
- Searle, J. B., S. Fedyk, K. Fredga, J. Hausser and V. T. Volobouev, 1991. Nomenclature for the chromosomes of the common shrew (Sorex araneus). Mém. Soc. vaud. Sci. Nat. 19 13–22. [Google Scholar]
- Sharman, G. B., 1956. Chromosomes of the common shrew. Nature 177 941–942. [DOI] [PubMed] [Google Scholar]
- Sherman, J. D., and S. M. Stack, 1995. Two-dimensional spreads of synaptonemal complexes from solanaceous plants. VI. High-resolution recombination nodule map for tomato (Lycopersicon esculentum). Genetics 141 683–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, F., M. Oliver-Bonet, T. Liehr, H. Starke, E. Ko et al., 2004. Human male recombination maps for individual chromosomes. Am. J. Hum. Genet. 74 521–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, F., K. Trpkov, A. Rademaker, E. Ko and R.H. Martin, 2005. Variation in meiotic recombination frequencies among human males. Hum. Genet. 116 172–178. [DOI] [PubMed] [Google Scholar]
- Sun, F., M. Oliver-Bonet, T. Liehr, H. Starke, P. Turek et al., 2006. a Variation in MLH1 distribution in recombination maps for individual chromosomes from human males. Hum. Mol. Genet. 15 2376–2391. [DOI] [PubMed] [Google Scholar]
- Sun, F., M. Oliver-Bonet, T. Liehr, H. Starke, P. Turek et al., 2006. b Analysis of non-crossover bivalents in pachytene cells from 10 normal men. Hum. Reprod. 21 2335–2339. [DOI] [PubMed] [Google Scholar]
- Tease, C., G. M. Hartshorne and M. A. Hultén, 2002. Patterns of meiotic recombination in human fetal oocytes. Am. J. Hum. Genet. 70 1469–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov, A. E., 1998. Genome size and GC-percent in vertebrates as determined by flow cytometry: the triangular relationship. Cytometry 31 100–109. [DOI] [PubMed] [Google Scholar]
- Wallace, B. M. N., and J. B. Searle, 1990. Synaptonemal complex studies of the common shrew (Sorex araneus). Comparison of Robertsonian heterozygotes and homozygotes by light microscopy. Heredity 65 359–367. [Google Scholar]
- Wójcik, J. M., and J. B. Searle, 1988. The chromosome complement of Sorex granarius—the ancestral karyotype of the common shrew (Sorex araneus)? Heredity 61 225–229. [DOI] [PubMed] [Google Scholar]
- Wójcik, J. M., P. M. Borodin, S. Fedyk, K. Fredga, J. Hausser et al., 2003. The list of the chromosome races of the common shrew Sorex araneus (updated 2002). Mammalia 67 169–178. [Google Scholar]
- Ye, J., L. Biltueva, L. Huang, W. Nie, J. Wang et al., 2006. Cross-species chromosome painting unveils cytogenetic signatures for the Eulipotyphla and evidence for the polyphyly of Insectivora. Chromosome Res. 14 151–159. [DOI] [PubMed] [Google Scholar]
- Zickler, D., and N. Kleckner, 1999. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 33 603–754. [DOI] [PubMed] [Google Scholar]
- Zima, J., L. Lukáčová and M. Macholán, 1998. Chromosomal evolution in shrews, pp. 175–218 in Evolution of Shrews, edited by J. M. Wójcik and M. Wolsan. Mammal Research Institute, Polish Academy of Sciences, Bialowieza, Poland.
- Zima, J., S. Fedyk, K. Fredga, J. Hausser, A. Mishta et al., 1996. The list of the chromosome races of the common shrew Sorex araneus. Hereditas 125 97–107. [Google Scholar]