Abstract
Rapid larval growth is essential in the development of most metazoans. In this article, we show that bene, a gene previously identified on the basis of its oogenesis defects, is also required for larval growth and viability. We show that all bene alleles disrupt gatA, which encodes the Drosophila homolog of glutamyl-tRNA(Gln) amidotransferase subunit A (GatA). bene alleles are now referred to as gatA. GatA proteins are highly conserved throughout eukaryotes and many prokaryotes. These enzymes are required for proper translation of the proteins encoded by the mitochondrial genome and by many eubacterial genomes. Mitotic and endoreplicating tissues in Drosophila gatA loss-of-function mutants grow slowly and never achieve wild-type size, and gatA larvae die before pupariation. gatA mutant eye clones exhibit growth and differentiation defects, indicating that gatA expression is required cell autonomously for normal growth. The gatA gene is widely expressed in mitotic and endoreplicating tissues.
FOR most metazoans, the energy stores available during embryogenesis are sufficient to implement the basic body plan, but not to attain the necessary size for reproductive development. Free-living larvae consume and expend enormous resources to attain appropriate size for adulthood. Larvae grow by increasing cell size and/or cell number in a manner consistent with the needs of the organism and the availability of metabolic factors, including amino acids and energy.
Drosophila larvae increase their mass ∼200-fold during the 4-day larval period (Lilly and Duronio 2005). Some larval tissues, including the central nervous system and the imaginal discs, grow via mitotic division, but most larval growth is due to increasing cell size in endoreplicating tissues. Cells in some of these tissues, such as the salivary gland and fat body, increase their DNA content to hundreds of times that of a normal diploid cell and achieve gigantic size. Adult cells that require rapid growth, such as the nurse cells of the ovary, also rely on endoreplication (Edgar and Orr-Weaver 2001).
Multiple mechanisms in the fly control tissue and organism size by coordinating cell growth and cell cycle regulation with developmental programs and with nutritional state. Most of the major intercellular signaling pathways, including Wnt, BMP, Notch, and Hedgehog, locally affect the size of mitotically dividing tissues via control of cell division and/or cell death, but they do not regulate the growth of the endoreplicating tissues that contribute most to larval growth (Britton et al. 2002). Organ size is also under the control of the hippo pathway, which regulates cell cycle and apoptosis (Saucedo and Edgar 2007). Drosophila size is also regulated by the cell growth/cell cycle regulators dMyc and cyclin D-Cdk4 and by the insulin–Tor-signaling pathway, which acts to coordinate anabolic metabolism and cell growth with nutrient supply (Weinkove and Leevers 2000; Edgar 2006).
Several metabolic factors are essential for normal growth in Drosophila. The translation initiation factor EIF4A regulates Drosophila growth in a dose-dependent manner (Galloni and Edgar 1999). EIF4A was also recently shown to act downstream of the Drosophila BMP homolog, Dpp, in regulating cell growth in the amnioserosa (Li and Li 2006). The Minute genes, which have long been known to mediate cell growth and cell competition, encode many cytoplasmic ribosomal proteins (Lambertsson 1998). Mutations in three other cytoplasmic ribosomal proteins, Pixie, RpL5, and RpL38, disrupt wing development downstream of the insulin pathway (Coelho et al. 2005). Defects in Bonsai, a mitochondrial ribosomal protein, result in small larvae that exhibit cell proliferation defects (Galloni and Edgar 1999; Galloni 2003). Whether these mutants grow poorly simply due to lack of permissive factors (e.g., efficient translation or energy production) or to instructive signals regulating energy allocation to cell growth is an open question (see discussion).
Mutants in benedict (bene) were first isolated in a clonal screen in the Drosophila ovary for oogenesis defects (Morris et al. 2003). All three bene alleles isolated in the clonal screen are lethal in trans to each other or the deficiency. Egg chambers with homozygous bene mutant germ cells arrest in mid-oogenesis. The nurse cell chromosomes in these clones contain less DNA than those in similarly aged heterozygous egg chambers, and they fail to properly transition from polytene to polyploid chromosomal morphology. Instead, the nurse cell chromosomes appear tightly condensed well into mid-oogenesis. bene oocyte chromosomes appear stringy and fragmented in contrast with the condensed, spherical karyosome morphology of wild-type oocyte chromosomes (Morris et al. 2003). The germ cell clone defects in the ovary were cell autonomous; mutant polyploid nurse cells and meiotic oocytes displayed growth and chromosome morphology phenotypes and other tissues appeared wild type (Morris et al. 2003).
In this article, we molecularly identify bene as the gene encoding Drosophila glutamyl-tRNA(Gln) amidotransferase subunit A (GatA), a protein required in many prokaryotes and in mitochondria for proper translation of glutamine codons. We therefore rename the 50-40, 112-38, and 145-30 alleles isolated in the clonal screen as gatA50, gatA112, and gatA145, respectively. We show that gatA mutant larvae exhibit several growth and maturation defects, including molting delays and decreased size of mitotic and endoreplicating tissues. The larvae die before pupariation. Consistent with an essential role in mitochondrial function, we show that gatA is expressed widely in many tissues in larvae and adults. We also show by mosaic analysis that gatA is required in eyes, as it is in egg chambers, for normal growth.
