Abstract
The temperature-sensitive phenotypes of yku70Δ and yku80Δ have provided a useful tool for understanding telomere homeostasis. Mutating the helicase domain of the telomerase inhibitor Pif1 resulted in the inactivation of cell cycle checkpoints and the subsequent rescue of temperature sensitivity of the yku70Δ strain. The inactivation of Pif1 in yku70Δ increased overall telomere length. However, the long G-rich, single-stranded overhangs at the telomeres, which are the major cause of temperature sensitivity, were slightly increased. Interestingly, the rescue of temperature sensitivity in strains having both pif1-m2 and yku70Δ mutations depended on the homologous recombination pathway. Furthermore, the BLM/WRN helicase yeast homolog Sgs1 exacerbated the temperature sensitivity of the yku70Δ strain. Therefore, the yKu70-80 heterodimer and telomerase maintain telomere size, and the helicase activity of Pif1 likely also helps to balance the overall size of telomeres and G-rich, single-stranded overhangs in wild-type cells by regulating telomere protein homeostasis. However, the absence of yKu70 may provide other proteins such as those involved in homologous recombination, Sgs1, or Pif1 additional access to G-rich, single-stranded DNA and may determine telomere size, cell cycle checkpoint activation, and, ultimately, temperature sensitivity.
Ku protein was originally identified as an autoantigen recognized by sera from patients with rheumatic disorders (Mimori and Hardin 1986; Reeves 1987). Biochemical analyses of the Ku70-Ku80 heterodimer protein demonstrated that it bound in a sequence-nonspecific fashion to virtually all double-stranded DNA ends, including 5′- or 3′-protruding ends, blunt ends (Mimori and Hardin 1986), and duplex DNA ending in stem-loop structures (Falzon et al. 1993). Ku's DNA-binding activity is believed to function by both holding together broken double-stranded DNA, at least in mammals through its interaction with DNA-PKcs (Downs and Jackson 2004; Spagnolo et al. 2006), and protecting against end-to-end fusions at telomeres (Riha et al. 2002; Jaco et al. 2004; Myung et al. 2004). Inactivation experiments in different species demonstrated that the Ku70-Ku80 heterodimer is important for the protection of DNA ends during both DNA repair and telomere maintenance (Nussenzweig et al. 1996; Gu et al. 1997; Ouyang et al. 1997; Boulton and Jackson 1998; Nugent et al. 1998; Bailey et al. 1999; Hsu et al. 1999; Samper et al. 2000; Friesland et al. 2003; Downs and Jackson 2004; Jaco et al. 2004; Myung et al. 2004).
Telomeres are specific DNA structures at the ends of chromosomes that secure genetic information by protecting chromosomes from degradation with the help of many telomere-associated proteins (Lingner and Cech 1998; De Lange 2002). In Saccharomyces cerevisiae, telomeric DNA is 250–400 bp long with a simple repeat tract, C1-3A/TG1-3 (Vega et al. 2003). A G-rich, single-stranded tail is generated at the ends of the telomeres in late S-phase, presumably by the nuclease activity (Wellinger et al. 1993; Maringele and Lydall 2002; Bertuch and Lundblad 2004). In wild-type cells, telomerase extends the G-rich strand followed by general DNA replication that fills in the opposite C-strand and, therefore, the G-rich, single-stranded tail is not detected in other phases of the cell cycle. Telomere length is affected by many factors, including DNA replication, telomere synthesis by telomerase, and the level of degradation protection provided by telomere maintenance proteins (Lingner and Cech 1998; De Lange 2002; Vega et al. 2003). Mutations in yKU70 or yKU80 decreased overall telomere length and led to an elongation of G-rich, single-stranded tails in all cell cycles (Porter et al. 1996; Tsukamoto et al. 1997; Boulton and Jackson 1998; Nugent et al. 1998). In addition to the perturbation of telomeric structures, the mutation of yKU70 or yKU80 altered telomere position effect, which was defined as a suppression of gene expression in a subtelomeric region (Boulton and Jackson 1998; Evans et al. 1998; Laroche et al. 1998; Nugent et al. 1998).
yku70Δ or yku80Δ mutations cause a growth defect at 37° (Feldmann and Winnacker 1993; Boulton and Jackson 1996; Barnes and Rio 1997). This temperature sensitivity is the result of telomere homeostasis defects that activate cell cycle checkpoints, and not the result of deficiencies in DNA double-strand break repair pathways, such as nonhomologous end joining (NHEJ) (Nugent et al. 1998; Teo and Jackson 2001; Lewis et al. 2002; Maringele and Lydall 2002). In these studies, the temperature sensitivity of yku70Δ or yku80Δ strains was rescued either by mutations in checkpoint genes or in EXO1 or by the overexpression of telomerase subunits, including EST2, TLC1, or EST1. Interestingly, the overexpression of telomerase subunits did not change telomere length or the G-rich, single-stranded overhangs at telomeres, despite the suppression of cell cycle checkpoint activation elicited by the yku mutation at 37° (Nugent et al. 1998; Teo and Jackson 2001; Lewis et al. 2002). How cell cycle checkpoints are repressed by telomerase overexpression remains unclear.
Mutations in yeast S. cerevisiae PIF1, which encodes a DNA helicase, were identified in a screen to detect telomere maintenance proteins (Schulz and Zakian 1994). Yeast PIF1 encodes a transcript that makes two alternatively translated proteins controlled by two distinct initiating methionine codons (Lahaye et al. 1991; Schulz and Zakian 1994). Both the nuclear and mitochondrial Pif1 proteins have a 5′-3′ DNA helicase activity (Lahaye et al. 1991; Schulz and Zakian 1994). The pif1-m2 allele, which inactivates only the nuclear function of Pif1, results in elongated telomeres, the addition of telomeres at telomere seed sequences placed at subtelomeric sites, and an increased rate of spontaneous gross chromosomal rearrangements (Schulz and Zakian 1994; Zhou et al. 2000; Myung et al. 2001a). On the other hand, the pif1-m1 allele, which abrogates the mitochondrial Pif1, results in a petite phenotype.
The temperature-sensitive phenotype of yku70Δ has been a useful tool for determining which proteins are required for the maintenance of telomeric structures. In this study, we found that the temperature-sensitive phenotype of yku70Δ was suppressed by the mutation of the telomerase inhibitor Pif1. The suppression of the temperature-sensitive phenotype of yku70Δ was due to the active role of homologous recombination (HR)-dependent end protection.
