Abstract
Genetic differences between azuki bean (Vigna angularis var. angularis) and its presumed wild ancestor (V. angularis var. nipponensis) were resolved into QTL for traits associated with adaptation to their respective distinct habits. A genetic linkage map constructed using progenies from a cross between Japanese cultivated and wild azuki beans covers 92.8% of the standard azuki bean linkage map. A reciprocal translocation between cultivated and wild azuki bean parents was identified on the basis of the linkage map having a pseudolinkage group and clustering of seed productivity-related QTL with large effect near the presumed breakpoints. In total, 162 QTL were identified for 46 domestication-related traits. Domestication of azuki bean has involved a trade-off between seed number and seed size: fewer but longer pods and fewer but larger seeds on plants with shorter stature in cultivated azuki bean being at the expense of overall seed yield. Genes found related to germination and flowering time in cultivated azuki bean may confer a selective advantage to the hybrid derivatives under some ecological conditions and may explain why azuki bean has evolved as a crop complex in Japan.
UNDERSTANDING the genetics of plant adaptation to different habitats is of fundamental importance to crop improvement. The most fundamental changes that all crops have undergone are in relation to adaptation to “the domus” to suit the needs of humans. The genetics of traits related to fitness of species adapted to wild habitats differ from those of the same species adapted to man-made environments. For grain legumes, such as azuki beans [Vigna angularis (Willd.) Ohwi & Ohashi], among key traits that distinguish the wild variety, var. nipponensis, from the cultivated form, var. angularis, are those related to reproductive traits particularly seed number, seed size, seed dispersal, and seed dormancy.
Azuki bean is the second most important legume in Japan after soybean. The presumed wild ancestor of cultivated azuki bean is V. angularis var. nipponensis (Yamaguchi 1992). This wild species is distributed across a wide area from Japan, the Korean peninsula, and China to Nepal and Bhutan (Tomooka et al. 2002). It is not known where azuki bean was domesticated, however, azuki bean exists as a crop complex in Japan where cultivated, wild, and weedy azuki bean can be found (Vaughan et al. 2004). In addition, carbonized azuki bean seeds have been found from archaeological sites in Japan dated ≈4000 years ago (Maeda 1987; Yano et al. 2004), predating archaeobotanical remains of azuki bean in China and Korea (Crawford 2006). Thus Japan is one possible place where azuki bean was domesticated.
In comparison with its presumed wild ancestor, cultivated azuki bean shows numerous differences in morphological and physiological traits probably associated with human selection during domestication. These differences, collectively called the domestication syndrome, result from selection over several thousands of years for adaptation to cultivated environments and human nutritional requirements and preferences (Hawkes 1983).
Azuki bean was one of the first crops subjected to scientific plant breeding in Japan but landraces of azuki bean are still grown in Japan and are predominantly large, red-seeded genotypes. Azuki bean is grown throughout Japan but most production is in Hokkaido prefecture. Apart from in Hokkaido, Japanese farmers usually grow azuki beans on a small scale in kitchen gardens for home consumption. Wild azuki bean is not present in Hokkaido but is commonly found on the other main islands of Japan—Honshu, Kyushu, and Shikoku. Wild azuki bean is a climbing, annual, herbaceous plant generally having black-mottled seeds. It grows in disturbed habitats such as riverbanks, edges of paddy fields, and roadside verges. Landrace varieties across Japan where wild azuki bean grows might have accumulated alleles as a result of natural introgression and farmer selection.
Gene flow in a crop complex contributes to gene exchange between wild and cultivated gene pools. In various parts of Japan, where wild and cultivated azuki beans are sympatric, plants with variable phenotype are commonly found (Kaga et al. 2004). Although wild and cultivated azuki beans are mainly self-pollinating, outcrossing between wild and cultivated azuki bean has been observed and its rate estimated at 1% (Yamamoto et al. 2006). Some plants in wild populations have been shown to have genes from cultivated azuki bean (Wang et al. 2004). These facts suggest that natural crossing among components of this crop complex is a regular occurrence. The population dynamics of introgressed wild populations is largely affected by the fitness of hybrids and their derivatives in natural conditions (Ellstrand et al. 1999).
During the process of domestication wild plants and selections from wild populations likely hybridized and resulting variation was subjected to further cycles of human selection (Harlan 1966). Through human selection wild plants became crops adapted to cultivation and these crops exhibit reduced fitness when growing in natural habitats. However, it is not known to what extent and what factors contribute to crop genes being a disadvantage to plants in natural conditions and why weedy forms with many cultivated genes can persist in natural conditions (Gressel 2005). The persistence or extinction of crop genes in wild populations is dependent on whether crop genes confer advantages or disadvantages to hybrid fitness outside the cultivated environment. In addition, genetic drift may also play a role in the survival of crop genes in wild populations.
Previously we have reported the genetics of many traits related to the domestication syndrome in azuki bean based on a cross between wild and cultivated accessions from different geographic regions, Nepal and Japan, respectively (Han et al. 2005; Isemura et al. 2007). The first objective of the study reported here was to construct a new linkage map of azuki bean using wild and cultivated Japanese accessions to represent the azuki bean crop complex of Japan. The second objective was to analyze a broad set of traits related to the genetic dichotomy between wild and cultivated azuki beans. We particularly focus on the identification of genetic loci associated with adaptation and fitness in the distinct habitats of wild and cultivated azuki bean. These genetic loci offer insights into the dynamics of the azuki bean crop complex when pleiotropy, allometry, and epistasis of quantitative and qualitative loci are taken into consideration.
MATERIALS AND METHODS
Mapping population:
An F2 mapping population was developed from the cross between a wild azuki bean accession (JP110658) collected in Yamanashi prefecture, Japan (35° 36′N, 138° 42′E), and an azuki bean cultivar (JP109685 cv. Kyoto-Dainagon) that is widely grown in Kyoto prefecture, Japan (35°N, 135° 42′E). The cultivated parent represents a pure line selection from a landrace and was female in the cross. The F2 population consisted of 188 plants. The parental accessions used in the cross were obtained from the National Institute of Agrobiological Sciences Genebank (Tsukuba, Japan).
DNA extraction:
Total genomic DNA in F2 plants was extracted from 200 mg of fresh leaf tissue using the DNeasy plant mini kit (QIAGEN, Valencia, CA). DNA concentration was adjusted to 1 and 25 ng/μl for SSR and AFLP analyses by comparing with known concentrations of standard λ-DNA on 1.5% agarose gel, respectively.
SSR analysis and construction of linkage map:
The SSR analysis in the F2 population was carried out according to the method of Han et al. (2005). In this study, 316 SSR primer pairs for azuki bean (Wang et al. 2004), 170 from common bean (Yu et al. 2000; Gaitan-Solis et al. 2002; Blair et al. 2003; Guerra-Sanz 2004) and 45 from cowpea (Li et al. 2001) were screened to detect polymorphism between the two parents.
The linkage map was constructed according to the method of Han et al. (2005). JoinMap ver. 3.0 (Van Ooijen and Voorrips 2001) was used to construct a linkage map. The recombination frequencies were converted into map distance (cM) using the Kosambi mapping function (Kosambi 1944). After building the framework map by codominant SSR markers, dominant SSR and AFLP markers were integrated into the framework map. The resulting map revealed a cluster of SSR markers from nonhomologous linkage groups 4 and 6. To distinguish between linkage groups 4 and 6, QuadMap was used (Durrant et al. 2006). Linkage maps were constructed with 1000 marker data files in which marker order in an original file was randomized, and then marker-pair distances, variances in marker-pair distance, and marker-pair frequencies among the 1000 maps were summarized. Among the three parameters, a cutoff score of 910 was applied to a minimum marker-pair frequency to find groups of markers. The filtered marker pairs were observed more than 910 times among 1000 linkage maps. Cutoff scores for the other two parameters, marker-pair distance and the variance, were 100 cM, which is a nonconstraint condition.
Bulked segregant analysis–amplified fragment-length polymorphism analysis:
To fill a large region on linkage group 9 lacking markers, bulked segregant analysis was conducted using AFLP markers. AFLP was analyzed following the protocol of Vos et al. (1995) using AFLP core reagent kit (Invitrogen, Carlsbad, CA). On the basis of six codominant SSR loci on linkage groups 9, 14, and 16, individuals were selected from 188 F2 plants. Total genomic DNA from 14 F2 plants with cultivated-parent homozygous alleles and 16 F2 plants with wild-parent homozygous alleles for all six SSR loci were combined to form two lots of bulk DNA and compared with parents using 384 primer combinations (32 EcoRI primers and 64 MseI primers in various combinations) with three selective nucleotides at the 3′-end. All the PCR products were amplified using GeneAmp 9700 (Applied Biosystems, Foster City, CA). EcoRI primers, labeled with one of the following four fluorescent dyes, 6-FAM, VIC, NED, or PET (Applied Biosystems), were used. Two microliters of PCR products were mixed with 8 μl of Hi-Di formamide containing 0.6 μl GeneScan 500 LIZ size standard (Applied Biosystems) and run on an ABI Prism 3100 genetic analyzer (Applied Biosystems). Using GENEMAPPER ver. 3.0 software (Applied Biosystems), PCR products were separated into respective fragments and their sizes determined. Fragments showing polymorphisms between the two parents and between the two bulked DNA samples were identified on the basis of the presence or absence of clear and unambiguous fragments. The F2 population was analyzed using the primer pairs showing polymorphism, and the bands of interest were scored as dominant/recessive in the F2 individuals. The segregating AFLPs were named according to the primer combination name with the estimated molecular weight of the fragment. To fill a large region on LG9 lacking markers, bulk segregant analysis was conducted using AFLPs. Target AFLP fragments were converted into PCR-based markers (supplemental data 1 at http://www.genetics.org/supplemental/).
Trait measurement:
A total of 46 traits related to fitness and domestication were evaluated (Table 1). Of these, 43 were treated as quantitative traits and 3, epicotyl color, seed-coat color, and black mottle of seed coat, were treated as qualitative traits. The F2 population of 188 plants, together with 10 plants of each parent, were grown in the field at the National Institute of Agrobiological Sciences (NIAS), Tsukuba, Japan, 36° 2′N, 140° 8′E, from July to November, in 2003. For the F2:3 population, 188 lines of 10 plants per line, together with 10 plants of each parent, were grown in the field at NIAS in 2004.
TABLE 1.