MATERIALS AND METHODS
Drosophila stocks, culturing and scoring of larvae, and genetic and molecular mapping:
Stocks were maintained on standard Drosophila medium at 18°. All fly crosses and stocks for experiments were grown at 25°. The Bloomington Drosophila Stock Center, the Drosophila Stock Center in Szeged, Hungary, and the Lehmann, Treisman, and Johnston labs all provided stocks. The following stocks were used: gatA50/TM3,Sb, gatA112/TM3,Sb, gatA145/TM3,Sb, Df(3R)cha7,red1/TM6B, hs-hid/TM3,Sb,kr-GFP, P{neoFRT}82B, P{lacW}l(3)S092902/TM3,Sb, Df(3R)Exel6182/TM6B, Df(3R)Exel6183/TM6B, w, ey-FLP, Gla-lacZ; FRT Rps3, P{ubi-GFP, and w+}/ TM6B.
All gatA alleles and Df(3R)cha7,red1 were balanced over TM3,Sb,kr-GFP using hs-hid/TM3,Sb,kr-GFP. Genetic mapping was carried out by complementation testing for lethality. Point mutants were further complementation tested for larval growth defects. For phenotypic analysis, gatA/TM3,Sb,kr-GFP alleles were crossed to each other or to Df(3R)cha7,red1 and permitted to lay eggs for 2–3 hr at 25°. Embryos were then permitted to develop at 25° for 1–6 days. Larvae were scored as gatA/+, gatA/gatA, or gatA/Df by the presence or absence of GFP in fluorescence microscopy. Larvae were staged by mouth-hook morphology under transmitted light microscopy and photographed using a Nikon Eclipse 50i microscope and a Photometric Coolsnap EZ camera.
Mapping of the P{neoFRT}82B P{lacW}l(3)S092902 insertion point by inverse PCR was carried out as described (Bellen et al. 2004). PCR templates were sequenced (Fisher) and analyzed using DNAStar software. Sequences were compared to the Drosophila genome by BLAST (Altschul et al. 1990) and aligned with ClustalW (Chenna et al. 2003).
Eye clones:
w, ey-FLP, Gla-lacZ; FRT Rps3, P{ubi-GFP, w+}/TM6B females were crossed by FRT e gatA50/TM3, Sb males, FRT e gatA112/TM3, Sb males, or FRT +/TM3, Sb males. We submerged adult offspring in 100% ethanol and photographed the eyes with a Zeiss Stemi SV11 Apo microscope and a Zeiss Axio Cam HRc camera.
Immunofluorescence and DAPI staining:
Larvae were dissected in Ringers or 1× insect PBS and fixed in 4% formaldehyde in PBS. Subsequent washes were carried out in PBSTr (1× PBS with 1% triton) with the following exceptions: when antibody was used, primary blocking and incubation and subsequent washes were in PBSTrB (PBSTr with 1% BSA). Secondary blocking and incubation was in PBSTrB-S (PBSTrB with 5% normal donkey serum from Jackson Immunoresearch Laboratories). Two micrograms of DAPI was added to the penultimate wash. We photographed samples using a Nikon Eclipse 50i microscope and a Photometric Coolsnap EZ camera.
Allele sequencing and sequence analysis:
PCR templates for sequencing of the gatA- and NP15.6-coding regions were generated using the Expand Hi-Fidelity PCR kit (Boehringer Mannheim, Indianapolis). Products of PCR reactions were purified from agarose gels using the QiaexII kit (QIAGEN, Valencia, CA) and sequenced with gene-specific primers (Fisher). Sequences were assembled and analyzed using the Lasergene DNAStar programs Editseq and Seqman. ClustalW sequence alignments were generated using the DNAStar Megalign program.
Northern blot:
We crossed gatA50/TM3,Sb kr-GFP by gatA112/TM3,Sb kr-GFP. Five days after egg laying (AEL) we used the GFP balancer to sort gatA50/gatA112 larvae from gatA/TM3, Sb kr-GFP. Total RNA was purified using TRIZOL (Invitrogen, San Diego) and mRNA was purified using the MAG mRNA purification kit (Ambion), using 115 μg total RNA from both larval classes. We ran and blotted the formaldehyde gel using standard techniques and probed the blot sequentially with random-primed 32P probes from PCR templates of NK15.6 and RP49.
RT–PCR:
Wild-type larvae were dissected 5 days AEL and the following tissues were isolated: gut (entire), imaginal discs (mixed eye-antennal, leg, and wing), fat body, salivary gland (with some associated fat body), and brain (including optic lobes and ventral ganglia). We also dissected the ovaries from adult females. We purified Trizol (Invitrogen), and we carried out first-strand cDNA synthesis with the Superscript II kit (Invitrogen) and poly(dT) primers. We used the following PCR primers that flank the third intron of gatA: sense primer (GACTAAGAACATCTGGAGCG) and antisense primer (CAGATGTAACCTGGTAAAAGGC).