MATERIALS AND METHODS
General genetic methods:
Media for propagating yeast strains used in this study are yeast extract–peptone–dextrose (YPD) media containing 1% (w/v) yeast extract, 2% (w/v) peptone, and 2% (w/v) dextrose, synthetic drop-out (SD) media containing 0.67% (w/v) yeast nitrogen base without amino acids, and 2% (w/v) dextrose with appropriate amino acids depending on the strains used. To make solid media, 1.5% (w/v) agar for YPD plates and 2.3% (w/v) for SD plates were added. All S. cerevisiae strains were propagated at 25°, 30°, or 37° as indicated. All S. cerevisiae strains used in this study were derived from the S288c parental strain RDKY3023 [MATa, ura3-52, leu2Δ1, trp1Δ63, his3Δ200, lys2ΔBgl, hom3-10, ade2Δ1, ade8] and detail genotypes or strains used in this study are listed in Table 1. Strains used in this study were generated by standard polymerase chain reaction (PCR)-based gene-disruption methods, and correct gene disruptions were verified by PCR as described (Myung et al. 2001c). The sequences of primers used to generate disruption cassettes and confirm disruption of indicated genes are available upon request. Yeast transformation, yeast chromosomal DNA isolation, and PCR were performed as previously described (Myung et al. 2001c; Smith et al. 2004). The overexpression of the RAD52 gene was achieved with a multicopy Yep13 plasmid carrying the RAD52 open reading frame with 4 kb upstream of the translation initiation codon.
TABLE 1.
S. cerevisiae strains used in this study
| Strains | Relevant genotype | Source |
|---|---|---|
| RDKY3023 | Wild type | Chen and Kolodner (1999) |
| RDKY3615 | Wild type, RDKY3023 + hxt13∷URA3 | Chen and Kolodner (1999) |
| RDKY3633 | RDKY3615 + mre11∷HIS3 | Chen and Kolodner (1999) |
| RDKY3636 | RDKY3615 + rad51∷HIS3 | Chen and Kolodner (1999) |
| RDKY3638 | RDKY3615 + rad57∷HIS3 | Chen and Kolodner (1999) |
| RDKY4343 | RDKY3615 + pif1-m2 | Myung et al. (2001a) |
| RDKY4393 | RDKY3615 + pif1-m1 | Myung et al. (2001a) |
| RDKY4399 | RDKY3615 + pif1∷KAN | Myung et al. (2001a) |
| RDKY4421 | RDKY3615 + rad52∷HIS3 | Myung et al. (2001a) |
| RDKY4423 | RDKY3615 + rad59∷TRP1 | Myung et al. (2001a) |
| RDKY4425 | RDKY3615 + rdh54∷TRP1 | Myung et al. (2001a) |
| YKJM245 | RDKY3615 + yku70∷TRP1 | This study |
| YKJM1589 | RDKY3615 + yku70∷HIS3 | This study |
| YKJM1763 | RDKY3615 + pif1-m2, yku70∷HIS3, rad52∷TRP1 | This study |
| YKJM2042 | RDKY3023 + pif1-K264A | This study |
| YKJM2056 | RDKY3023 + pif1-K264A, yku70∷TRP1 | This study |
| YKJM2133 | RDKY3615 + rad54∷TRP1 | This study |
| YKJM2189 | RDKY3615 + pif1-m2, yku70∷HIS3 + pRS314 | This study |
| YKJM2191 | RDKY3615 + pif1-m2, yku70∷HIS3 + pRS314-yPIF1 | This study |
| YKJM2381 | RDKY3615 + pif1-m2, yku70∷HIS3, lif1∷TRP1 | This study |
| YKJM2402 | RDKY3615 + pif1-m2, yku70∷HIS3, rad51∷TRP1 | This study |
| YKJM2404 | RDKY3615 + pif1-m2, yku70∷HIS3, rad54∷TRP1 | This study |
| YKJM2406 | RDKY3615 + pif1-m2, yku70∷HIS3, rad57∷TRP1 | This study |
| YKJM2408 | RDKY3615 + pif1-m2, yku70∷HIS3, rad59∷TRP1 | This study |
| YKJM2410 | RDKY3615 + pif1-m2, yku70∷HIS3, rdh54∷TRP1 | This study |
| YKJM2544 | RDKY3615 + yku70∷TRP1 + Yep13-RAD52 | This study |
| YKJM2548 | RDKY3615 + yku70∷TRP1 + Yep13 | This study |
| YKJM2503 | RDKY3615 + pif1-m2, yku70∷HIS3, mre11∷TRP1 | This study |
| YKJM2598 | RDKY3615 + pif1-m2, yku70∷HIS3, nej1∷TRP | This study |
| YKJM2777 | RDKY3615 + pif1-m2, lig4∷HIS3, yku70∷TRP1 | This study |
| YKJM3303 | RDKY3023 + pif1-m2, yku70∷TRP1 | This study |
| YKJM3955 | RDKY3023 + pif1-m2, cdc13-1 | This study |
| YKJM3957 | RDKY3023 + cdc13-1 | This study |
All strains are isogenic with the wild-type strains, RDKY3023 [ura3-52, leu2Δ1, trp1Δ63, his3Δ200, lys2ΔBgl, hom3-10, ade2Δ1, ade8], or RDKY3615 [ura3-52, leu2Δ1, trp1Δ63, his3Δ200, lys2ΔBgl, hom3-10, ade2Δ1, ade8, hxt13∷URA3] that are derived from the Winston 288c background with the exception of the indicated mutations followed by a plus sign.
Determination of length of telomeres and G-rich, single-stranded DNA at telomeres:
Telomere lengths were determined by XhoI digestion of chromosomal DNA, followed by size fractionation and Southern hybridization with a TG repeat probe as described (Banerjee and Myung 2004). The amount of single-stranded DNA at telomeres was determined by a comparison of telomeric signals by Southern hybridization with a TG repeat probe in nondenaturing in-gel hybridization with signals from denaturing conditions.