Domestication- and fitness-related traits examined in the cross between cultivated azuki bean and its presumed wild ancestor
| General attribute | Organ | Trait | Trait abbreviation | Evaluation method |
|---|---|---|---|---|
| Seed dormancy | Seed | Germination in field (%) | SDG | Germinated seedlings at 10th day after sowing in field |
| Days to germination in field (days) | SDDG | No. of days from sowing to germination in field | ||
| Seed-coat permeability (%) | SDP | Percentage of imbibed seeds at 21 days after sowing at 25° in incubator | ||
| Winter survival seed in soil (%) | SDWS | No. of germinated seeds and survived seeds in the soil in the time from sowing in field in winter (December 2004) to next spring (May 2005) | ||
| Days to germination of winter survival seed in field (day) | SDDGWS | No. of days from sowing to germination of winter survival seed in field | ||
| Water content (g) | SDWC | Difference between weight of 50 seeds before and after desiccation at 105° for 3 days | ||
| Pod dehiscence | Pod | No. of twists (count) | PDT | No. of twists along the length of the shattered pod |
| Gigantism | Seed | 100 seed weight (g) | SD100WT | Weight of 100 seeds |
| Length (mm) | SDL | Maximum distance from top to bottom of the seed | ||
| Width (mm) | SDW | Maximum distance from hilum to its opposite side | ||
| Thickness (mm) | SDT | Maximum distance between both sides of the hilum | ||
| Pod | Length (cm) | PDL | Length of straight pod | |
| Width (mm) | PDW | Maximum width | ||
| Stem | Thickness (mm) | STT | Stem diameter under the primary leaf | |
| Leaf | Primary leaf length (mm) | LFPL | Distance from pulvinus to leaf tip | |
| Primary leaf width (mm) | LFPW | Maximum width | ||
| Maximum leaflet length (mm) | LFML | Length of the largest terminal leaflet on leaves between node on first trifoliate leaf and node on tenth trifoliate leaf | ||
| Maximum leaflet width (mm) | LFMW | Width of the largest terminal leaflet on leaves between node on first trifoliate leaf and node on tenth trifoliate leaf | ||
| Plant type | Epicotyl | Length (cm) | ECL | Length from cotyledon to primary leaf |
| Stem | Internode length (first to tenth) (cm) | ST1–10I | Length from node on primary leaf to each node | |
| Length (cm) | STL | Length from node on primary leaf to node on tenth trifoliate leaf | ||
| Twining (%) | STTW | Rate of twining plants that main stem upper tenth internode twined | ||
| Branch | No. (count) | BRN | No. of branches on main stem from node on first trifoliate leaf to node on tenth trifoliate leaf | |
| Position of ith branch (ith node) | BRP | Position of first branch on main stem from node on first trifoliate leaf to node on tenth trifoliate leaf | ||
| Earliness | Flower | Days to first flower (day) | FLD | No. of days from sowing to flowering of first flower |
| Pod | Days to maturity of 25% pods (day) | PDDM25 | No. of days from flowering of first flower to maturity of 25% pods | |
| Days to maturity of 50% pods (day) | PDDM50 | No. of days from flowering of first flower to maturity of 50% pods | ||
| Days to maturity of 75% pods (day) | PDDM75 | No. of days from flowering of first flower to maturity of 75% pods | ||
| Days to maturity of 100% pods (day) | PDDM100 | No. of days from flowering of first flower to maturity of 100% pods | ||
| Yield potential | Seed | Total weight (g) | SDTWT | Total weight of harvested seeds |
| Total no. (seed) | SDTN | Total no. of harvested seeds | ||
| No. of seeds per pod (seed/pod) | SDNPPD | No. of seeds per pod | ||
| Pod | Total no. (pod) | PDTN | Total no. of harvested pods | |
| Pigmentation | Pod | Color (degree of darkness) | PDC | Pod color, divided into six classes (0, white; 5, black) according to the degree of pigmentation |
| Epicotyl | Color | ECC | Red or green | |
| Seed | Seed-coat color | SDC | Ivory or red | |
| Seed-coat color black mottle color | SDCBM | Present or absent |
The seedling traits primary leaf length (LFPL), primary leaf width (LFPW), epicotyl color (ECC), and epicotyl length (ECL) were recorded when the first trifoliate leaf opened and the vegetative traits maximum leaflet length (LFML), maximum leaflet width (LFMW), stem internode length (ST1I–ST10I), stem length (STL), stem twining (STTW), stem thickness (STT), branch number (BRN), and branch position (BRP) were recorded when the tenth trifoliate leaf was fully developed. STTW was evaluated on the basis of twining above the tenth internode. LFPL, LFPW, LFML, LFMW, and STT were evaluated in the F2 and F2:3 populations. All other traits were evaluated in either the F2 or F2:3 population (see Table 2 for details).
TABLE 2.
The mean, standard deviation, and heritability values for parents, the F2 and F2:3 populations of the cross between cultivated and wild azuki bean
| Cultivated azuki bean
|
Wild ancestor
|
F2 or F2:3b
|
||||||
|---|---|---|---|---|---|---|---|---|
| Traita | Pop.b | Mean | SDc | Mean | SDc | Mean | SDc | Heritability (%) |
| PDT (count) | F2 | 0.6 | 0.03 | 2.8 | 0.13 | 2.1 | 0.85 | 98.9 |
| PDL (cm) | F2 | 11.1 | 0.16 | 5.8 | 0.14 | 7.1 | 1.21 | 98.5 |
| PDW (mm) | F2 | 12.9 | 0.51 | 5.4 | 0.42 | 8.2 | 0.70 | 56.2 |
| STT (mm) | F2 | 8.5 | 0.71 | 4.2 | 0.26 | 6.4 | 0.86 | 61.1 |
| LFPL (mm) | F2 | 52.0 | 4.10 | 23.2 | 4.19 | 41.6 | 6.02 | 52.6 |
| LFPW (mm) | F2 | 51.2 | 4.67 | 18.2 | 2.14 | 37.5 | 5.35 | 53.9 |
| LFML (mm) | F2 | 113.9 | 8.40 | 58.4 | 2.16 | 83.0 | 12.80 | 77.0 |
| LFMW (mm) | F2 | 101.2 | 8.97 | 43.0 | 4.40 | 72.1 | 9.90 | 49.1 |
| FLD (day) | F2 | 70.9 | 2.80 | 84.3 | 3.59 | 81.7 | 8.82 | 86.7 |
| PDDM25 (day) | F2 | 79.0 | 7.18 | 45.3 | 5.56 | 60.0 | 9.27 | 52.0 |
| PDDM50 (day) | F2 | 88.0 | 10.53 | 51.5 | 4.93 | 66.1 | 9.87 | 30.7 |
| PDDM75 (day) | F2 | 95.2 | 11.41 | 58.5 | 5.20 | 72.6 | 10.56 | 29.5 |
| PDDM100 (day) | F2 | 112.3 | 2.88 | 79.3 | 8.34 | 89.5 | 12.17 | 73.7 |
| PDTN (pod) | F2 | 104.2 | 43.98 | 608.3 | 228.44 | 362.9 | 163.45 | — |
| SDTWT (g) | F2 | 81.7 | 40.73 | 104.9 | 40.93 | 115.7 | 53.70 | 42.2 |
| SDTN (seed) | F2 | 350.2 | 173.47 | 4036.0 | 1522.49 | 1424.2 | 738.14 | — |
| SDNPPD (seed/pod) | F2 | 6.0 | 0.19 | 8.5 | 0.19 | 6.3 | 1.65 | 98.6 |
| SDG (%) | F2 | 73.3 | 5.77 | 0.0 | 0.00 | 31.6 | 22.44 | 96.7 |
| SDDG (day) | F2 | 11.7 | 0.68 | 19.6 | 2.48 | 13.5 | 2.82 | 58.4 |
| SDP (%) | F2 | 90.0 | 14.14 | 15.0 | 7.07 | 41.8 | 27.51 | 83.5 |
| SDWS (%) | F2 | 20.8 | — | 70.8 | — | 20.4 | 19.17 | — |
| SDDGWS (day) | F2 | 130.2 | 11.92 | 134.2 | 8.86 | 125.2 | 7.84 | — |
| SDWC (g) | F2 | 1.02 | 0.22 | 0.12 | 0.00 | 0.38 | 0.09 | — |
| SD100WT (g) | F2 | 24.0 | 1.41 | 2.5 | 0.08 | 8.9 | 2.10 | 77.3 |
| SDL (mm) | F2 | 9.8 | 0.58 | 4.0 | 0.44 | 7.0 | 0.70 | 44.7 |
| SDW (mm) | F2 | 6.4 | 0.22 | 3.0 | 0.14 | 4.8 | 0.33 | 70.2 |
| SDT (mm) | F2 | 5.8 | 0.23 | 2.6 | 0.04 | 4.3 | 0.35 | 76.3 |
| STT (mm) | F2:3 | 7.9 | 0.61 | 2.8 | 0.11 | 5.6 | 0.78 | 68.6 |
| LFPL (mm) | F2:3 | 54.2 | 1.42 | 25.5 | 1.47 | 39.0 | 3.64 | 84.2 |
| LFPW (mm) | F2:3 | 53.1 | 1.89 | 19.5 | 0.80 | 34.8 | 3.80 | 85.4 |
| LFML (mm) | F2:3 | 106.0 | 3.06 | 60.3 | 2.30 | 85.5 | 8.41 | 89.6 |
| LFMW (mm) | F2:3 | 88.2 | 0.97 | 49.5 | 0.91 | 71.7 | 7.54 | 98.5 |
| ECL (cm) | F2:3 | 7.3 | 0.93 | 1.5 | 0.25 | 3.6 | 0.98 | 51.3 |
| ST1I (cm) | F2:3 | 2.1 | 0.13 | 0.7 | 0.13 | 1.3 | 0.37 | 87.9 |
| ST2I (cm) | F2:3 | 1.8 | 0.13 | 1.1 | 0.07 | 1.4 | 0.30 | 88.5 |
| ST3I (cm) | F2:3 | 2.1 | 0.19 | 1.3 | 0.08 | 1.5 | 0.30 | 77.4 |
| ST4I (cm) | F2:3 | 2.5 | 0.14 | 2.0 | 0.11 | 2.0 | 0.47 | 93.2 |
| ST5I (cm) | F2:3 | 2.9 | 0.14 | 3.5 | 0.16 | 2.7 | 0.65 | 94.6 |
| ST6I (cm) | F2:3 | 3.1 | 0.13 | 4.6 | 0.21 | 3.4 | 0.75 | 94.7 |
| ST7I (cm) | F2:3 | 3.3 | 0.06 | 6.1 | 0.07 | 3.8 | 0.82 | 99.4 |
| ST8I (cm) | F2:3 | 3.3 | 0.06 | 7.0 | 0.16 | 4.1 | 0.96 | 98.5 |
| ST9I (cm) | F2:3 | 3.3 | 0.07 | 8.5 | 0.20 | 4.4 | 1.21 | 98.4 |
| ST10I (cm) | F2:3 | 3.5 | 0.11 | 10.6 | 0.12 | 4.8 | 1.56 | 99.5 |
| STL (cm) | F2:3 | 27.7 | 0.68 | 45.5 | 0.70 | 29.4 | 6.26 | 98.8 |
| STTW (%) | F2:3 | 0.0 | 0.00 | 100.0 | 0.00 | 45.3 | 36.59 | 100.0 |
| BRN (count) | F2:3 | 3.5 | 0.17 | 4.6 | 0.09 | 3.7 | 0.94 | 97.9 |
| BRP (ith) | F2:3 | 2.3 | 0.22 | 1.1 | 0.25 | 2.0 | 0.63 | 86.4 |
| PDC (degree) | F2 | 0.0 | 0.00 | 5.0 | 0.00 | 2.6 | 1.44 | 100.0 |
| ECC | F2 | Green | Purple | G:P = 50:138 (χ2 = 0.255) | ||||
| SDC | F2 | Red | No red | R:N = 48:140 (χ2 = 0.028) | ||||
| SDCBM | F2 | Absent | Present | A:P = 54:134 (χ2 = 1.390) | ||||
Trait abbreviations are shown in Table 1.