RESULTS
gatA alleles:
Of ∼10,000 independent lines generated in an ethyl methanesulfonate (EMS) clonal screen for ovary defects, three alleles, previously denoted 112-38, 50-40, and 145-30, compose the “benedict” (bene) complementation group (Morris et al. 2003). We now denote the alleles gatA112, gatA50, and gatA145 in light of our identification of gatA as the gene disrupted in these mutants (see below). We identified several Deficiencies in the Bloomington collection that failed to complement these alleles, including Df(3R)cha7 red1, Df(3R)Exel6182, and Df(3R)Exel6183. Complementation tests against the Szeged collection of lethal P insertions that had been mapped to or close to the region defined by these Deficiencies identified a single insertion line, l(3)S092902, that failed to complement gatA112, gatA50, and gatA145. We now denote that insertion allele gatAS.
gatA larval growth and lethality:
We used gatA112, gatA50, and gatAS and the deficiency, Df(3R)cha7 red1, for most of our phenotypic analysis, and we observed no differences in the phenotypes of gatAS homozygotes or of gatA112, gatA50, or gatAS in trans to Df(3R)cha7 red1. We carried out 2-hr egg lays of the cross gatA/TM3,Sb,kr-GFP × Df/TM3,Sb,kr-GFP and sorted sibling gatA/+ (including Df/+) from gatA/Df larvae by the presence or absence of GFP. gatA/Df larvae do not exhibit any obvious patterning defect, although they grow more slowly than gatA/+. By 3–4 days AEL, gatA/Df larvae are smaller than their wild-type siblings. By 5 days AEL, some of the gatA/Df larvae are dead, and the living larvae are obviously smaller than their siblings (Figure 1A). Wild-type larvae pupariate by the end of the fifth day AEL but the gatA/Df larvae do not. gatA/Df larvae continue to forage actively on the surface of the food throughout their lives, although in contrast with their wild-type siblings, they rarely burrow into the food. The mutant larvae do not grow significantly beyond day 3 AEL, although some of them live 9 days or longer.
Figure 1.—
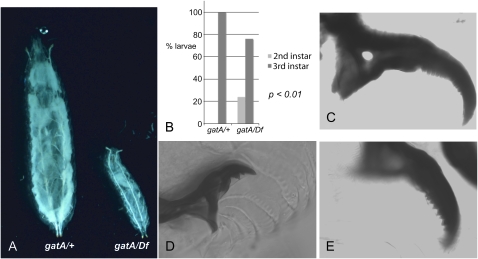
gatA growth and maturation defects. All larvae are siblings hatched from eggs collected from the same vial after a 2-hr egg lay. (A) gatA larvae are much smaller than wild-type 5 days AEL. (Left) gatA/+. (Right) gatA112/Df. (B) Graph showing delayed molting into third instar larvae of gatA mutants. (Left bar) gatA/+. (Right bars) gatA112/Df. (C–E) mouth-hook morphology 5 days AEL. (C) Wild-type third instar larva. (D) gatA112/Df second instar larva. (E) gatA112/Df third instar larva.
To determine if the larvae were developmentally arrested, developmentally delayed, or merely slow growing, we examined their mouth hooks. By 4 days AEL, the wild-type larvae all had the characteristic mouth-hook morphology of third instar larvae (Figure 1, B and C). In contrast, even on day 5 AEL, only 76% of gatA/Df larvae were third instar larvae, and the remaining 24% were second instar larvae (P < 0.01) (Figure 1, B–E). The first larval molt is also delayed in gatA mutants (not shown). We conclude that mutations in gatA arrest growth, delay larval molts, and permit development only as far as the third larval stage. These observations are consistent with the fact that timing of larval molting and pupariation depends on nutrition and growth rates (Edgar 2006).
gatA organ growth defects:
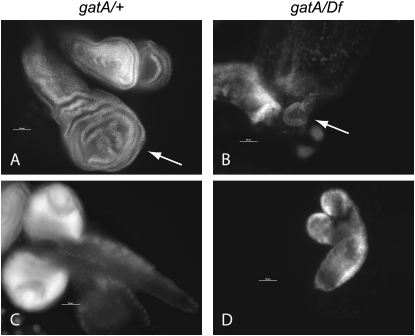
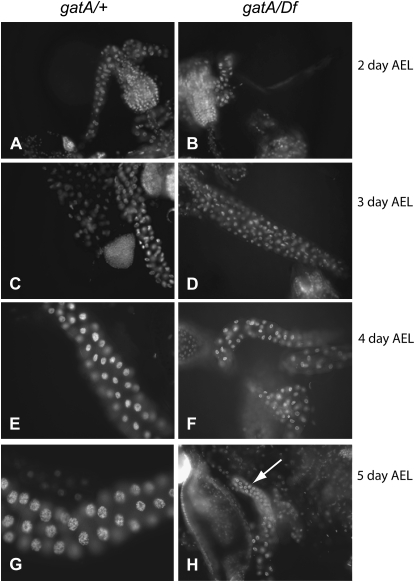
Because we had originally identified gatA mutants on the basis of growth and DNA replication defects in mutant nurse cells, we were interested in learning how the sizes of individual tissues were affected in gatA mutants. Dissection and DAPI staining of larvae revealed dramatic differences between gatA/+ and gatA/Df or gatA/gatA larvae. Diploid tissues, including imaginal discs (Figure 2, A and B) and brains (Figure 2, C and D), are much smaller in gatA/Df 5-day-old larvae than in their wild-type siblings of the same age. Endoreplicating tissues also show strong growth defects. By 3–4 days AEL, salivary gland cells in wild-type larvae are much larger and contain more DNA than salivary gland cells in their gatA/Df siblings. One day later, this difference is even more dramatic (Figure 3, A–H). We observed similar differences between wild-type and mutant larvae in all larval tissues (not shown). We observed no obvious defects in chromosome condensation in gatA salivary glands in contrast to the chromosome morphology defects that we observed in nurse cells and oocytes in our clonal screen (Morris et al. 2003), perhaps because wild-type salivary gland cell chromosomes retain their polyploid organization throughout larval development.