Northern hybridization:
RNA was prepared by using Gentra Systems Purescript RNA purification system from exponential cultures grown for an additional 4 hr at either 30° or 37° after a 1:50 dilution of overnight cultured yeast at 30°. RNA samples were boiled in the presence of ethidium bromide and formaldehyde, cooled, and then loaded onto a 1.2% agarose, 1× MOPS, 1% formaldehyde gel in 1× RNA gel loading dye. Electrophoresis was performed at 100 V for 1 hr. The RNA was transferred to a nitrocellulose membrane and crosslinked with UV. Prehybridization was carried out for 2 hr at 68° in 0.5 m sodium phosphate, 7% (w/v) SDS, 1 mm EDTA (pH 7.0), followed by hybridization with radiolabeled probe. DNA for making a probe to detect HUG1 expression was generated by PCR with the primers PRKJM 1107 (5′-GACCATGGACCAAGGCCTTAACCCAAAG-3′) and PRKJM1108 (5′-CAGAAAGACCGCCGCGACGTTCGACGGC-3′). The DNA for a control probe to detect ACT1 expression was made by PCR with the primers PRKJM959 (5′-CTCAATCCAAGAGAGGTATCTTGAC-3′) and PRKJM960 (5′-GTGGTGGAGAAAGAGTAACCACGTTC-3′). Both PCR products were then radiolabeled with [α-32P]dCTP by random priming as previously described (Banerjee and Myung 2004).
Western blotting:
Cell extracts were prepared from exponential cultures grown for an additional 4 hr at either 30° or 37° after 1:50 dilution of overnight cultured yeast at 30°. Cultures were harvested and washed with 20% trichloroacetic acid, glass beads were used to break the cell walls, and the collected cells were resuspended in 1× SDS loading buffer and 2 m Tris. Samples were boiled and centrifuged before loading onto a 7–12% SDS-PAGE (Bio-Rad, Hercules, CA). The protein was transferred to a PVDF membrane. The membranes were blocked in 1× Western blocking reagent (Roche) and then incubated with an anti-Rad53 antibody (Santa Cruz). After washing and incubation in the secondary anti-goat HRP antibody, detection was achieved using Western blocking detection reagents (GE Healthcare).
RESULTS
pif1 mutations rescued the temperature sensitivity of yku70Δ and cdc13-1 strains:
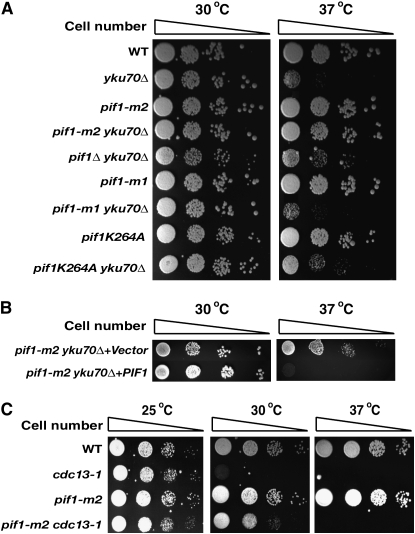
We recently observed that the overexpression of yKu70-yKu80 in the pif1-m2 strain caused cell cycle arrest at S-phase in a cell cycle checkpoint-dependent manner (Banerjee et al. 2006). Similarly, in the same study, we showed that the overexpression of Pif1, in either yku70Δ or yku80Δ strains, also caused growth arrest. Since the overexpression of Pif1 in yku70Δ generated a growth defect, we hypothesized that the inactivation of Pif1 could rescue the growth defect of yku70Δ at 37°. Wild-type, yku70Δ, pif1-m2 (which inactivates only nuclear Pif1), and pif1-m2 yku70Δ strains growing exponentially at 30° were serially diluted and spotted onto two YPD plates. One plate was incubated at 30° and the other at 37°. Similar to previous observations (Feldmann and Winnacker 1993; Boulton and Jackson 1996; Barnes and Rio 1997), the yku70Δ strain showed a growth defect at 37°, unlike wild type and pif1-m2 (Figure 1A).
Figure 1.—
The temperature-sensitive phenotype of yku70Δ is rescued by pif1 mutations. (A) pif1 mutations, including pif1-m2 (nuclear Pif1 defective), pif1Δ (complete deletion), and pif1K264A (ATPase and helicase defective), rescued the temperature sensitivity of yku70Δ, whereas the pif1-m1 mutation (mitochondrial Pif1 defective) could not. pif1K264A and pif1Δ exhibit a petite phenotype due to mitochondrial defects that caused these cells to grow more slowly compared to other clones. (B) The expression of full-length Pif1 in pif1-m2 yku70Δ restored the temperature-sensitive phenotype. (C) The temperature-sensitive phenotype of cdc13-1 is partially rescued by the pif1-m2 mutation.
In accordance with our hypothesis, the pif1-m2 mutation rescued the temperature-sensitive phenotype of yku70Δ. A similar rescue of growth defect occurred when the entire PIF1 gene was completely deleted in yku70Δ (Figure 1A). However, the pif1-m1 allele, which is defective only in mitochondrial Pif1, did not rescue the temperature-sensitive phenotype of yku70Δ. The K264A mutation, which disrupts Pif1's helicase activity, also rescued the temperature sensitivity of yku70Δ (Figure 1A), suggesting that the helicase activity of Pif1 is necessary for the temperature-sensitive phenotype of the yku70Δ strain. The expression of Pif1 in a single-copy plasmid under its own promoter in the pif1-m2 yku70Δ strain restored temperature sensitivity at 37° (Figure 1B).
The cdc13-1 strain has a mutation in the region of Cdc13, which interacts with telomerase and therefore generates large, G-rich, single-stranded overhangs similar to the yku70Δ strain (Weinert and Hartwell 1993; Maringele and Lydall 2002). The cdc13-1 strain also exhibits temperature sensitivity at 30° (Figure 1C). The pif1-m2 mutation partially suppressed the temperature sensitivity of the cdc13-1 strain at 30°, but not at 37°, similar to what has been previously reported (Figure 1C) (Downey et al. 2006). Therefore, although perhaps not exactly identical, the mechanism of temperature sensitivity suppression by the pif1 mutation appears to be similar in yku70Δ and cdc13-1.