Population of trait value is listed.
Standard deviation.
The seed-related traits were investigated using the seeds and pods from F2 plants. Seed germination was investigated under three different conditions. The first experiment, seed-coat permeability (SDP), was carried out in the laboratory. Ten unscarified seeds stored at 10° for 1 year were placed on wet filter paper, incubated in the dark at 25° for 21 days, and the number of seeds that imbibed water was counted daily. The second experiment, seed germination (SDG) and days to germination of seed (SDGD), was carried out in the field from mid June, 2004. Ten unscarified seeds were sown in the field and the date of germination and number of germinated seeds was recorded. The third experiment, seed winter survival (SDWS) and days to germination of winter survival seed (SDWSGD), was carried out in the field from winter to the following spring. Ten unscarified seeds were sown in the field in December, 2004, and the date and number of germinated seeds and nongerminated hard seeds in the soil was recorded in May, 2005. Seed dimensions (SDL, SDW, and SDT) were the average of 5 seeds. The 100-seed weight (SD100WT) was evaluated using intact seeds.
The pod traits—pod length (PDL), pod width (PDW), pod dehiscence, and number of twists along the length of the dehisced pod when kept at room temperature (PDT)—were based on 10 pods. Pod dehiscence was scored as dehiscent or indehiscent on the basis of whether seeds scattered from pods in harvest envelope or not. The PDT was used as index of pod structure. The pod color (PDC) was divided into six classes according to the degree of darkness (0, light tan; 1, pale brown; 2, brown; 3, dark brown; 4, blackish brown; and 5, black).
The number of days from sowing to first flowering (FLD) was recorded in the F2 population. The days to 25%, 50%, 75%, and 100% mature pods (PDDM25, PDDM50, PDDM75, and PDDM100) were defined as number of days from first flowering to harvesting of 25%, 50%, 75%, and 100% of pods of the total number of pods, respectively. After harvesting all pods, total pod number (PDTN), total seed number (SDTN), and total seed weight (SDTWT) were measured for each individual. The number of seeds per pod (SDNPPD) was measured using 10 pods.
Data analysis:
The mean, standard deviation, and broad sense heritability were calculated, and the frequency distribution of phenotypes in F2 and F2:3 populations were examined for each trait (supplemental data 2 at http://www.genetics.org/supplemental/). The correlation coefficient between each trait was also calculated. Path analysis, graphical linear statistical modeling, was used to further investigate the interaction between seed productivity and its related traits using Excel GM software (Ohmsha, Japan). The total seed number per plant was used as an index of seed productivity and was the dependent variable. Since stem length (the length from the eighth to tenth internodes), branch number, leaf size (the product of maximum length and width), 100-seed weight, pod length, total number of pods per plant, and seed number per pod had significant correlation with total seed number, these traits were selected as the independent variables.
QTL analysis:
The QTL analysis was conducted by using the software package MultiQTL ver. 2.6 according to procedures described by Peng et al. (2003). In brief, the entire genome was scanned for each trait using general interval mapping with the following approach. First, a single QTL model was fitted for each trait-chromosome (linkage group) combination. Chromosomewise statistical significance thresholds (α = 0.001) for declaring putative QTL were obtained by 10,000 runs of a permutation test (Churchill and Doerge 1994) and parameters (position, additive and dominance effect, and the percentage of explained variance) for significant QTL were obtained. Standard deviations were estimated on the basis of 10,000 bootstrap sampling per linkage group. When the LOD graph indicated the possibility of two QTL, a two-linked QTL model (Korol et al. 1998) was fitted for each trait-chromosome combination and putative two-linked QTL was declared at the same threshold as described above. Further, the hypothesis of two-linked QTL in the chromosome (H2) was compared with single QTL (H1) at the α = 0.01 level using parametric bootstrap (Ronin et al. 1999). For the traits evaluated over two years on the same population, the single or two-linked QTL models for multiple environments (Jansen et al. 1995) was fitted and tested using the same procedure.
Multiple interval mapping (MIM) (Kao et al. 1999) was then conducted to reduce the background variation by taking into account QTL effects from other chromosomes. On the basis of parameters defined for each putative QTL above, the chromosome was included or removed iteratively into/from the MIM model at the more stringent level of significance (α = 0.01) than default. The stepwise selection of chromosomes based on significance using a permutation test was repeated until the process converged when no QTL on the remaining chromosomes were found. The QTL effects were reevaluated by fitting all positive QTL in the order of their power and by a global permutation test (10,000 runs) to get more precise estimates of significance. To correct for multiple comparisons, experimentwise significance level for all QTL was estimated on the basis of the method of Benjamini and Hogberg (1995) and QTL significant at false discovery rate (FDR) = 0.05 are reported in this study.
A chi-square goodness-of-fit test for observed number of domestication- and fitness-related QTL to the expected number of QTL across each linkage group was applied to determine the random distribution of QTL with the assumption of independent gene action. To test whether or not QTL were randomly distributed along a linkage group, a Poisson distribution function P(x) = e−μμx/x!, where x is the number of QTL per 10-cM interval and μ is the average QTL density on linkage groups, was calculated.
RESULTS
Linkage map construction:
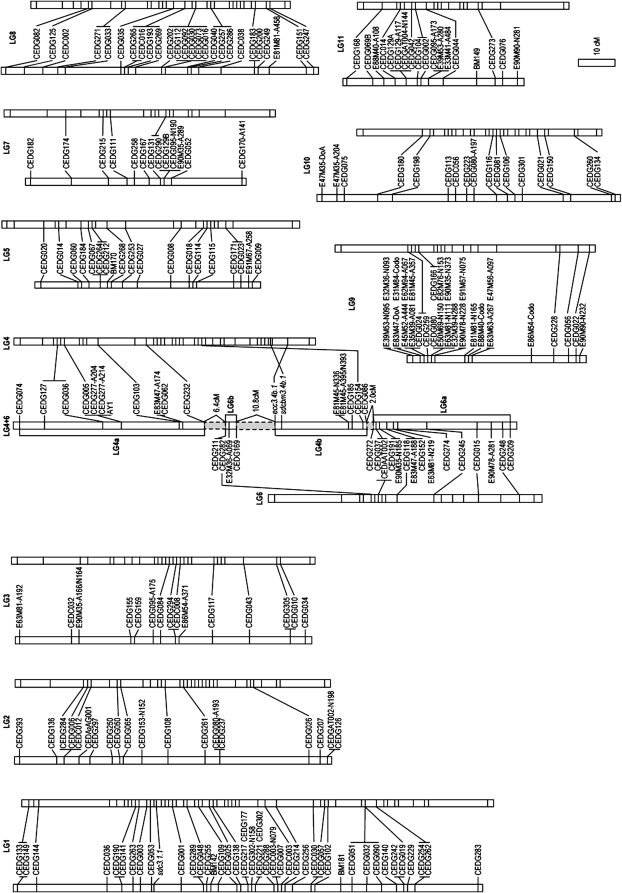
SSR markers were screened to reveal polymorphism between the cultivated and wild parents. Of 316 azuki bean SSR primer pairs, 176 revealed clear polymorphism. In addition, of 170 common bean SSR primer pairs, 5 (BM140, BM149, BM170, BM181, and AY1) revealed clear polymorphism. No clear polymorphism was detected in 45 cowpea primer pairs. All 191 marker loci derived from the 181 SSR primer pairs were assigned to 10 LGs less than the 11 haploid chromosome number of azuki bean. Although the marker order on every linkage group was highly conserved with the azuki bean linkage map developed by Han et al. (2005), one linkage group, named “LG4+6” (Figure1), was found to consist of markers from LG4 and LG6 of the previous map and other maps developed for related Vigna species by Somta et al. (2006) and T. Isemura and A. Kaga (unpublished results). In the middle of this linkage group, a cluster of three SSR markers, CEDG185, CEDG154, and CEDG086 and two morphological marker genes, epicotyl color (ecc3.4a.1) and black-mottled seed coat (sdcbm3.4a.1), from the linkage group 4 of the previous map (Han et al. 2005) was intermingled with a cluster of three SSR markers, CEDG211, CEDG282, and CEDG169, from linkage group 6 of the previous map in spite of all markers in this linkage group showing normal segregation ratios. These markers revealed genotypes related to each other and cannot be separated into two groups under the general LOD score 3.0 that is used for marker grouping but partially separated at LOD > 17. Livingstone et al. (2000) observed that markers near reciprocal translocation breakpoints clustered together with markers from both chromosomes into a single “pseudolinkage group.” By using QuadMap (Durrant et al. 2006), multiple maps were constructed to find groups of markers and the presumed reciprocal translocation was evaluated using the variance in marker-pair distance among the permutated maps. A segment from CEDG074 to CEDG232 of linkage group 4 and a segment from CEDG272 to CEDG209 of linkage group 6 were consistently found among 1000 permutated codominant SSR marker maps (supplemental data 3 at http://www.genetics.org/supplemental/). On the other hand, all intermingled markers did not consistently appear and were not clearly grouped; together these were represented as quadrivalent maps. Since the observed variance was almost 0 cM comparing to the larger variation ranging from 0 to 4283.9 cM reported by Durrant et al. (2006), it was not possible to distinguish two translocation segments but instead intermingled markers from the two interstitial segments by the marker-pair frequency. For further analysis, the two interstitial segments (LG4a and LG6a) and two tentative translocation segments (LG4b and LG6b) were grouped using information from other linkage maps to prevent artificial genotypes in interval QTL mapping between the segments (shaded area in Figure1).