Figure 2.—
Growth defects in mitotic tissues in 5-day-AEL gatA larvae. (A–D) 20×, DAPI staining; bars, 200 μm. (A) gatA/+ wing disc (arrow) and haltere discs (top right). (B) gatA112/Df wing disc (arrow). (C) gatA/+ brain (D) gatA112/Df brain.
Figure 3.—
Growth and DNA content of salivary glands in wild-type and gatA larvae. (A–H) 20×, DAPI staining. (A, C, E, and G) gatA/+. (B, D, F, and H) gatA112/Df. (A and B) 2 days AEL. (C and D) 3 days AEL. (E and F) 4 days AEL. (G and H) 5 days AEL. Arrow points to gatA/Df salivary gland.
Mapping and molecular identification of gatA:
The gatA112, gatA50, and gatA145 alleles had been previously mapped to a 300-kb region (91D1-F2) by complementation testing against the 3R deficiency mapping kit (Morris et al. 2003). We carried out further complementation testing and found two particularly informative Deficiencies that failed to complement all three gatA EMS alleles and the gatAs allele: Df(3R)Exel6182 and Df(3R)Exel6183.
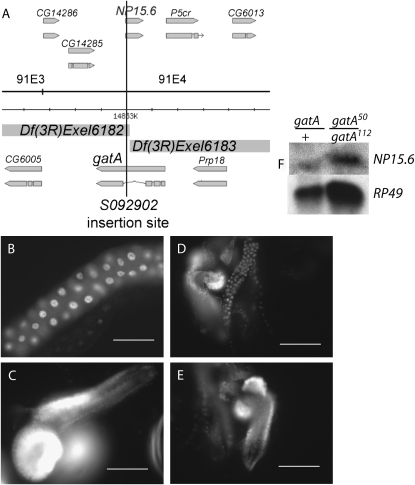
The right breakpoint of Df(3R)Exel6182 and the left breakpoint of Df(3R)Exel6183 fall at the same site in the genome (K. Cook, personal communication, FlyBase). Df(3R)Exel6182 removes the fourth exon of gatA (which encodes nearly two-thirds of the protein), as well as the 5′-UTR and approximately the first 10 codons of NP15.6. Df(3R)Exel6183 removes the first three exons of gatA and >90% of NP15.6 (Figure 4). NP15.6 and gatA fall in the densest cluster of unrelated genes in any characterized eukaryote (Misener and Walker 2000). On both the left and right, gatA is separated from adjacent genes by ∼100 bp. gatA has three introns; the first two introns are 53 and 60 bp long, and the third intron, which is 910 bp long, contains the 664-bp intronless gene, NP15.6 (Figure 4A).
Figure 4.—
Mapping and identification of gatA as the gene disrupted in all bene alleles. (A) Df(3R)Exel6182 and Df(3R)Exel6183 extend in opposite directions from a shared breakpoint at the insertion site of P{XP}d03824, which falls at bp 14,853,107 (K. Cook, FlyBase, personal communication). The P insertion P{lacW}l(3)S092902 was previously mapped by cytology to 91F1-F2. We mapped the insertion site molecularly, using inverse PCR (see materials and methods), to the third intron of gatA between codons 8 and 9 of NP15.6. Gene positions and splice forms adapted from FlyBase (Grumbling and Strelets 2006). (B–E) DAPI stainings of tissues in 5-day-AEL gatA/+ and gatA50/gatA112 trans-heterozygotes. (B and C) gatA/+. (D and E) gatA50/gatA112. (B and D) salivary gland. (C and E) Brain. (F) Northern blot showing strong NP15.6 expression in gatA/+ and gatA50/gatA112. RP49 is the loading control.
A number of transposon insertions had been mapped to the 300-kb region defined by our initial deficiency mapping. We tested these insertions and found that P{neoFRT}82B P{lacW}l(3)S092902 failed to complement the gatA alleles. We molecularly mapped the insertion site by inverse PCR amplification and sequencing of the genomic DNA flanking the P element (Bellen et al. 2004). The transposon separates the first three exons of gatA from the large fourth exon. The insertion also separates the first eight codons of NP15.6, including the only methionine codon, from the rest of the gene (Figure 4A).
Since deficiency mapping and molecular identification of the gatAs insertion site implicated gatA or NP15.6 in our mutants, we sequenced both genes in our EMS alleles. All three alleles showed no base changes in NP15.6, but each allele showed a single base change predicted to severely disrupt gatA (Figure 5 and see below). gatA50/gatA112 trans-heterozygotes show the same phenotypes as gatA/Df (Figure 4, B–E), indicating that point mutations in gatA cause the growth defects that we observed. To rule out the possibility that the gatA EMS alleles reduce NP15.6 expression in addition to disrupting gatA translation, we carried out Northern blot analysis on gatA/+ and gatA50/gatA112 larvae (Figure 4F). The trans-heterozygous larvae express NP15.6 strongly, indicating that these point mutants affect only gatA. Taken together, our mapping, allele sequencing, mRNA expression, and phenotypic analysis data show that mutations in gatA are responsible for the phenotypes that we observe in our gatA mutants. The similarity in phenotypes of the EMS and transposon alleles in trans to each other or to the deficiency, along with our molecular characterization of the alleles (see below), leads us to believe that the alleles are null.