The length of telomeres and the amount of G-rich, single-stranded overhangs are increased by the pif1-m2 mutation in yku70Δ:
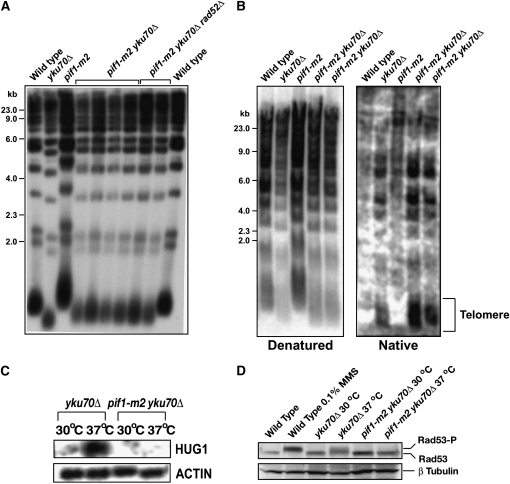
Telomere size is increased by pif1 mutations (Schulz and Zakian 1994; Zhou et al. 2000). We speculated that pif1 mutations would increase the telomere size of the yku70Δ strain to wild-type levels. When we compared the telomere sizes of wild-type, yku70Δ, pif1-m2, and pif1-m2 yku70Δ, we found that an additional pif1-m2 mutation in yku70Δ did increase overall telomere length but not to that of wild type at either 30° or 37° (Figure 2A and data not shown). Four independent clones carrying pif1-m2 yku70Δ mutations all showed an increase in telomere length compared to the yku70Δ strain, similar to what has been recently observed in pif1Δ yku70Δ strains (Vega et al. 2007). Mutation of yKU70 also increases the length of G-rich, single-stranded overhangs at telomeres in all phases of the cell cycle (Polotnianka et al. 1998). It has been suggested that the G-rich, single-stranded overhang in yku70Δ is a major signal for the activation of cell cycle checkpoints and ultimately temperature sensitivity (Maringele and Lydall 2002). However, we did not find any significant decrease of G-rich, single-stranded overhangs by the pif1-m2 mutation in yku70Δ measured by native in-gel hybridization (Figure 2B). We did observe even a slight increase in the amount of G-rich, single-stranded overhangs in two independent pif1-m2 yku70Δ clones and concluded that the inactivation of the Pif1 helicase increased the length of telomeres and the G-rich, single-stranded overhangs.
Figure 2.—
Cell cycle checkpoint activation is abrogated by the pif1-m2 mutation in yku70Δ at 37°. (A) Telomere sizes from wild-type, yku70Δ, pif1-m2, pif1-m2 yku70Δ, and pif1-m2 yku70Δ rad52Δ strains were compared. (B) Chromosomal DNAs from wild-type, yku70Δ, pif1-m2, and pif1-m2 yku70Δ strains were hybridized with telomeric probe by a native in-gel hybridization to detect G-rich, single-stranded overhangs at telomeres. (C) The activation of cell cycle checkpoints in yku70Δ at 37° demonstrated by the HUG1 mRNA expression disappeared by the pif1-m2 mutation. (D) The phosphorylation of Rad53 in yku70Δ at 37° was abrogated by the pif1-m2 mutation. MMS-treated yeast cell extract (lane 2) was used to demonstrate Rad53 phosphorylation as a positive control.
The overexpression of telomerase subunits suppressed the activation of cell cycle checkpoints although it did not change the length of telomeres or G-rich, single-stranded overhangs (Nugent et al. 1998; Teo and Jackson 2001; Lewis et al. 2002). To determine whether the pif1-m2 mutation also suppresses cell cycle checkpoint activation in yku70Δ, the expression of HUG1 was monitored when cells were cultured at 37°. HUG1 is a small open reading frame whose expression is induced by different kinds of DNA damage in a cell cycle checkpoint-dependent manner (Basrai et al. 1999). Whereas the expression of HUG1 was induced when yku70Δ cells were shifted to 37°, the pif1-m2 yku70Δ strain displayed no induction of HUG1 expression (Figure 2C). Rad53 phosphorylation, another indication of cell cycle checkpoint activation, was not observed in pif1-m2 yku70Δ, unlike yku70Δ, at 37° (Figure 2D). Therefore, both the overexpression of telomerase subunits (Nugent et al. 1998; Teo and Jackson 2001; Lewis et al. 2002) and the inactivation of Pif1 rescued the temperature-sensitive phenotype of yku70Δ and suppressed the activation of cell cycle checkpoints.
The rescue of yku70Δ temperature sensitivity by the inactivation of Pif1 requires HR proteins:
The rescue of the temperature sensitivity of yku70Δ by the exo1Δ mutation suggested that the production of G-rich, single-stranded overhangs at telomeres by Exo1 in the absence of Ku is essential for both checkpoint activation and temperature sensitivity (Maringele and Lydall 2002). Therefore, it is possible that these overhangs could be amended to prohibit cell cycle checkpoint activation in pif1-m2 yku70Δ without significantly altering the length of telomeres.
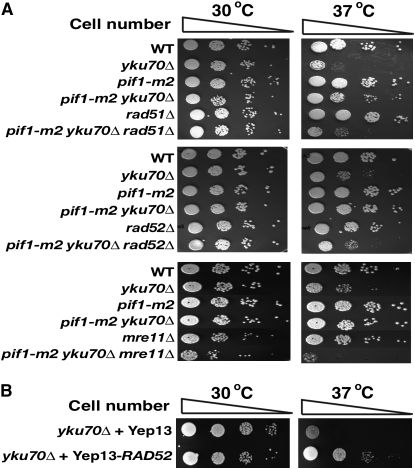
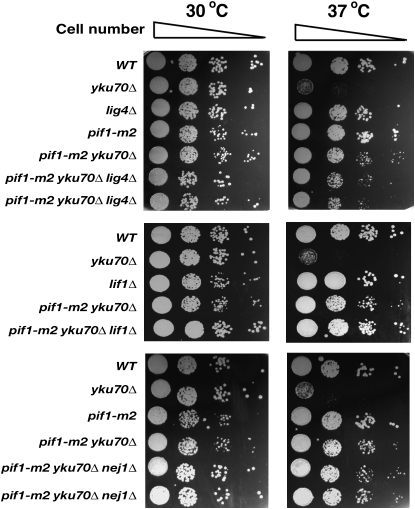
There are at least two distinct DNA repair mechanisms for the alteration of DNA structures: HR and NHEJ (Aylon and Kupiec 2004; Krogh and Symington 2004). To determine whether these repair machineries are responsible for the temperature sensitivity rescue of yku70Δ by the inactivation of Pif1, genes responsible for HR were deleted in the pif1-m2 yku70Δ strain. Interestingly, the rad51Δ pif1-m2 yku70Δ triple mutant resulted in the restoration of a temperature-sensitive phenotype at 37° (Figure 3A). Similar results were observed when a rad52Δ, mre11Δ, or rad54Δ mutation was combined with the pif1-m2 yku70Δ strain (Figure 3A; supplemental Figure 1 at http://www.genetics.org/supplemental/). Mutating RAD51, RAD52, MRE11, or RAD54 in the pif1-m2 strain did not result in temperature sensitivity at 37° (data not shown). The mre11Δ mutation in the pif1-m2 yku70Δ strain was slightly growth retarded even at 30° (Figure 3A), consistent with previous results that mutations in both mre11Δ and yku70Δ cause growth defects and severe temperature sensitivity (Maringele and Lydall 2002). Furthermore, the overexpression of Rad52 in the yku70Δ strain rescued the temperature sensitivity of yku70Δ (Figure 3B). However, the rad57Δ, rad59Δ, and rdh54Δ mutations did not affect the temperature-resistant phenotype of pif1-m2 yku70Δ (supplemental Figure 1). In contrast to HR, mutations of NHEJ genes, including LIG4, LIF1, or NEJ1, did not affect the temperature-resistant phenotype of pif1-m2 yku70Δ (Figure 4).