Figure 1.—
A comparative genetic linkage map of azuki bean based on common SSR markers for crosses between Japanese azuki bean cv. Kyoto Dianagon and wild azuki bean from Japan (left) and between Japanese azuki bean cv. Tokushima landrace and wild azuki bean from Nepal (right). The linkage map on the right is modified from Han et al. (2005). Shading in linkage group LG4+6 indicates the segment that was not included in QTL analysis.
In addition, this map included a large gap in the middle of LG9 of ∼40 cM. To find markers to fill this gap, bulked segregant analysis was carried out on the basis of AFLP analysis. Polymorphism between parents and two samples of bulked DNA of selected F2 individuals (see materials and methods) were screened with 384 AFLP primer pairs. In total 13,085 bands were detected, 446 (3.4%) were polymorphic between the parents. Of 446 polymorphic bands, only 21 bands from 21 primer pairs were polymorphic between the two samples of bulked F2 DNA. The entire F2 population was analyzed using these 21 primer pairs. Forty-one bands including the 21 bands expected to be located on LG9 segregated and were integrated into the SSR linkage map. As expected, the 21 polymorphic bands detected by bulked segregation analysis were mapped on LG9. However, only five AFLP markers were mapped in the middle region with a large gap.
Six AFLP markers on LG9 (E31M84-A329, E31M84-N331, E83M47-A125, E88M40-A123, E86M54-N075) and LG10 (E47M35-A151) were converted into STS markers. The AFLP markers E31M84-A329 and E31M84-N331 on LG9 were putatively codominant given their segregation (1:2:1) in this mapping population. The sequence of DNA in the AFLP band derived from the cultivated parent had 2 bases inserted compared to that of the wild parent. A primer pair was designed on the basis of sequence information so that this polymorphic region could be amplified and mapped as a codominant marker (E31M84-Codo). The region adjacent to AFLP marker E88M40-A123 could be amplified by anchor PCR from both parents. Comparison of the sequences of both parents revealed an insertion of 19 bases in DNA of the AFLP band derived from the cultivated parent. This polymorphic sequence was mapped as a codominant marker (E88M40-Codo). For the AFLP marker E86M54-N075, the adjacent region of this marker was characterized in both parents. Comparison of the sequences of both parents reveals that there was a restriction enzyme site for EcoRI in the sequence of wild parent. PCR product amplification using a nested gene-specific primer and a nested adapter primer was treated with EcoRI to detect polymorphisms in the F2 population. The AFLP marker E86M54-N075 was converted into a CAPS marker and mapped as a codominant marker (E86M54-Codo). The AFLP markers E83M47-A125 (LG9) and E47M35-A151 (LG10), since the adjacent region in the wild parent could not be amplified by PCR using nested gene-specific and adapter primers, were mapped on the linkage map as dominant SCAR markers (E83M47-DoA, E47M35-DoA).
As a result, a total of 233 markers, 191 SSR, 2 STS, 1 CAPS, 2 SCAR, 36 AFLP marker(s), and three morphological genes were mapped on 10 linkage groups (Figure 1). This map spans a total length of 771.9 cM with an average marker distance of 3.48 cM, and the coverage was 92.8% of the azuki bean map reported by Han et al. (2005).
Field data analysis:
The means, standard deviation, and heritability of traits in the parental lines F2 and F2:3 populations are shown in Table 2. The means of the cultivated parent were higher than those of the wild parent in SDG and SDP, whereas the means of the wild parent were higher than those of the cultivated parent in SDWS. In the cultivated parent the SDDG (summer) and SDDGWS (spring) were shorter than those of the wild parent. The PDT of the wild parent was more than that of the cultivated parent. The size of leaf, stem, seed, and pod in the cultivated parent were larger than those of the wild parent. For stem length-related traits, ECL and the ST1I–ST4I in the cultivated parent were longer than those in the wild parent, whereas the ST5I–ST10I and STL in the cultivated parent were shorter than those in the wild parent. The FLD of the cultivated parent was shorter than that for the wild parent, whereas the PDDM25–PDDM100 of the cultivated parent was longer than that for the wild parent. The SDTWT, SDTN, PDTN, and SDNPPD in the wild parent were greater than those in the cultivated parent.
The F2 and F2:3 populations showed a high degree of morphological and physiological variation (Table 2). High heritability (>70%) was observed for many traits. The means of F2 plants and F2:3 lines fell between the means of cultivated and wild parents for all traits except for SDWS, SDDGWS, and ST4I and ST5I. Most traits showed a nearly normal distribution among lines (or plants) in these populations (supplemental data 2). Transgressive segregation was observed in FLD, SDWS, SDDGWS, SDTWT, SDNPPD, ST4I, and ST5I. The PDT and FLD showed nearly binomial distribution among lines (or plants) in these populations.
In general, there were significant positive correlations between similar or related traits such as between stem length and each internode length, between seed size-related traits and between yield-related traits at P ≤ 0.05 (supplemental data 4 at http://www.genetics.org/supplemental/). Seed, pod, stem- and leaf-size-related traits, and days to pod maturity were positively correlated with each other. On the other hand, these traits were negatively correlated with the days to flowering and seed productivity-related traits. For seed germination-related traits, SDG and SDP were positively correlated with each other but negatively correlated with SDWS at P ≤ 0.001. PDTN and SDNPPD were highly correlated with upper internode (ST8I–ST10I) and PDL, respectively. SDTWT was highly correlated (r = 0.86, P ≤ 0.001) with the value of SDTN multiplied by SD100WT, despite these traits being evaluated separately.
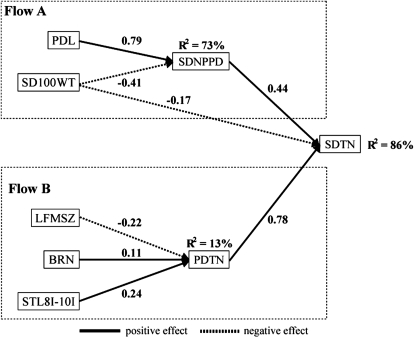
The seed productivity trait, SDTN, is an important fitness-related trait. The relationship between seed productivity and related traits was investigated by path analysis (Figure 2). PDTN and SDNPPD showed positive direct effects (0.78 and 0.44, P ≤ 0.05, respectively) on SDTN. The eighth to tenth upper internode length and BRN showed positive direct effects (0.24 and 0.11, P ≤ 0.10, respectively) on PDTN, whereas leaf size (product of length and width) showed negative direct effects (−0.22, P ≤ 0.05). PDL showed positive direct effect (0.79, P ≤ 0.05) on SDNPPD, whereas SD100WT showed negative direct effects (−0.41, P ≤ 0.05). Hence longer upper internode length and many branches via total pod number indirectly increase the total seed number. Longer pod via seed number per pod increases total seed number, whereas large leaf via total pod number and large seed via seed number per pod decrease total seed number.
Figure 2.—
Path diagram by graphical modeling showing traits that directly and indirectly effect total seed number per plant. The value shown along arrows is the path coefficient. R2, coefficient of determination ST8I–10I:length from the eighth internode to the tenth internode; LFMSZ, maximum leaf size (product of length and width).
QTL for each trait:
The results of QTL analyses for each trait in each population are shown in Table 3, Figure 3, and in supplemental data 5 at http://www.genetics.org/supplemental in detail. In total, 162 QTL and three morphological marker genes are reported here for 46 domestication- and fitness-related traits. This number of QTL overestimates the total number of QTL due to the measurement of correlated traits. Generally 1–9 QTL were detected for each trait at the level of significance (α = 0.01) except for BRN for which no QTL was detected.
TABLE 3.