Figure 5.—
ClustalW alignment of Drosophila GatA protein sequence with GatA sequences from other species. Accession numbers are as follows: fly—NP_650775.2; human—CAB66614; yeast (S. cerevisiae)—Q03557; bacteria (the cyanobacteria, Prochlorococcus marinus)—YP_291459. Arrows signify lesions in gatA point mutants. gatA145, W145STOP; gatA112, W154STOP; gatA50, T-A substitution in intron donor sequence, predicted to disrupt splicing between 174Y and 175A.
gatA sequence analysis:
All three gatA EMS alleles showed single base changes in gatA. None showed a change in NP15.6. gatA145 encodes a nonsense mutation at 145W. gatA112 encodes a nonsense mutation at 154W. The gatA50 mutation is a T-to-A substitution in the donor splice site of intron 3 (Figure 5). If the intron fails to splice, an in-frame translational stop occurs at the 46th codon within the intron. It is possible that the mutant transcript may use a cryptic splice site, but we would not predict protein translated from the mutant allele to have significant activity, since the allele is genetically null. The GatA amino acid sequence is highly conserved, including extensive regions that fall after the stop codons in the mutants. Drosophila and human gatA share 53% amino acid identity. Drosophila and yeast gatA share 29% identity. Drosophila and cyanobacteria gatA share 39% identity. The EMS alleles and the P insertion are therefore strong candidates for molecular null alleles.
gatA expression and cell autonomy:
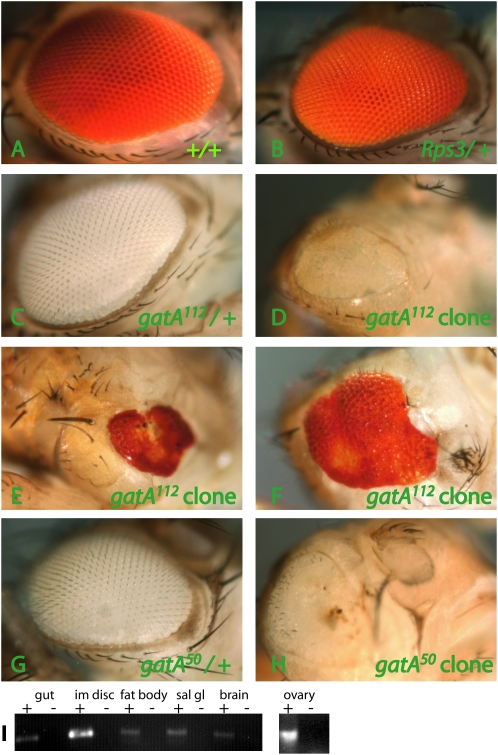
gatA is the only obvious candidate for the mitochondrial glutamyl-tRNA(Gln) amidotransferase subunit A gene in the Drosophila genome. The genome includes several other amidotransferases, but their sequences disqualify them as candidate gatA orthologs. We would therefore predict gatA to be required in all cells. We generated gatA mutant clones in eye imaginal discs by crossing w, ey-FLP; FRT, Rps3, P{ubi-GFP, w+}/TM6B females by FRT e gatA/TM3, Sb, kr-GFP males. The Rps3 allele is recessive cell-lethal and has a subtle dominant growth phenotype (Figure 6, A and B). gatA/TM6B eyes are wild type, as expected (Figure 6, C and G), but gatA112 and gatA50 mutant clones show several defects (Figure 6, D, E, F, and H). Eyes with gatA clones are small and misshapen and show various differentiation defects, including cuticle scars, ectopic bristles, rough eyes, and failure to develop ommatidia. The gatA growth defects that we observed in larvae are therefore not likely to be due to defects in a single organ (e.g., the brain), which would arrest growth in the whole organism. Instead, mutant tissues autonomously exhibit growth defects. Consistent with our prediction that every tissue would require gatA activity, RT–PCR analysis of wild-type tissues shows gatA expression in a wide range of mitotic and endoreplicating tissues, including larval gut, imaginal disc, fat body, salivary gland, and adult ovary (Figure 6I)
Figure 6.—
Mosaic analysis and expression of gatA. w, ey-FLP, Gla-lacZ; FRT Rps3, P{ubi-GFP, w+}/TM6B females were crossed with FRT e gatA50/TM3, Sb males, FRT e gatA112/TM3, Sb males or FRT +/TM3, Sb males. (A–H) Adult eyes. The FRT Rps3 chromosome carries a w+ transgene. Rps3 is recessive cell lethal. Therefore, in males, cells that have not undergone recombination are yellow. Cells that have recombined are white (homozygous for + or gatA, depending on the cross). (A, B, E, and F) Female eyes are w+, aiding phenotypic analysis, but making it difficult to determine the genotype of cells. (C, D, G, and H) Male eyes are w−, permitting easy genotyping of eye clones. (A) Wild-type clone shows normal growth and development. (B) Rps3/+ eyes (no clones induced) are slightly smaller than wild type. (C) gatA112/+ eyes (no clones induced) and (G) gatA50/+ eyes (no clones induced) are normal. (D–F) gatA112 clones and (H) gatA50 clones cause small, misshapen eyes with cuticle scars and ectopic bristles. There is a small, pale yellow region in the top portion of the eye in D, because ey-FLP induced mitotic recombination somewhat late in this eye. (I) RT–PCR of gatA from RNA extracts from larval gut (lanes 1 and 2), imaginal discs (lanes 3 and 4), fat bodies (lanes 5 and 6), salivary gland (lanes 7 and 8), brain (lanes 9 and 10), and from adult ovary (separate gel, lanes 11 and 12). “+” and “−” refer to treatment of extracts with reverse transcriptase.