Figure 3.—
HR proteins actively engage at telomeres to stabilize telomeric structure to rescue temperature sensitivity in the pif1-m2 yku70Δ strain. (A) Mutations of HR genes, including RAD51, RAD52, and MRE11, made pif1-m2 yku70Δ sensitive to high temperature. (B) The overexpression of Rad52 partially suppressed the temperature-sensitive phenotype of yku70Δ.
Figure 4.—
Nonhomologous end joining proteins, Lig4, Lif1, and Nej1 are not required to rescue the temperature sensitivity in the pif1-m2 yku70Δ strain.
Although mutation of RAD52 in the pif1-m2 yku70Δ strain restored temperature sensitivity, we found no detectable alteration in telomere length (Figure 2A). These data suggest that, while telomere length restoration may contribute, HR events are required for the temperature-resistant phenotype of pif1-m2 yku70Δ.
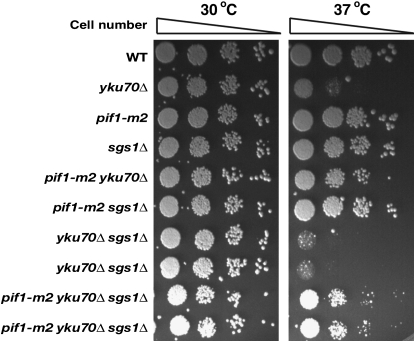
An additional sgs1 mutation enhanced temperature sensitivity in the yku70Δ strain as well as in the pif1-m2 yku70Δ strain (Figure 5). Sgs1 is a yeast homolog of human WRN and BLM helicases (Gangloff et al. 1994; Watt et al. 1995). Therefore, in addition to HR proteins, Sgs1 also functions to prevent cell cycle checkpoint activation in yku70Δ.
Figure 5.—
The sgs1Δ mutation enhanced temperature sensitivity in yku70Δ and pif1-m2 yku70Δ.
DISCUSSION
The helicase activity of Pif1 has been suggested to block the interaction between telomeric DNA and the RNA subunit of telomerase (Boule et al. 2005). In addition to directly inhibiting telomerase, Pif1 also has a role in general DNA replication. Pif1 is required for replication fork pausing at the Fob1-dependent replication fork barrier in the rDNA repeat array (Ivessa et al. 2000) and could promote the formation of the long flap structure during RNA removal by Dna2 on the basis of the observation of the rescue of the lethal phenotype of the DNA2 deletion by the pif1 mutation (Budd et al. 2006). The rescue of the temperature-sensitive phenotype of the yku70Δ strain could be caused by the absence of any one of Pif1's functions. The temperature sensitivity in the absence of the Ku proteins seems to be largely due to defects in telomere maintenance, since temperature sensitivity could be partially suppressed by the overexpression of telomerase subunits (Nugent et al. 1998; Teo and Jackson 2001; Lewis et al. 2002) and an additional mutation in any telomerase subunit in the yku70 strain caused an almost synthetic lethal phenotype (Gravel et al. 1998; Nugent et al. 1998; Polotnianka et al. 1998). The pif1-m2 mutation also rescued the temperature-sensitive phenotype of cdc13-1, which also exhibits telomere defects (Figure 1C) (Weinert and Hartwell 1993; Maringele and Lydall 2002). Therefore, the rescue of the temperature-sensitive phenotype by the pif1-m2 mutation in yku70Δ is likely due to the restoration of telomere functions. However, we could not exclude completely the possibility that the inactivation of the general DNA replication function of Pif1 could indirectly lead to the rescue of temperature sensitivity of yku70Δ.
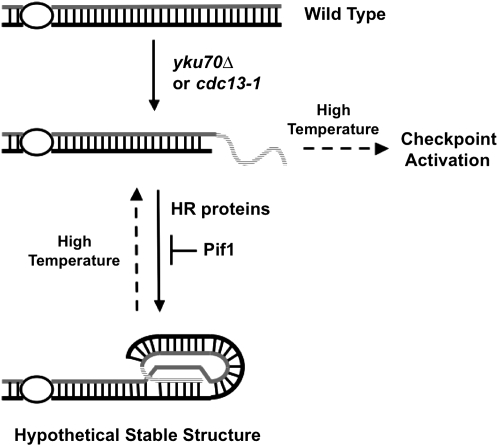
The absence of Ku hinders the recruitment of telomerase to telomeres completely in G1-phase and significantly in late S-phase (Fisher et al. 2004). The inactivation of Pif1 consequently could allow telomerase to be enriched at telomeres. Neither Pif1 inactivation by the pif1-m2 mutation (Figure 2A) (Vega et al. 2007) nor the overexpression of telomerase subunits, which also rescues the temperature sensitivity of yku70Δ (Nugent et al. 1998; Teo and Jackson 2001; Lewis et al. 2002), was able to restore the length of telomere or C-strand synthesis to a wild-type level. Therefore, any enrichment of telomerase does not appear to restore DNA replication at telomeres. In the absence of Pif1, single-stranded overhangs might form a stable structure and block the activation of cell cycle checkpoints by HR proteins (Figure 6). Although we do not know the exact structure stabilized by HR proteins, this structure is not what has been observed in survivors that display substantial recombination-dependent amplification of telomeres because the pif1 mutation did not change telomere size similar to those observed in survivors (Figure 2A). We speculate that HR proteins promote the generation of stable G-strand loops similar to the t-loop observed in mammals.