QTL detected in the populations derived from the cross between cultivated azuki bean and its presumed wild ancestor
| F2
|
F2:3
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | QTL name | LG | LOD | P-value | Loci (cM) | PEV (%) | Additive effect | Dominant effect | D/|A| | LG | LOD | P-value | Loci (cM) | PEV (%) | Additive effect | Dominant effect | D/|A| |
| SDGa | Sdg3.1.1+b | 1 | 6.1 | 0.0027 | 33.7 | 8.8 | 3.40 | 0.00 | 0.00 | ||||||||
| Sdg3.1.2−(od) | 69.6 | 6.1 | −0.14 | −2.17 | −15.50 | ||||||||||||
| SDDG | Sddg3.9.1− | 9 | 4.1 | 0.0017 | 13.6 | 25.8 | −1.16 | −1.94 | −1.66 | ||||||||
| SDPa | Sdp3.1.1+b | 1 | 14.2 | 0.0001 | 33.8 | 25.9 | 11.32 | −2.20 | −0.19 | ||||||||
| Sdp3.6a.1− | 6a | 5.8 | 0.0001 | 0.5 | 9.9 | −2.23 | −3.24 | −1.45 | |||||||||
| SDWS | Sdws3.1.1+(od) | 1 | 4.8 | 0.0006 | 41.1 | 14.7 | −0.79 | 3.99 | 5.05 | ||||||||
| SDDGWS | Sddgws3.2.1− | 2 | 3.3 | 0.0088 | 62.3 | 35.3 | −4.54 | −7.11 | −1.57 | ||||||||
| SDWC | Sdwc3.1.1+ | 1 | 7.2 | 0.0001 | 91.4 | 6.9 | 0.03 | −0.01 | −0.30 | ||||||||
| Sdwc3.5.1+ | 5 | 12.6 | 0.0001 | 2.2 | 12.9 | 0.05 | 0.01 | 0.19 | |||||||||
| Sdwc3.6a.1+(od) | 6a | 19.2 | 0.0001 | 0.5 | 20.0 | 0.02 | 0.08 | 4.53 | |||||||||
| Sdwc3.9.1+ | 9 | 14.0 | 0.0001 | 18.6 | 19.5 | 0.05 | −0.02 | −0.39 | |||||||||
| Sdwc3.10.1+ | 10 | 4.9 | 0.0001 | 47.8 | 4.0 | 0.03 | 0.00 | 0.17 | |||||||||
| Sdwc3.11.1+ | 11 | 6.4 | 0.0001 | 9.1 | 6.5 | 0.03 | 0.02 | 0.82 | |||||||||
| PDTa | Pdt3.7.1−b | 7 | 49.7 | 0.0001 | 7.5 | 74.3 | −0.85 | 0.75 | 0.87 | ||||||||
| SD100WTa | Sd100wt3.1.1+b | 1 | 9.5 | 0.0001 | 90.1 | 6.4 | 0.75 | −0.14 | −0.18 | ||||||||
| Sd100wt3.2.1+b | 2 | 10.7 | 0.0001 | 47.6 | 8.2 | 0.83 | 0.27 | 0.33 | |||||||||
| Sd100wt3.5.1+b | 5 | 14.6 | 0.0001 | 11.2 | 10.6 | 0.93 | 0.34 | 0.36 | |||||||||
| Sd100wt3.6a.1+(od) | 6a | 26.8 | 0.0001 | 0.5 | 23.3 | 0.46 | 1.91 | 4.12 | |||||||||
| Sd100wt3.9.1+b | 9 | 15.9 | 0.0001 | 19.9 | 15.3 | 1.15 | −0.21 | −0.18 | |||||||||
| Sd100wt3.10.1+ | 10 | 10.8 | 0.0001 | 53.5 | 6.5 | 0.75 | 0.15 | 0.20 | |||||||||
| Sd100wt3.11.1+ | 11 | 8.5 | 0.0001 | 30.3 | 5.5 | 0.55 | 0.61 | 1.10 | |||||||||
| SDLa | Sdl3.1.1+b | 1 | 12.9 | 0.0001 | 92.2 | 14.4 | 0.38 | −0.08 | −0.21 | ||||||||
| Sdl3.5.1+ | 5 | 4.1 | 0.0010 | 32.9 | 4.0 | 0.20 | 0.00 | −0.02 | |||||||||
| Sdl3.6a.1+(od) | 6a | 28.8 | 0.0001 | 0.3 | 34.6 | 0.13 | 0.81 | 6.17 | |||||||||
| Sdl3.9.1+b | 9 | 9.4 | 0.0001 | 19.1 | 12.8 | 0.36 | −0.03 | −0.07 | |||||||||
| SDWa | Sdw3.1.1+b | 1 | 6.4 | 0.0001 | 90.6 | 5.6 | 0.11 | −0.05 | −0.48 | ||||||||
| Sdw3.2.1+b | 2 | 6.2 | 0.0001 | 47.0 | 6.1 | 0.11 | 0.05 | 0.42 | |||||||||
| Sdw3.4a.1+ | 4a | 5.7 | 0.0001 | 27.2 | 4.0 | 0.09 | 0.03 | 0.34 | |||||||||
| Sdw3.5.1+ | 5 | 9.0 | 0.0001 | 9.0 | 7.7 | 0.13 | 0.02 | 0.16 | |||||||||
| Sdw3.6a.1+(od) | 6a | 18.6 | 0.0001 | 0.5 | 19.7 | 0.06 | 0.28 | 4.50 | |||||||||
| Sdw3.9.1+b | 9 | 10.5 | 0.0001 | 9.0 | 9.5 | 0.14 | 0.02 | 0.15 | |||||||||
| Sdw3.10.1+ | 10 | 7.0 | 0.0001 | 53.5 | 5.3 | 0.10 | 0.05 | 0.49 | |||||||||
| Sdw3.11.1+ | 11 | 9.4 | 0.0001 | 10.1 | 4.1 | 0.05 | 0.10 | 1.92 | |||||||||
| Sdw3.11.2+ | 40.0 | 4.6 | 0.09 | 0.00 | 0.04 | ||||||||||||
| SDTa | Sdt3.1.1+b | 1 | 5.6 | 0.0001 | 84.2 | 5.2 | 0.09 | −0.09 | −0.92 | ||||||||
| Sdt3.2.1+b | 2 | 8.7 | 0.0001 | 52.9 | 8.1 | 0.14 | 0.05 | 0.35 | |||||||||
| Sdt3.4a.1+ | 4a | 3.3 | 0.0051 | 27.2 | 2.5 | 0.08 | 0.01 | 0.13 | |||||||||
| Sdt3.5.1+b | 5 | 13.0 | 0.0001 | 4.6 | 15.4 | 0.18 | 0.10 | 0.55 | |||||||||
| Sdt3.6a.1+(od) | 6a | 16.1 | 0.0001 | 0.0 | 14.3 | 0.02 | 0.26 | 14.11 | |||||||||
| Sdt3.9.1+b | 9 | 12.1 | 0.0001 | 17.5 | 13.8 | 0.18 | −0.04 | −0.23 | |||||||||
| Sdt3.11.1+ | 11 | 9.5 | 0.0001 | 6.2 | 4.6 | 0.09 | 0.06 | 0.72 | |||||||||
| Sdt3.11.2+(od) | 33.1 | 3.7 | 0.01 | 0.12 | 14.35 | ||||||||||||
| PDLa | Pdl3.1.1+b | 1 | 8.0 | 0.0001 | 88.9 | 6.5 | 0.41 | −0.02 | −0.04 | ||||||||
| Pld3.5.1+ | 5 | 12.7 | 0.0001 | 18.1 | 9.0 | 0.42 | −0.36 | −0.87 | |||||||||
| Pld3.6a.1−(od) | 6a | 26.1 | 0.0001 | 0.5 | 25.9 | 0.21 | −1.13 | −5.40 | |||||||||
| Pdl3.7.1+b | 7 | 13.4 | 0.0001 | 4.5 | 11.5 | 0.48 | −0.40 | −0.84 | |||||||||
| Pdl3.9.1+b | 9 | 10.7 | 0.0001 | 17.4 | 9.5 | 0.45 | 0.30 | 0.67 | |||||||||
| Pdl3.10.1−b | 10 | 5.5 | 0.0001 | 53.4 | 3.8 | −0.31 | −0.07 | −0.23 | |||||||||
| Pdl3.11.1+ | 11 | 6.5 | 0.0001 | 0.0 | 5.1 | 0.30 | 0.31 | 1.04 | |||||||||
| PDWa | Pdw3.1.1+ | 1 | 11.0 | 0.0001 | 91.0 | 12.7 | 0.33 | −0.12 | −0.37 | ||||||||
| Pdw3.4a.1+ | 4a | 9.1 | 0.0001 | 34.2 | 11.0 | 0.29 | −0.19 | −0.67 | |||||||||
| Pdw3.6a.1+(od) | 6a | 7.3 | 0.0001 | 0.5 | 6.9 | 0.09 | 0.33 | 3.64 | |||||||||
| Pdw3.7.1+b | 7 | 10.4 | 0.0001 | 6.6 | 11.6 | 0.28 | −0.24 | −0.84 | |||||||||
| Pdw3.9.1+b | 9 | 10.3 | 0.0001 | 15.1 | 12.5 | 0.34 | −0.03 | −0.09 | |||||||||
| Pdw3.10.1+ | 10 | 6.6 | 0.0001 | 53.5 | 6.4 | 0.23 | 0.11 | 0.50 | |||||||||
| STTa | Stt3.2.1+ | 2 | 4.5 | 0.0054 | 80.2 | 6.9 | 0.20 | −0.34 | −1.70 | 2 | 4.5 | 0.0054 | 80.2 | 1.1 | 0.11 | −0.02 | −0.21 |
| Stt3.4a.1+b | 4a | 11.0 | 0.0001 | 31.5 | 7.0 | 0.30 | 0.12 | 0.38 | 4a | 11.0 | 0.0001 | 31.5 | 16.7 | 0.41 | 0.21 | 0.51 | |
| Stt3.9.1+b | 9 | 5.4 | 0.0005 | 1.4 | 5.6 | 0.27 | 0.09 | 0.35 | 9 | 5.4 | 0.0005 | 1.4 | 4.6 | 0.19 | 0.19 | 0.98 | |
| Stt3.11.1+ | 11 | 6.4 | 0.0003 | 20.2 | 5.2 | 0.27 | 0.05 | 0.18 | 11 | 6.4 | 0.0003 | 20.2 | 7.7 | 0.26 | 0.20 | 0.75 | |
| LFPLa | Lfpl3.1.1+b | 1 | 7.3 | 0.0001 | 87.8 | 1.0 | 0.82 | 0.11 | 0.14 | 1 | 7.3 | 0.0001 | 87.8 | 10.4 | 1.60 | −0.58 | −0.36 |
| Lfpl3.5.1 | 5 | 4.5 | 0.0040 | 3.9 | 0.7 | −0.29 | 0.88 | 3.07 | 5 | 4.5 | 0.0040 | 3.9 | 6.5 | 1.26 | 0.45 | 0.36 | |
| Lfpl3.6a.1 | 6a | 14.4 | 0.0001 | 0.5 | 1.1 | −0.34 | −1.15 | −3.37 | 6a | 14.4 | 0.0001 | 0.5 | 19.6 | 0.99 | 2.89 | 2.92 | |