DISCUSSION
gatA function is required cell-autonomously in the germ cells of the ovary for endoreplication-associated growth and DNA accumulation and for proper chromosome morphology. We have shown here that the gatA112, gatA145, and gatA50 EMS alleles and the gatAS transposon insertion allele are all predicted to result in early stop codons or splicing defects in the gatA gene. Homozygous gatAS mutations and all mutant gatA alleles in trans to the deficiency cause similar phenotypes; the alleles therefore fulfill the criteria for genetic and molecular nulls. Both the mitotic and endoreplicating tissues in gatA larvae are small, and the larvae die before pupariation. The endoreplicating nuclei exhibit much lower DNA contents but appear otherwise normal, suggesting that the mutant endocycle is slower or less efficient. As in the germ cells of the ovary, gatA is required autonomously in the cells of the eye for normal growth and differentiation, and gatA is expressed very broadly throughout the larva, as we would expect of a gene required for mitochondrial function.
Function of gatA:
In the cytoplasm of eukaryotic cells, different tRNA synthetases are used to charge the appropriate tRNAs with each of the 20 amino acids encoded in the genome. In many eubacteria, as well as in mitochondria and chloroplasts and many archeabacteria, there is no dedicated tRNA synthetase for Gln tRNAs analogous to the Gln tRNA synthetases present in eukaryotic genomes. Instead, in what is believed to be the more ancient process, both Gln and Glu tRNAs are initially charged with glutamic acid. The glutamic acid misacylated to Gln tRNA [Glu-tRNA(Gln)] then undergoes an amidotransferase reaction catalyzed by glutamyl-tRNA(Gln) amidotransferase (RajBhandary 1997). The A and B subunits of this heterodimeric enzyme are encoded by gatA and gatB, respectively (Curnow et al. 1997). In archeabacteria, GatD and GatE carry out the amidotransferase reactions on Gln tRNAs (Feng et al. 2005). Although GatA proteins are highly conserved in prokaryotes, fungi, and animals, the gatA alleles described here are the first reported eukaryotic gatA mutants. The only other characterized eukaryotic mutants in this pathway are defective in Saccharomyces cerevisiae pet112. Intriguingly, pet112 cells are small, but viable. The petite phenotype can be complemented by Bacillus subtilis gatB (Kim et al. 1997). This suggests that the gatA growth/cell cycle defects that we observed in Drosophila may reflect an evolutionarily conserved requirement for normal mitochondrial physiology for cell cycle and growth control.
Mitochondrial mutant phenotypes:
Mitochondria are complex organelles responsible for the production of NADH, FADH2, and ATP via the Krebs cycle and the transfer of electrons from NADH to oxygen via the electron transport chain. In addition, mitochondria regulate transport across their membranes, replicate their genomes, and transcribe and translate their own mRNAs and proteins. Although mitochondria require hundreds of proteins to carry out their diverse functions, almost all animal mitochondrial proteins are encoded in the nuclear genome and translated in the cytoplasm. The 13 protein-encoding genes in the animal mitochondrial genome all participate in the various complexes involved in electron transfer (Table 1).
TABLE 1.
Polypeptides encoded by the Drosophila mitochondrial genome
| Protein | Composition | Complex |
|---|---|---|
| ND1 | 5 Gln/312 aa | I |
| ND2 | 7 Gln/341 aa | I |
| ND3 | 1 Gln/117 aa | I |
| ND4 | 6 Gln/446 aa | I |
| ND4L | 1 Gln/96 aa | I |
| ND5 | 10 Gln/574 aa | I |
| ND6 | 3 Gln/174 aa | I |
| CytB | 8 Gln/378 aa | III |
| COX1 | 10 Gln/511 aa | IV |
| COX2 | 7 Gln/228 aa | IV |
| COX3 | 7 Gln/262 aa | IV |
| ATP6 | 4 Gln/224 aa | V |
| ATP8 | 1 Gln/53 aa | V |
GatA is one of the mitochondrial proteins encoded in the nuclear genome. gatA loss of function could result in one of two possible outcomes, either of which would affect all 13 of the proteins encoded in the mitochondrial genome. One possible outcome would be that all glutamines would be mistranslated as glutamates, which, for some of the highly conserved proteins that contain multiple glutamine codons, would likely result in inactive proteins (Table 1). Another possibility, suggested by data from chloroplast translation, would be that translation of all the proteins would simply fail. Misacylated Glu-tRNA(Gln) in chloroplasts is not brought to the ribosome by Ef-Tu, so it cannot participate in translation (Stanzel et al. 1994). It is not known whether mitochondrial translation functions similarly. Either likely effect of gatA loss of function would render aerobic respiration impossible.
A number of interesting genetic and pharmacological studies of flies compromised in aerobic respiration have been published. Drosophila grown under hypoxic conditions grow poorly, although they upregulate glycolysis sufficiently to survive (Digregorio et al. 2001; Frazier et al. 2001; Frei et al. 2005). Consistent with our observations of growth defects in gatA mutants, loss of function in the mitochondrial protein translocator component gene, tim50, causes growth and cell cycle defects in Drosophila larvae (Sugiyama et al. 2007). bonsai mutants, like gatA mutants, exhibit growth defects, although even bonsai null animals can develop into small adults (Galloni 2003). Possibly, loss of bonsai, which encodes a mitochondrial ribosomal protein, is less deleterious than loss of gatA. Arguing in favor of this model, bonsai expression after early embryonic development appears restricted to the gut, suggesting that most tissues do not require high levels of Bonsai (Galloni 2003).