Figure 6.—
A model for how telomeric structures are dynamically regulated.
During the review of our manuscript, a study was published suggesting that the pif1Δ mutation rescued the temperature sensitivity of the yku70Δ strain by increasing telomere size (Vega et al. 2007). Similarly, we also observed an increase of telomere size by the pif1-m2 mutation (Figure 2A). However, because the addition of the rad52Δ mutation in the pif1-m2 yku70Δ strain restored the temperature-sensitive phenotype (Figure 3) but did not further increase or decrease telomere size (Figure 2A), it is likely that the observed increase of telomere size by the pif1-m2 mutation is not the sole factor determining temperature resistance.
The stabilization of telomeric structures by HR proteins in yku70 requires Rad51, Rad52, Rad54, and Mre11 but not Rad55, Rad57, Rdh54, and Rad59 (Figure 3A; supplemental Figure 1 at http://www.genetics.org/supplemental/). The Rad55–Rad57 complex is required to stabilize Rad51 filament formation and also to make Rad51 coated single-stranded DNA homologous pairing (Sugawara et al. 2003). It is possible that this activity might not be required at telomeres or that Rad55-Rad57 may not be required for HR at 37° since the radiation sensitivity and recombination-defect phenotypes of rad55Δ and rad57Δ were observed only at cold temperatures (Lovett and Mortimer 1987). Rad59 and Rdh54 have roles in Rad51-independent recombination (Krogh and Symington 2004) and are presumably not required for the stabilization of the G-rich, single-stranded DNA at telomeres in the absence of yKu and Pif1.
The sgs1 mutation aggravated temperature sensitivity of yku70Δ and pif1-m2 yku70Δ (Figure 5). Sgs1 and its human homolog, the BLM helicase, unwind different DNA structures. Hyperrecombination phenotypes such as increase of sister-chromatid exchange rate or heteroallelic recombination in mitotic cells caused by mutations in these genes suggest that Sgs1 and HR proteins operate in the same HR repair pathway (Onoda et al. 2000; Myung et al. 2001b; Onoda et al. 2001; Hickson 2003). The aggravating phenotype by the sgs1 mutation in the pif1-m2 ku70 strain could be explained by the lack of HR repair pathway for temperature resistance.
Sgs1 and BLM can unwind G-quadruplex structures (Sun et al. 1999; Han et al. 2000; Li et al. 2001; Huber et al. 2002). Intriguingly, the pif1-m2 yku70Δ strain became sensitive to high temperature in the presence of N-methyl mesoporphyrin IX that specifically binds to the G-quadruplex structure and blocks its unwinding (supplemental Figure 2 at http://www.genetics.org/supplemental/) (Han et al. 2000; Li et al. 2001; Wu and Maizels 2001; Joyce and McGown 2004). At high temperatures, the G-rich, single-stranded overhangs could form a new DNA structure and cause lethality in a yku70Δ background. It has been proposed that G-quadruplex structures form at telomeres under certain conditions (Williamson et al. 1989; Zahler et al. 1991). Therefore, if Ku proteins are absent, G-rich, single-stranded overhangs are modified to structures, which not only are sensitive to NMM at 37° but also are unwound by Sgs1. The persistence of this structure might be the cause of cell cycle checkpoint activation. The removal of Pif1 would then hypothetically allow the recruitment of HR proteins, forming another more stable telomeric structure.
Both Sgs1 and Pif1 encode helicases functioning at telomeres, at least in certain conditions. Although the substrate specificities for these helicases are different, there is a possibility that Sgs1 and Pif1 might compete to, respectively, unwind and stabilize stable telomeres structures. Recently, competitive functions of Sgs1 and Pif1 during DNA replication have been suggested (Wagner et al. 2006). However, since HR proteins are required to restore temperature resistance in yku70Δ when Pif1 is absent (Figure 3 and supplemental Figure 1), we speculate that Pif1 removes HR proteins from telomeres in cells lacking the Ku protein.
In this study, we uncovered a mechanism that prevents the activation of the cell cycle checkpoint at high temperatures in the yku70Δ strain, resulting in cell survival. The traditional means for achieving temperature resistance in this strain required either telomerase or a mutation resulting in telomere lengthening. We found that, in the absence nuclear Pif1, the telomeres of yku70Δ were lengthened, temperature resistance was restored, and cell cycle checkpoints were no longer activated. Our data, however, show that a HR-dependent pathway is ultimately more important to temperature resistance than telomere length.
Acknowledgments
We thank S. Lee (University of Texas at San Antonio) and M. Lichten of the National Institutes of Health (NIH) for helpful discussions; J. Haber (Brandeis University), W. Heyer (University of California at Davis), S. Jackson (University of Cambridge), N. Sugawara (Brandeis University), P. Sung (Yale University), and S. Teo (University of Cambridge) for providing plasmids, strains, and antibodies; and B. Berman at the National Human Genome Research Insitute (NHGRI), S. Dunaway (Drew University), E. Hendrickson (University of Minnesota), H. Liaw (NHGRI), and A. Zhang (NHGRI) for comments on the manuscript. K. Myung thanks E. Cho for great support. This work was supported by the intramural research program of the NHGRI at the NIH (to K.M.).