| Lfpl3.9.1 | 9 | 5.1 | 0.0006 | 11.7 | 2.6 | −0.45 | 1.81 | 3.99 | 9 | 5.1 | 0.0006 | 11.7 | 5.8 | 1.23 | −0.10 | −0.08 | |
| Lfpl3.11.1+ | 11 | 13.7 | 0.0001 | 6.2 | 14.2 | 2.63 | 0.54 | 0.21 | 11 | 13.7 | 0.0001 | 6.2 | 4.2 | 0.99 | −0.03 | −0.03 | |
| Lfpl3.11.2+(od) | 36.2 | 8.1 | 1.86 | 0.83 | 0.45 | 36.2 | 6.7 | 0.30 | 1.74 | 5.74 | |||||||
| LFPWa | Lfpw3.1.1+ | 1 | 7.5 | 0.0001 | 87.8 | 0.8 | 0.45 | −0.52 | −1.17 | 1 | 7.5 | 0.0001 | 87.8 | 9.6 | 1.60 | −0.45 | −0.28 |
| Lfpw3.5.1+ | 5 | 4.9 | 0.0018 | 4.1 | 0.1 | 0.21 | −0.17 | −0.84 | 5 | 4.9 | 0.0018 | 4.1 | 6.5 | 1.31 | 0.41 | 0.31 | |
| Lfpw3.6a.1 | 6a | 12.8 | 0.0001 | 0.5 | 1.8 | 0.16 | −1.23 | −7.90 | 6a | 12.8 | 0.0001 | 0.5 | 15.3 | 0.57 | 2.81 | 4.95 | |
| Lfpw3.8.1+b | 8 | 5.4 | 0.0013 | 30.2 | 5.9 | 1.29 | 1.34 | 1.04 | 8 | 5.4 | 0.0013 | 30.2 | 3.8 | 1.01 | −0.25 | −0.24 | |
| Lfpw3.9.1+(od)b | 9 | 8.1 | 0.0001 | 6.8 | 0.7 | −0.10 | 0.74 | 7.46 | 9 | 8.1 | 0.0001 | 6.8 | 10.2 | 1.66 | 0.36 | 0.21 | |
| Lfpw3.11.1+b | 11 | 13.3 | 0.0001 | 8.4 | 14.0 | 1.63 | 2.10 | 1.29 | 11 | 13.3 | 0.0001 | 8.4 | 5.6 | 1.17 | −0.33 | −0.28 | |
| Lfpw3.11.2+(od)b | 31.9 | 1.3 | 0.53 | 0.47 | 0.89 | 31.9 | 6.5 | 0.36 | 1.74 | 4.79 | |||||||
| LFMLa | Lfml3.1.1 | 1 | 9.8 | 0.0001 | 45.8 | 0.4 | −1.13 | −0.03 | −0.03 | 1 | 9.8 | 0.0001 | 45.8 | 10.0 | 3.48 | 1.92 | 0.55 |
| Lfml3.1.2−(od) | 95.2 | 2.9 | 2.77 | 2.17 | 0.78 | 95.2 | 2.2 | 0.93 | −1.96 | −2.11 | |||||||
| Lfml3.2.1+ | 2 | 13.2 | 0.0001 | 15.5 | 9.6 | 4.09 | −4.36 | −1.06 | 2 | 13.2 | 0.0001 | 15.5 | 3.0 | 2.02 | −0.09 | −0.04 | |
| Lfml3.2.2+ | 80.2 | 6.1 | 3.24 | −3.34 | −1.03 | 80.2 | 0.3 | 0.62 | 0.41 | 0.65 | |||||||
| Lfml3.5.1+b | 5 | 14.1 | 0.0001 | 15.7 | 3.5 | 3.92 | 0.47 | 0.12 | 5 | 14.1 | 0.0001 | 15.7 | 8.0 | 2.86 | 1.40 | 0.49 | |
| Lfml3.5.2 | 55.7 | 2.9 | −3.59 | −0.36 | −0.10 | 55.7 | 4.7 | 2.30 | 0.04 | 0.02 | |||||||
| Lfml3.7.1−(od) | 7 | 5.1 | 0.0015 | 0.6 | 2.7 | 1.43 | −3.44 | −2.40 | 7 | 5.1 | 0.0015 | 0.6 | 3.7 | 2.28 | 0.75 | 0.33 | |
| Lfml3.9.1+ | 9 | 15.7 | 0.0001 | 11.0 | 9.5 | 4.66 | 3.58 | 0.77 | 9 | 15.7 | 0.0001 | 11.0 | 14.2 | 4.53 | 1.01 | 0.22 | |
| Lfml3.11.1+ | 11 | 11.5 | 0.0001 | 8.9 | 6.7 | 3.50 | 1.87 | 0.53 | 11 | 11.5 | 0.0001 | 8.9 | 1.2 | 2.52 | 0.23 | 0.09 | |
| Lfml3.11.2+ | 35.7 | 6.3 | 2.63 | 3.53 | 1.35 | 35.7 | 4.1 | 0.50 | 0.15 | 0.30 | |||||||
| LFMWa | Lfmw3.1.1+(od) | 1 | 15.5 | 0.0001 | 38.3 | 2.4 | −0.04 | 2.94 | 67.49 | 1 | 15.5 | 0.0001 | 38.3 | 7.1 | 2.63 | 1.05 | 0.40 |
| Lfmw3.1.2+ | 88.7 | 9.5 | 4.23 | −0.88 | −0.21 | 88.7 | 2.7 | 1.50 | −0.90 | −0.60 | |||||||
| Lfmw3.2.1+ | 2 | 9.3 | 0.0001 | 21.5 | 7.0 | 3.28 | −2.18 | −0.66 | 2 | 9.3 | 0.0001 | 21.5 | 4.2 | 2.26 | 0.03 | 0.01 | |
| Lfmw3.5.1+b | 5 | 18.3 | 0.0001 | 17.3 | 7.3 | 4.01 | 2.58 | 0.64 | 5 | 18.3 | 0.0001 | 17.3 | 7.6 | 2.59 | 0.54 | 0.21 | |
| Lfmw3.5.2 | 55.7 | 3.2 | −2.74 | 0.81 | 0.30 | 55.7 | 7.3 | 2.54 | 0.27 | 0.10 | |||||||
| Lfmw3.9.1+b | 9 | 17.6 | 0.0001 | 18.1 | 11.6 | 4.51 | 1.73 | 0.38 | 9 | 17.6 | 0.0001 | 18.1 | 16.1 | 4.25 | 1.70 | 0.40 | |
| Lfmw3.11.1+ | 11 | 16.1 | 0.0001 | 4.7 | 3.2 | 1.98 | −0.80 | −0.41 | 11 | 16.1 | 0.0001 | 4.7 | 3.8 | 1.89 | −0.48 | −0.25 | |
| Lfmw3.11.2+(od) | 25.1 | 12.0 | 3.43 | 3.23 | 0.94 | 25.1 | 2.1 | 0.79 | 1.62 | 2.05 | |||||||
| ECL | Ecl3.1.1+ | 1 | 7.9 | 0.0001 | 39.2 | 17.9 | 0.57 | −0.13 | −0.23 | ||||||||
| Ecl3.9.1+ | 9 | 3.1 | 0.0069 | 37.2 | 6.8 | 0.33 | −0.19 | −0.59 | |||||||||
| ST1Ia | St1i3.1.1+ | 1 | 7.4 | 0.0001 | 42.8 | 14.1 | 0.16 | −0.07 | −0.44 | ||||||||
| St1i3.2.1+b | 2 | 4.3 | 0.0008 | 48.8 | 8.6 | 0.12 | 0.07 | 0.53 | |||||||||
| St1i3.4b.1+ | 4b | 3.5 | 0.0012 | 0.0 | 5.7 | 0.09 | 0.07 | 0.78 | |||||||||
| St1i3.9.1+ | 9 | 4.7 | 0.0001 | 38.1 | 9.7 | 0.13 | −0.05 | −0.33 | |||||||||
| ST2Ia | St2i3.2.1+b | 2 | 3.2 | 0.0079 | 48.8 | 7.5 | 0.11 | 0.04 | 0.32 | ||||||||
| St2i3.4b.1+ | 4b | 3.3 | 0.0014 | 0.0 | 6.3 | 0.08 | 0.10 | 1.20 | |||||||||
| St2i3.9.1+ | 9 | 4.6 | 0.0002 | 38.9 | 11.3 | 0.11 | −0.12 | −1.04 | |||||||||
| ST3Ia | St3i3.2.1+b | 2 | 3.4 | 0.0065 | 48.8 | 9.2 | 0.12 | 0.02 | 0.19 | ||||||||
| St3i3.4b.1+ | 4b | 2.7 | 0.0053 | 0.0 | 5.9 | 0.09 | 0.06 | 0.68 | |||||||||
| ST4Ia | St4i3.4b.1+ | 4b | 3.6 | 0.0006 | 0.0 | 8.2 | 0.18 | 0.05 | 0.29 | ||||||||
| St4i3.8.1− | 8 | 3.3 | 0.0056 | 15.2 | 7.3 | −0.16 | 0.09 | 0.58 | |||||||||
| ST5Ia | St5i3.4b.1+ | 4b | 3.8 | 0.0006 | 0.0 | 7.6 | 0.24 | 0.02 | 0.07 | ||||||||
| St5i3.8.1− | 8 | 3.0 | 0.0014 | 14.9 | 5.6 | −0.19 | 0.11 | 0.55 | |||||||||
| St5i3.10.1− | 10 | 4.6 | 0.0005 | 25.4 | 11.1 | −0.25 | 0.22 | 0.88 | |||||||||
| ST6Ia | St6i3.7.1−b | 7 | 3.1 | 0.0082 | 10.7 | 6.3 | −0.19 | 0.23 | 1.22 | ||||||||
| St6i3.10.1− | 10 | 4.6 | 0.0003 | 23.5 | 11.3 | −0.27 | 0.27 | 1.02 | |||||||||
| ST7Ia | St7i3.9.1−b | 9 | 4.0 | 0.0010 | 3.6 | 9.0 | −0.29 | 0.20 | 0.68 | ||||||||
| St7i3.10.1− | 10 | 4.4 | 0.0005 | 38.5 | 9.7 | −0.34 | 0.00 | −0.01 | |||||||||
| ST8Ia | St8i3.1.1− | 1 | 3.4 | 0.0075 | 66.3 | 6.4 | −0.27 | −0.28 | −1.04 | ||||||||
| St8i3.2.1− | 2 | 3.8 | 0.0033 | 12.8 | 7.7 | −0.30 | −0.30 | −1.01 | |||||||||
| St8i3.9.1−b | 9 | 6.0 | 0.0001 | 3.6 | 11.0 | −0.41 | 0.21 | 0.51 | |||||||||
| St8i3.10.1− | 10 | 5.4 | 0.0001 | 39.8 | 9.9 | −0.41 | −0.04 | −0.09 | |||||||||
| ST9Ia | St9i3.1.1− | 1 | 3.9 | 0.0028 | 66.3 | 7.1 | −0.33 | −0.39 | −1.15 | ||||||||
| St9i3.2.1− | 2 | 3.7 | 0.0031 | 12.6 | 7.1 | −0.39 | −0.26 | −0.67 | |||||||||
| St9i3.9.1− | 9 | 6.7 | 0.0001 | 3.6 | 12.5 | −0.55 | 0.20 | 0.36 | |||||||||
| St9i3.10.1− | 10 | 4.7 | 0.0001 | 46.0 | 8.3 | −0.47 | 0.05 | −0.11 | |||||||||
| ST10Ia | St10i3.9.1−b | 9 | 8.4 | 0.0001 | 3.6 | 20.0 | −0.92 | 0.16 | 0.17 | ||||||||
| STLa | Stl3.9.1−b | 9 | 4.5 | 0.0003 | 3.6 | 10.1 | −2.51 | 1.18 | 0.47 | ||||||||
| Stl3.10.1− | 10 | 3.9 | 0.0014 | 42.9 | 8.7 | −2.44 | 0.31 | 0.13 | |||||||||