In contrast to the growth defects observed in gatA and tim50 mutants, later or more subtle defects in aerobic respiration primarily affect nerve and muscle function. Late exposure to electron transport inhibitors such as the complex I inhibitor, rotenone, or the complex III inhibitor, antimycin, causes nervous system defects (Miwa and Brand 2003; Coulom and Birman 2004; Frei et al. 2005). Similarly, weak loss-of-function mutations in, for example, the mitochondrial ribosomal protein, tko (Royden et al. 1987), or in the complex V component, ATP6 (Celotto et al. 2006), permit flies to reach adulthood with significant muscle and/or nervous system defects. The fact that null alleles of the mitochondria-associated ubiquitin E3 protein ligase, parkin, cause adult muscular defects rather than larval lethality and growth defects (Greene et al. 2003) may suggest that parkin is not required for mitochondrial activity in all tissues.
One interesting question is how gatA larvae manage to survive and grow as well as they do, given the importance and broad expression pattern of gatA. Possibly, the early gatA survival should be attributed to the fly's remarkable capacity to rely on glycolysis for most of its energy requirements (Digregorio et al. 2001). Very likely, gatA embryos and early larvae also benefit from maternal rescue via maternally contributed mitochondria and gatA mRNA, consistent with our detection of gatA expression in ovaries (Figure 6I).
Permissive vs. instructive role for mitochondrial mutants in growth:
The mitochondrial genes required for normal growth, including gatA, may act permissively, merely providing energy required for increasing cell numbers and/or cell size. Alternatively, the growth genes may play an instructive role, such that cells experiencing metabolic defects actively reallocate resources away from growth and toward pathways and behaviors appropriate to survival under suboptimal conditions. We do not yet know whether gatA mutants specifically downregulate growth pathways, although several lines of evidence suggest that they might. Mitochondrial malfunction has been shown to arrest growth by interfering specifically with cell cycle and cell size regulators. Cells in flies grown under hypoxic conditions or in the presence of the electron transport chain inhibitor, cyanide, arrest at specific points in the cell cycle (Digregorio et al. 2001). Diploid cells homozygous for null mutations in tenured, which encodes the ATP synthase component, Cox Va, arrest at the G1-S checkpoint by repressing CycE in a p53-dependent manner (Mandal et al. 2005). Since control of CycE is sufficient to regulate the endocycle (Lilly and Duronio 2005), this mitochondrial checkpoint might explain growth defects in endoreplicating tissues as well. CycD/Cdk4 control of growth also depends on mitochondrial genes. CycD/Cdk4 activation stimulates mitochondrial activity and induces Hif-1 prolyl hydroxylase to signal cell growth. Both of these functions require the mitochondrial ribosomal protein mRpL12 (Frei et al. 2005).
In addition to the mitochondrial protein–cyclin interactions described above, the specificity of the gatA phenotypes argues that the growth defects in gatA larvae should not be attributed simply to running out of energy. gatA mutant larvae continue to move and forage for days after they cease growing, suggesting mitochondrial dysfunction in gatA mutants may instruct the cell to reallocate resources away from growth before the animal has entirely depleted its energy stores. In the ovary, chromosome condensation and chromosome morphology are defective in gatA mutants long before the egg chambers die, possibly indicating cell cycle or DNA repair defects in gatA mutants (Morris et al. 2003). It would be interesting to learn if these defects are related to chromosome condensation defects reported in Drosophila embryos that were grown under hypoxic conditions (Foe and Alberts 1985) and if other mutations that cause chromosome morphology defects in egg chambers are also defective in aerobic respiration.
Acknowledgments
The authors gratefully acknowledge R. Lehmann and J. Treisman for their generosity in giving us access to their microscopes and fly kitchen; Vanessa Flores, Christopher O'Connor, and Jane Mueller for assistance with experiments; and L. Gilboa, C. Navarro, and M. Morris for their feedback and support. We thank Kevin Legent for technical advice concerning the eye clones experiment. We also thank the R. Lehmann, J. Treisman, and L. Johnston labs and the Bloomington and Szeged stock centers for providing the fly stocks used in this research. This work was supported by National Institutes of Health R15 Award GM074735-01 and by a Fordham Faculty Research Grant.