References
- Aylon, Y., and M. Kupiec, 2004. DSB repair: the yeast paradigm. DNA Repair 3 797–815. [DOI] [PubMed] [Google Scholar]
- Bailey, S. M., J. Meyne, D. J. Chen, A. Kurimasa, G. C. Li et al., 1999. DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc. Natl. Acad. Sci. USA 96 14899–14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, S., and K. Myung, 2004. Increased genome instability and telomere length in the elg1-deficient Saccharomyces cerevisiae mutant are regulated by S-phase checkpoints. Eukaryot. Cell 3 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, S., S. Smith and K. Myung, 2006. Suppression of gross chromosomal rearrangements by yKu70-yKu80 heterodimer through DNA damage checkpoints. Proc. Natl. Acad. Sci. USA 103 1816–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, G., and D. Rio, 1997. DNA double-strand-break sensitivity, DNA replication, and cell cycle arrest phenotypes of Ku-deficient Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94 867–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basrai, M. A., V. E. Velculescu, K. W. Kinzler and P. Hieter, 1999. NORF5/HUG1 is a component of the MEC1-mediated checkpoint response to DNA damage and replication arrest in Saccharomyces cerevisiae. Mol. Cell. Biol. 19 7041–7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertuch, A. A., and V. Lundblad, 2004. EXO1 contributes to telomere maintenance in both telomerase-proficient and telomerase-deficient Saccharomyces cerevisiae. Genetics 166 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boule, J. B., L. R. Vega and V. A. Zakian, 2005. The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature 438 57–61. [DOI] [PubMed] [Google Scholar]
- Boulton, S. J., and S. P. Jackson, 1996. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 15 5093–5103. [PMC free article] [PubMed] [Google Scholar]
- Boulton, S. J., and S. P. Jackson, 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd, M. E., C. C. Reis, S. Smith, K. Myung and J. L. Campbell, 2006. Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta. Mol. Cell. Biol. 26 2490–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., and R. D. Kolodner, 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 23 81–85. [DOI] [PubMed] [Google Scholar]
- de Lange, T., 2002. Protection of mammalian telomeres. Oncogene 21 532–540. [DOI] [PubMed] [Google Scholar]
- Downey, M., R. Houlsworth, L. Maringele, A. Rollie, M. Brehme et al., 2006. A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell 124 1155–1168. [DOI] [PubMed] [Google Scholar]
- Downs, J. A., and S. P. Jackson, 2004. A means to a DNA end: the many roles of Ku. Nat. Rev. Mol. Cell Biol. 5 367–378. [DOI] [PubMed] [Google Scholar]
- Evans, S. K., M. L. Sistrunk, C. I. Nugent and V. Lundblad, 1998. Telomerase, Ku, and telomeric silencing in Saccharomyces cerevisiae. Chromosoma 107 352–358. [DOI] [PubMed] [Google Scholar]
- Falzon, M., J. W. Fewell and E. L. Kuff, 1993. EBP-80, a transcription factor closely resembling the human autoantigen Ku, recognizes single- to double-strand transitions in DNA. J. Biol. Chem. 268 10546–10552. [PubMed] [Google Scholar]
- Feldmann, H., and E. L. Winnacker, 1993. A putative homologue of the human autoantigen Ku from Saccharomyces cerevisiae. J. Biol. Chem. 268 12895–12900. [PubMed] [Google Scholar]
- Fisher, T. S., A. K. Taggart and V. A. Zakian, 2004. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat. Struct. Mol. Biol. 11 1198–1205. [DOI] [PubMed] [Google Scholar]
- Friesland, S., L. Kanter-Lewensohn, R. Tell, E. Munck-Wikland, R. Lewensohn et al., 2003. Expression of Ku86 confers favorable outcome of tonsillar carcinoma treated with radiotherapy. Head Neck 25 313–321. [DOI] [PubMed] [Google Scholar]
- Gangloff, S., J. P. McDonald, C. Bendixen, L. Arthur and R. Rothstein, 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolg: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14 8391–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel, S., M. Larrivee, P. Labrecque and R. J. Wellinger, 1998. Yeast Ku as a regulator of chromosomal DNA end structure. Science 280 741–744. [DOI] [PubMed] [Google Scholar]
- Gu, Y., S. Jin, Y. Gao, D. T. Weaver and F. W. Alt, 1997. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc. Natl. Acad. Sci. USA 94 8076–8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, H., R. J. Bennett and L. H. Hurley, 2000. Inhibition of unwinding of G-quadruplex structures by Sgs1 helicase in the presence of N,N′-bis[2-(1-piperidino)ethyl]-3,4,9,10-perylenetetracarboxylic diimide, a G-quadruplex-interactive ligand. Biochemistry 39 9311–9316. [DOI] [PubMed] [Google Scholar]
- Hickson, I. D., 2003. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer 3 169–178. [DOI] [PubMed] [Google Scholar]
- Hsu, H. L., D. Gilley, E. H. Blackburn and D. J. Chen, 1999. Ku is associated with the telomere in mammals. Proc. Natl. Acad. Sci. USA 96 12454–12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, M. D., D. C. Lee and N. Maizels, 2002. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 30 3954–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa, A. S., J. Q. Zhou and V. A. Zakian, 2000. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 100 479–489. [DOI] [PubMed] [Google Scholar]
- Jaco, I., P. Munoz and M. A. Blasco, 2004. Role of human Ku86 in telomere length maintenance and telomere capping. Cancer Res. 64 7271–7278. [DOI] [PubMed] [Google Scholar]
- Joyce, M. V., and L. B. McGown, 2004. Detection of G-quartet structure in a DNA aptamer stationary phase using a fluorescent dye. Appl. Spectrosc. 58 831–835. [DOI] [PubMed] [Google Scholar]
- Krogh, B. O., and L. S. Symington, 2004. Recombination proteins in yeast. Annu. Rev. Genet. 38 233–271. [DOI] [PubMed] [Google Scholar]
- Lahaye, A., H. Stahl, D. Thines-Sempoux and F. Foury, 1991. PIF1: a DNA helicase in yeast mitochondria. EMBO J. 10 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche, T., S. G. Martin, M. Gotta, H. C. Gorham, F. E. Pryde et al., 1998. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol. 8 653–656. [DOI] [PubMed] [Google Scholar]
- Lewis, L. K., G. Karthikeyan, J. W. Westmoreland and M. A. Resnick, 2002. Differential suppression of DNA repair deficiencies of yeast rad50, mre11 and xrs2 mutants by EXO1 and TLC1 (the RNA component of telomerase). Genetics 160 49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. L., R. J. Harrison, A. P. Reszka, R. M. Brosh, Jr., V. A. Bohr et al., 2001. Inhibition of the Bloom's and Werner's syndrome helicases by G-quadruplex interacting ligands. Biochemistry 40 15194–15202. [DOI] [PubMed] [Google Scholar]