| STTWa | Sttw3.9.1−b | 9 | 9.4 | 0.0001 | 3.6 | 21.9 | −21.74 | −0.01 | 0.00 | ||||||||
| BRPa | Brp3.6a.1− | 6a | 3.8 | 0.0009 | 5.7 | 10.0 | −0.25 | −0.18 | −0.70 | ||||||||
| FLDa | Fld3.2.1− | 2 | 5.0 | 0.0001 | 64.1 | 6.0 | −2.94 | −0.92 | −0.31 | ||||||||
| Fld3.3.1+ | 3 | 5.5 | 0.0001 | 10.2 | 5.4 | 2.22 | 2.58 | 1.16 | |||||||||
| Fld3.4a.1−b | 4a | 32.4 | 0.0001 | 21.7 | 43.7 | −6.40 | 7.18 | 1.12 | |||||||||
| Fld3.5.1+ | 5 | 9.7 | 0.0001 | 19.6 | 8.8 | 3.57 | 1.09 | 0.31 | |||||||||
| Fld3.11.1− | 11 | 6.5 | 0.0001 | 20.9 | 5.8 | −2.98 | 0.14 | 0.05 | |||||||||
| PDDM25 | 25Pddm3.2.1+ | 2 | 8.7 | 0.0001 | 58.8 | 11.2 | 4.04 | 1.31 | 0.32 | ||||||||
| 25Pddm3.3.1−(od) | 3 | 5.2 | 0.0001 | 10.2 | 7.0 | −1.74 | −3.95 | −2.26 | |||||||||
| 25Pddm3.4a.1+ | 4a | 19.9 | 0.0001 | 24.2 | 26.1 | 4.89 | −5.71 | −1.17 | |||||||||
| 25Pddm3.6a.1+(od) | 6a | 11.2 | 0.0001 | 0.5 | 10.2 | 2.04 | 4.80 | 2.35 | |||||||||
| 25Pddm3.11.1+ | 11 | 8.6 | 0.0001 | 10.0 | 9.5 | 3.72 | 1.21 | 0.32 | |||||||||
| PDDM50 | 50Pddm3.2.1+ | 2 | 7.7 | 0.0001 | 62.6 | 11.4 | 4.43 | −0.06 | −0.01 | ||||||||
| 50Pddm3.3.1−(od) | 3 | 5.0 | 0.0001 | 14.0 | 5.1 | −1.58 | −3.54 | −2.24 | |||||||||
| 50Pddm3.4a.1+ | 4a | 18.5 | 0.0001 | 32.3 | 27.4 | 6.01 | −4.72 | −0.79 | |||||||||
| 50Pddm3.6a.1+(od) | 6a | 9.7 | 0.0001 | 0.5 | 9.7 | 1.85 | 5.14 | 2.77 | |||||||||
| 50Pddm3.11.1+ | 11 | 6.9 | 0.0001 | 10.4 | 8.1 | 3.69 | 0.91 | 0.25 | |||||||||
| PDDM75 | 75Pddm3.2.1+ | 2 | 6.0 | 0.0001 | 63.1 | 9.0 | 4.21 | −0.12 | −0.03 | ||||||||
| 75Pddm3.3.1−(od) | 3 | 4.6 | 0.0005 | 12.5 | 5.5 | −1.96 | −3.75 | −1.92 | |||||||||
| 75Pddm3.4a.1+ | 4a | 16.4 | 0.0001 | 31.8 | 25.2 | 6.07 | −5.12 | −0.84 | |||||||||
| 75Pddm3.6a.1+(od) | 6a | 11.5 | 0.0001 | 0.5 | 12.5 | 1.62 | 6.66 | 4.11 | |||||||||
| 75Pddm3.11.1+ | 11 | 6.2 | 0.0001 | 10.4 | 7.8 | 3.85 | 1.14 | 0.30 | |||||||||
| PDDM100 | 100Pddm3.4a.1+ | 4a | 8.5 | 0.0001 | 32.3 | 17.4 | 6.35 | −4.53 | −0.71 | ||||||||
| 100Pddm3.6a.1+(od) | 6a | 10.5 | 0.0001 | 0.5 | 17.2 | 1.14 | 9.88 | 8.70 | |||||||||
| 100Pddm3.11.1+ | 11 | 3.6 | 0.0020 | 9.1 | 6.7 | 4.16 | 2.13 | 0.51 | |||||||||
| SDTWT | Sdtwt3.5.1+ | 5 | 4.9 | 0.0004 | 54.1 | 11.5 | 22.18 | −7.62 | −0.34 | ||||||||
| Sdtwt3.9.1− | 9 | 3.0 | 0.0090 | 3.6 | 6.6 | −15.95 | 9.48 | 0.59 | |||||||||
| SDTN | Sdtn3.4a.1− | 4a | 5.6 | 0.0001 | 17.2 | 10.0 | −326.95 | −17.78 | −0.05 | ||||||||
| Sdtn3.6a.1−(od) | 6a | 7.6 | 0.0001 | 1.5 | 11.2 | −42.92 | −484.40 | −11.29 | |||||||||
| Sdtn3.9.1− | 9 | 9.6 | 0.0001 | 3.6 | 14.9 | −386.91 | 135.99 | 0.35 | |||||||||
| Sdtn3.10.1− | 10 | 6.5 | 0.0001 | 58.8 | 11.6 | −302.29 | −253.74 | −0.84 | |||||||||
| SDNPPD | Sdnppd3.5.1+ | 5 | 5.3 | 0.0002 | 10.5 | 4.1 | 0.43 | −0.32 | −0.74 | ||||||||
| Sdnppd3.6a.1−(od) | 6a | 43.2 | 0.0001 | 0.0 | 55.9 | −0.12 | −2.52 | −21.03 | |||||||||
| Sdnppd3.7.1− | 7 | 4.9 | 0.0003 | 53.0 | 5.0 | −0.51 | −0.24 | −0.48 | |||||||||
| Sdnppd3.10.1− | 10 | 3.7 | 0.0031 | 53.5 | 2.8 | −0.37 | −0.21 | −0.56 | |||||||||
| Sdnppd3.11.1+ | 11 | 3.5 | 0.0025 | 0.0 | 3.0 | 0.39 | 0.20 | 0.51 | |||||||||
| PDTN | Pdtn3.4b.1+(od) | 4b | 3.0 | 0.0034 | 5.8 | 6.4 | −9.02 | 77.17 | 8.56 | ||||||||
| Pdtn3.9.1− | 9 | 8.2 | 0.0001 | 3.6 | 18.2 | −93.01 | 5.89 | 0.06 | |||||||||
| PDC | Pdc3.4b.1− | 4b | 41.5 | 0.0001 | 6.7 | 53.6 | −1.19 | 1.32 | 1.11 | ||||||||
| Pdc3.5.1− | 5 | 16.3 | 0.0001 | 55.7 | 14.4 | −0.67 | 0.58 | 0.86 | |||||||||
| Pdc3.7.1− | 7 | 5.2 | 0.0001 | 4.9 | 3.9 | −0.31 | 0.38 | 1.24 | |||||||||
Trait abbreviation was studied by Isemura et al. (2007).
QTL found at a similar location in study by Isemura et al. (2007).
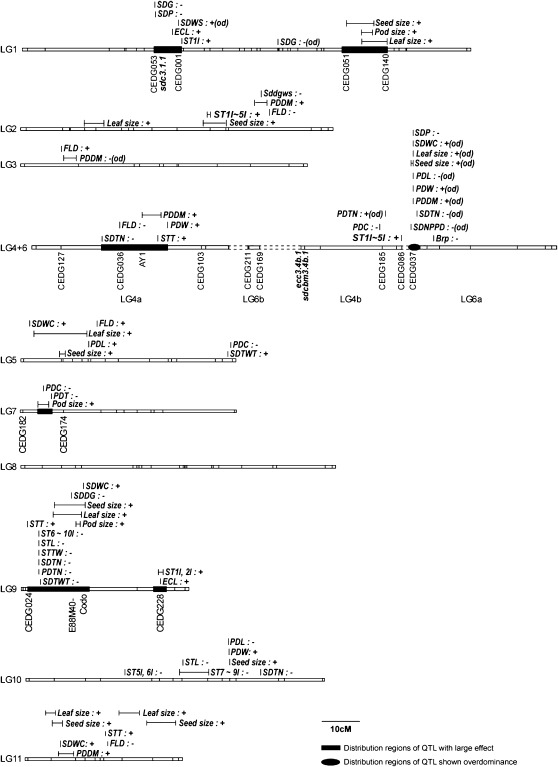
Figure 3.—
A summary of the domestication- and fitness-related QTLs detected on each linkage group in the population derived from the cross between cultivated and wild azuki bean. The signs “+” and “−” after trait name indicate positive and negative effect of allele from cultivated azuki bean on the trait. QTLs of traits expressing overdominance are indicated with “od”. Seed size, SD100WT, SDL, SDW, and SDT; leaf size, LFPL, LFPW, LFML, and LFMW; pod size, PDL and PDW; pod days to maturity (PDDM), PDDM25, PDDM50, PDDM75, and PDDM100.
Seed germination and seed survival (SDP, SDG, SDDG, SDWS, SDDGWS):
One of the most important changes that occurred during the domestication of azuki bean was selection for reduced seed dormancy and increased seed germination after planting. It was expected that the alleles from the cultivated parent would increase the SDG, increase SDP, and decrease SDWS in field conditions. For SDG in the field, two QTL (Sdg3.1.1 and Sdg3.1.2) were found on the same linkage group 1. As expected, the allele from the cultivated parent on Sdg3.1.1 increased SDG whereas Sdg3.1.2 revealed greater overdominance than additive effect. Only one QTL (Sddg3.9.1) for SDDG with a relatively high contribution (PEV 25.8%) was found on LG9. The cultivated-parent allele accelerated germination date and was dominant over the wild-parent allele. On the basis of the laboratory assay for SDP, two QTL (Sdp3.1.1 and Sdp3.6a.1) were detected on LG1 and LG6a, respectively. Sdp3.1.1 on LG1 explained 25.9% of the phenotypic variation. The alleles of the cultivated parent additively increased SDP at Sdp3.1.1 on LG1, while alleles from the cultivated parent dominantly decreased seed-coat permeability at Sdp3.6a.1 on LG6a near the presumed reciprocal translocation breakpoint. On the basis of the field assay of SDWS, only one QTL (Sdws3.1.1, PVE 14.7%) was found near SSR marker CEDG001 on LG1. Sdws3.1.1 had greater overdominance than additive effect. The SDDGWS was controlled by a major QTL (Sddgws3.2.1, PEV 35.3%) on LG2. The cultivated-parent allele resulted in earlier germination and was dominant over the wild-parent allele.