References
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Bellen, H. J., R. W. Levis, G. Liao, Y. He, J. W. Carlson et al., 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton, J. S., W. K. Lockwood, L. Li, S. M. Cohen and B. A. Edgar, 2002. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell 2 239–249. [DOI] [PubMed] [Google Scholar]
- Celotto, A. M., A. C. Frank, S. W. McGrath, T. Fergestad, W. A. Van Voorhies et al., 2006. Mitochondrial encephalomyopathy in Drosophila. J. Neurosci. 26 810–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson et al., 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31 3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho, C. M., B. Kolevski, C. D. Walker, I. Lavagi, T. Shaw et al., 2005. A genetic screen for dominant modifiers of a small-wing phenotype in Drosophila melanogaster identifies proteins involved in splicing and translation. Genetics 171 597–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulom, H., and S. Birman, 2004. Chronic exposure to rotenone models sporadic Parkinson's disease in Drosophila melanogaster. J. Neurosci. 24 10993–10998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curnow, A. W., K. Hong, R. Yuan, S. Kim, O. Martins et al., 1997. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc. Natl. Acad. Sci. USA 94 11819–11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGregorio, P. J., J. A. Ubersax and P. H. O'Farrell, 2001. Hypoxia and nitric oxide induce a rapid, reversible cell cycle arrest of the Drosophila syncytial divisions. J. Biol. Chem. 276 1930–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, B. A., 2006. How flies get their size: genetics meets physiology. Nat. Rev. Genet. 7 907–916. [DOI] [PubMed] [Google Scholar]
- Edgar, B. A., and T. L. Orr-Weaver, 2001. Endoreplication cell cycles: more for less. Cell 105 297–306. [DOI] [PubMed] [Google Scholar]
- Feng, L., K. Sheppard, D. Tumbula-Hansen and D. Soll, 2005. Gln-tRNAGln formation from Glu-tRNAGln requires cooperation of an asparaginase and a Glu-tRNAGln kinase. J. Biol. Chem. 280 8150–8155. [DOI] [PubMed] [Google Scholar]
- Foe, V. E., and B. M. Alberts, 1985. Reversible chromosome condensation induced in Drosophila embryos by anoxia: visualization of interphase nuclear organization. J. Cell Biol. 100 1623–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier, M. R., H. A. Woods and J. F. Harrison, 2001. Interactive effects of rearing temperature and oxygen on the development of Drosophila melanogaster. Physiol. Biochem. Zool. 74 641–650. [DOI] [PubMed] [Google Scholar]
- Frei, C., M. Galloni, E. Hafen and B. A. Edgar, 2005. The Drosophila mitochondrial ribosomal protein mRpL12 is required for cyclin D/Cdk4-driven growth. EMBO J. 24 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloni, M., 2003. Bonsai, a ribosomal protein S15 homolog, involved in gut mitochondrial activity and systemic growth. Dev. Biol. 264 482–494. [DOI] [PubMed] [Google Scholar]
- Galloni, M., and B. A. Edgar, 1999. Cell-autonomous and non-autonomous growth-defective mutants of Drosophila melanogaster. Development 126 2365–2375. [DOI] [PubMed] [Google Scholar]
- Greene, J. C., A. J. Whitworth, I. Kuo, L. A. Andrews, M. B. Feany et al., 2003. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl. Acad. Sci. USA 100 4078–4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumbling, G., and V. Strelets, 2006. FlyBase: anatomical data, images and queries. Nucleic Acids Res. 34 D484–D488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. I., N. Stange-Thomann, O. Martins, K. W. Hong, D. Soll et al., 1997. A nuclear genetic lesion affecting Saccharomyces cerevisiae mitochondrial translation is complemented by a homologous Bacillus gene. J. Bacteriol. 179 5625–5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertsson, A., 1998. The minute genes in Drosophila and their molecular functions. Adv. Genet. 38 69–134. [DOI] [PubMed] [Google Scholar]
- Li, J., and W. X. Li, 2006. A novel function of Drosophila eIF4A as a negative regulator of Dpp/BMP signalling that mediates SMAD degradation. Nat. Cell Biol. 8 1407–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly, M. A., and R. J. Duronio, 2005. New insights into cell cycle control from the Drosophila endocycle. Oncogene 24 2765–2775. [DOI] [PubMed] [Google Scholar]
- Mandal, S., P. Guptan, E. Owusu-Ansah and U. Banerjee, 2005. Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev. Cell 9 843–854. [DOI] [PubMed] [Google Scholar]
- Misener, S. R., and V. K. Walker, 2000. Extraordinarily high density of unrelated genes showing overlapping and intraintronic transcription units. Biochim. Biophys. Acta 1492 269–270. [DOI] [PubMed] [Google Scholar]
- Miwa, S., and M. D. Brand, 2003. Mitochondrial matrix reactive oxygen species production is very sensitive to mild uncoupling. Biochem. Soc. Trans. 31 1300–1301. [DOI] [PubMed] [Google Scholar]
- Morris, J. Z., C. Navarro and R. Lehmann, 2003. Identification and analysis of mutations in bob, Doa and eight new genes required for oocyte specification and development in Drosophila melanogaster. Genetics 164 1435–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RajBhandary, U. L., 1997. Once there were twenty. Proc. Natl. Acad. Sci. USA 94 11761–11763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royden, C. S., V. Pirrotta and L. Y. Jan, 1987. The tko locus, site of a behavioral mutation in D. melanogaster, codes for a protein homologous to prokaryotic ribosomal protein S12. Cell 51 165–173. [DOI] [PubMed] [Google Scholar]
- Saucedo, L. J., and B. A. Edgar, 2007. Filling out the Hippo pathway. Nat. Rev. Mol. Cell Biol. 8 613–621. [DOI] [PubMed] [Google Scholar]
- Stanzel, M., A. Schon and M. Sprinzl, 1994. Discrimination against misacylated tRNA by chloroplast elongation factor Tu. Eur. J. Biochem. 219 435–439. [DOI] [PubMed] [Google Scholar]
- Sugiyama, S., S. Moritoh, Y. Furukawa, T. Mizuno, Y. M. Lim et al., 2007. Involvement of the mitochondrial protein translocator component tim50 in growth, cell proliferation and the modulation of respiration in Drosophila. Genetics 176 927–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinkove, D., and S. J. Leevers, 2000. The genetic control of organ growth: insights from Drosophila. Curr. Opin. Genet. Dev. 10 75–80. [DOI] [PubMed] [Google Scholar]