- Lingner, J., and T. R. Cech, 1998. Telomerase and chromosome end maintenance. Curr. Opin. Genet. Dev. 8 226–232. [DOI] [PubMed] [Google Scholar]
- Lovett, S. T., and R. K. Mortimer, 1987. Characterization of null mutants of the RAD55 gene of Saccharomyces cerevisiae: effects of temperature, osmotic strength and mating type. Genetics 116 547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringele, L., and D. Lydall, 2002. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev. 16 1919–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori, T., and J. A. Hardin, 1986. Mechanism of interaction between Ku protein and DNA. J. Biol. Chem. 261 10375–10379. [PubMed] [Google Scholar]
- Myung, K., C. Chen and R. D. Kolodner, 2001. a Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411 1073–1076. [DOI] [PubMed] [Google Scholar]
- Myung, K., A. Datta, C. Chen and R. D. Kolodner, 2001. b SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 27 113–116. [DOI] [PubMed] [Google Scholar]
- Myung, K., A. Datta and R. D. Kolodner, 2001. c Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104 397–408. [DOI] [PubMed] [Google Scholar]
- Myung, K., G. Ghosh, F. J. Fattah, G. Li, H. Kim et al., 2004. Regulation of telomere length and suppression of genomic instability in human somatic cells by Ku86. Mol. Cell. Biol. 24 5050–5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent, C. I., G. Bosco, L. O. Ross, S. K. Evans, A. P. Salinger et al., 1998. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 8 657–660. [DOI] [PubMed] [Google Scholar]
- Nussenzweig, A., C. Chen, V. da Costa Soares, M. Sanchez, K. Sokol et al., 1996. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature 382 551–555. [DOI] [PubMed] [Google Scholar]
- Onoda, F., M. Seki, A. Miyajima and T. Enomoto, 2000. Elevation of sister chromatid exchange in Saccharomyces cerevisiae sgs1 disruptants and the relevance of the disruptants as a system to evaluate mutations in Bloom's syndrome gene. Mutat. Res. 459 203–209. [DOI] [PubMed] [Google Scholar]
- Onoda, F., M. Seki, A. Miyajima and T. Enomoto, 2001. Involvement of SGS1 in DNA damage-induced heteroallelic recombination that requires RAD52 in Saccharomyces cerevisiae. Mol. Gen. Genet. 264 702–708. [DOI] [PubMed] [Google Scholar]
- Ouyang, H., A. Nussenzweig, A. Kurimasa, V. C. Soares, X. Li et al., 1997. Ku70 is required for DNA repair but not for T cell antigen receptor gene recombination in vivo. J. Exp. Med. 186 921–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polotnianka, R. M., J. Li and A. J. Lustig, 1998. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol. 8 831–834. [DOI] [PubMed] [Google Scholar]
- Porter, S. E., P. W. Greenwell, K. B. Ritchie and T. D. Petes, 1996. The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 24 582–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves, W. H., 1987. Antinuclear antibodies as probes to explore the structural organization of the genome. J. Rheumatol. 14(Suppl. 13): 97–105. [PubMed] [Google Scholar]
- Riha, K., J. M. Watson, J. Parkey and D. E. Shippen, 2002. Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J. 21 2819–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samper, E., F. A. Goytisolo, P. Slijepcevic, P. P. van Buul and M. A. Blasco, 2000. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 1 244–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, V., and V. A. Zakian, 1994. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell 76 145–155. [DOI] [PubMed] [Google Scholar]
- Smith, S., J. Y. Hwang, S. Banerjee, A. Majeed, A. Gupta et al., 2004. Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101 9039–9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnolo, L., A. Rivera-Calzada, L. H. Pearl and O. Llorca, 2006. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol. Cell 22 511–519. [DOI] [PubMed] [Google Scholar]
- Sugawara, N., X. Wang and J. E. Haber, 2003. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell 12 209–219. [DOI] [PubMed] [Google Scholar]
- Sun, H., R. J. Bennett and N. Maizels, 1999. The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs. Nucleic Acids Res. 27 1978–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo, S. H., and S. P. Jackson, 2001. Telomerase subunit overexpression suppresses telomere-specific checkpoint activation in the yeast yku80 mutant. EMBO Rep. 2 197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto, Y., J. Kato and H. Ikeda, 1997. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature 388 900–903. [DOI] [PubMed] [Google Scholar]
- Vega, L. R., M. K. Mateyak and V. A. Zakian, 2003. Getting to the end: telomerase access in yeast and humans. Nat. Rev. Mol. Cell Biol. 4 948–959. [DOI] [PubMed] [Google Scholar]
- Vega, L. R., J. A. Phillips, B. R. Thornton, J. A. Benanti, M. T. Onigbanjo et al., 2007. Sensitivity of yeast strains with long G-tails to levels of telomere-bound telomerase. PLoS Genet. 3: e105. [DOI] [PMC free article] [PubMed]
- Wagner, M., G. Price and R. Rothstein, 2006. The absence of Top3 reveals an interaction between the Sgs1 and Pif1 DNA helicases in Saccharomyces cerevisiae. Genetics 174 555–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt, P. M., E. J. Louis, R. H. Borts and I. D. Hickson, 1995. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell 81 253–260. [DOI] [PubMed] [Google Scholar]
- Weinert, T. A., and L. H. Hartwell, 1993. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics 134 63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger, R. J., A. J. Wolf and V. A. Zakian, 1993. Origin activation and formation of single-strand TG1-3 tails occur sequentially in late S phase on a yeast linear plasmid. Mol. Cell. Biol. 13 4057–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson, J. R., M. K. Raghuraman and T. R. Cech, 1989. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell 59 871–880. [DOI] [PubMed] [Google Scholar]
- Wu, X., and N. Maizels, 2001. Substrate-specific inhibition of RecQ helicase. Nucleic Acids Res. 29 1765–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler, A. M., J. R. Williamson, T. R. Cech and D. M. Prescott, 1991. Inhibition of telomerase by G-quartet DNA structures. Nature 350 718–720. [DOI] [PubMed] [Google Scholar]
- Zhou, J., E. K. Monson, S. Teng, V. P. Schulz and V. A. Zakian, 2000. Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science 289 771–774. [DOI] [PubMed] [Google Scholar]