Pod dehiscence and twist number:
Pod dehiscence:
Loss of pod dehiscence is advantageous for harvesting seeds while diminishing the chance for seed dispersal in natural habitats. F2 progeny segregated for dehiscence type in the ratio 106 (dehiscent) to 34 (nondehiscent) with 48 individuals having ambiguous dehiscence type. Using data from plants with unambiguous dehiscence type a chi-square test was performed for goodness of fit to the 3:1 ratios (d.f. = 1, χ2 = 0.038), suggesting a single dominant gene controls pod dehiscence. The position of this gene was determined to be between markers CEDG182 and CEDG174 on LG7.
PDT:
PDT was measured as a quantitative trait and may represent the force of pod dehiscence. One QTL, Pdt3.7.1, with a high contribution to this trait (74.3%) was found at the same marker interval on LG7. As expected, the allele from the cultivated parent decreased PDT.
Increase in organ size:
Domestication of azuki bean has resulted in a 10-fold increase in seed weight and a 2-fold increase in pod length, stem diameter, and leaf size (Table 2).
Seed size (SD100WT, SDL, SDW, SDT):
Three to nine QTL were detected for seed size-related traits (SD100WT, SDL, SDW, and SDT) on LG1, LG2, LG5, LG6a, LG9, LG10, and LG11. As expected, all QTL alleles from the cultivated parent had an additive effect, increasing size except for one QTL on LG6a near the presumed reciprocal translocation breakpoint. Sd100wt3.6a.1 on LG6a revealed a greater dominant than additive effect.
Six QTL were detected for seed water content (SDWC) and most of the QTL were located near seed size-related QTL. All QTL alleles from the cultivated parent had the effect of increasing seed water content. Sdwc3.6a.1 on LG6a had an overdominance effect with the largest PEV.
Pod size (PDL, PDW):
Eight QTL for PDL and eight QTL for PDW were found. Those affecting both traits were mainly found on LG1, LG6a, LG7, LG9, and LG10. As expected, the alleles from the cultivated parent had increasing additive effect on both PDL and PDW at most QTL positions. Interestingly, the cultivated-parent allele at Pdl3.10.1 on LG10 had the effect of decreasing the additive effect on PDL. Also, Pdl3.6a.1 and Pdw3.6a.1 with high overdominance effect were identified near the presumed reciprocal translocation breakpoints on LG6a; only Pdl3.6a.1 had reduced effect when the genotype was a heterozygote. Two QTL, Pdl3.7.1 and Pdw3.7.1, for both pod length and width were situated close to Pdt3.7.1 for pod dehiscence on LG7.
Leaf size (LFPL, LFPW, LFML, LFMW):
Leaf size-related traits were evaluated in two generations, the F2 and F2:3. For the size of primary leaf (LFPL and LFPW), six QTL were mainly detected on LG1, LG5, LG6a, LG9, and LG11 (two QTL on LG11). The alleles from the cultivated parent had a positive effect on the size at most QTL positions. Lfpl3.11.1 and Lfpw3.11.1 on LG11 revealed the largest effect for both traits in both generations analyzed. Compared to the results in the F2 population, more QTL with a higher PEV were identified in the F2:3 population. A high positive correlation between seed size and primary leaf was observed only in the F2:3 population (supplemental data 4). These results suggest that the maternal effect of uniform seed size harvested from the F1 plant is the reason for the reduced variation in primary leaf size and lower PEV found in the F2 population.
QTL for the maximum leaf size (LFML and LFMW), were mainly detected on LG1, LG2, LG5, LG9, and LG11 (two QTL on LG1, LG5, and LG11). Among them, four QTL (Lfml3.2.1, Lfml3.5.1, Lfml3.9.1, and Lfml3.11.2) for LFML and four QTL (Lfmw3.2.1, Lfmw3.5.1, Lfmw3.9.1, and Lfml3.11.2) for LFMW are reproducible across generations and the others are conditional QTL judging from their PEV values. The alleles from the cultivated parent had a positive effect on leaf size at most QTL positions. Lfml3.9.1 and Lfmw3.9.1 on LG9 had the largest effect for both traits in both the F2 and F2:3 populations. Generally, QTL for primary leaf size (LFPL and LFPW) in the F2:3 population were situated close to the QTL for maximum leaf size on LG5, LG9, and LG11.
Plant type:
The lower stem of the cultivated parent including epicotyl grows longer and thicker than that of the wild parent (Table 2). In the middle stem, differences in the length of internodes between the cultivated and the wild parent are not marked but the wild parent has longer internodes than the cultivated parent in the upper stem. These differences have a large effect on the plant habit differences between cultivated and wild azuki bean.
STL, ECL, and ST1I–ST10I:
Two QTL (Stl3.9.1 and Stl3.10.1) for STL were found on LG9 and LG10. Alleles from the cultivated parent at both QTL had the effect of reducing stem length. However, stem growth at early, middle, and late stages is controlled by different QTL, based on a comparison of parents. When each internode was measured separately, more QTL were found. As expected from stem length QTL Stl3.9.1 and Stl3.10.1, and late stem-growth stages internode lengths (sixth to tenth) QTL (St7–10i3.9.1 and St5–10i3.9.1) were consistently located at a similar map position, despite the different extents of each QTL effect. On the other hand, QTL for the first to fifth internode lengths, early to middle stem-growth stage, were mainly found on LG1, LG2, LG4b, and LG9 and at these QTL positions, alleles from the cultivated parent increased internode length. QTL for the first to third internode lengths (ST1–3I) are located close to ECL. QTL for early- to middle-stage stem growth on LG4b are located close to the breakpoint.
STT:
Stem thickness was evaluated in two generations, the F2 and F2:3. Among four QTL identified, three QTL, Stt3.4a.1, Stt3.9.1, and Stt3.11.1 on LG4a, LG9, and LG11, are reproducible among generations. At these QTL, the alleles from the cultivated parent increased stem thickness. The QTL on LG4a and LG11 were located near flowering-time QTL.
BRN and first BRP:
The wild parent produced more branches on the main stem and initiated first branching at a lower internode than the cultivated parent. In the wild parent, primary and secondary branch development occurred at later developmental stages. Only one significant QTL, Brp3.6a.1, was identified at BRP whereas no significant QTL at α = 0.01 was found for BRN.
STTW:
Twining habit is a characteristic trait of the wild parent enabling it to spread in natural habitats. At evaluation in the F2 generation, the segregation ratio (149 twining:39 nontwining) fit a 3:1 inheritance ratio well (d.f. = 1, χ2 = 1.816), suggesting that a single dominant gene controls twining. The gene is located between CEDG166 and E88M40-Codo on LG9. Further phenotypic characterization in the F2:3 population revealed segregation among F3 individuals while some F2:3 lines derived from the twining type did not show twining behavior due to incomplete gene expression at the time of the evaluation. Using the number of twining individuals per line for QTL analysis, alleles from the cultivated parent at one QTL, Sttw3.9.1, on LG9 were found to be involved in the loss of twining ability. Sttw3.9.1 was localized to the same marker interval on LG9 as above and had a PEV of 21.9%.
Phenology:
Cultivated azuki bean has a long pod-filling period and thus flowers earlier but matures later than wild azuki bean.
FLD:
The first flowering of the cultivated parent occurred earlier than the wild parent, 71 days compared with 84 days (Table 2). On the basis of the frequency distribution in F2 plants, the trait days to first flowering is controlled by a few genes. A major QTL (Fld3.4a.1, PEV 43.7%) was identified on LG4a and the recessive allele from the cultivated parent hastened flowering time mode. On the other hand, four QTL with smaller effect were found on LG2, LG3, LG5, and LG11. These alleles of the cultivated parent hastened flowering at QTL on LG2 and LG11 but delayed flowering at the others on LG3 and LG5.
Percentage of mature pods (PDDM25, PDDM50, PDDM75, PDDM100):
In comparison with the wild parent, it took longer for all pods to mature in the cultivated parent. For the days to each maturity stage, three to five QTL were found on LG2, LG3, LG4a, LG6a, and LG11. The alleles from the cultivated parent that delayed maturity are on LG2, LG4a, LG6a, and LG11, while a QTL on LG3 hastened maturity. 25Pddm3.4a.1–100Pddm3.4a.1 on LG4a was consistently found as the QTL with the greatest effect at all stages and the recessive cultivated-parent allele delayed maturity like Fld3.4a. 25Pddm3.6a.1–100Pddm3.6a.1 were identified near the presumed reciprocal translocation breakpoint on LG6a.
Seed productivity (SDTN, PDTN, SDNPPD, SDTWT):
The wild parent produces more pods and seeds than the cultivated parent (Table 2). A QTL, Pdtn3.9.1 for PDTN, was identified at a similar position as a QTL for SDTN, Sdtn3.9.1 on LG9. The alleles from the cultivated parent at both QTL had a negative additive effect on these traits. On the other hand, another QTL, Pdtn3.4b.1, for PDTN near the presumed reciprocal translocation breakpoints on LG4b, had a stronger dominant effect than additive effect when the genotype was in the heterozygous state. Similarly, another QTL, Sdtn3.6a.1, for SDTN near the presumed reciprocal translocation breakpoints on LG6a, had an overdominance effect whereas the direction of the overdominance effect at Sdtn3.6a.1 was the reverse of Pdtn3.4b.1. For SDTN, an additional QTL was identified on LG10.
The wild parent has short pods but many seeds, whereas cultivated azuki bean has long pods but fewer seeds. Unexpectedly, a single QTL with a large effect on SDNPPD was identified near the presumed reciprocal translocation breakpoint on LG6a. The QTL, Sdnppd3.6a.1, had the largest effect (55.9%) and a stronger negative overdominant effect than additive effect. The location was close to Sdtn3.6a.1 with negative dominant effect for SDTN. At QTL on LG5 and LG11, alleles from the cultivated parent had a positive effect on SDNPPD.
Although seed size of the wild parent is small, the total weight of seeds was more than that of the cultivated parent. Two QTL, Sdtwt3.5.1 and Sdtwt3.9.1, for SDTWT were found on LG5 and LG9, respectively. The alleles from the cultivated parent at Sdtwt3.9.1 on LG9 decreased the total seed weight while at Sdtwt3.5.1 on LG5, they increased the total seed weight and had a larger effect (11.5%).
Pigmentation (PDC, ECC, SDC, SDCBM):
The PDC of the cultivated parent is light tan whereas the wild parent is black. Three QTL on LG4b, LG5, and LG7 are involved in PDC. The QTL, Pdc3.4b.1, with the largest effect was detected on LG4b (53.6%). Pdc3.7.1 on LG7 was found at a similar position to other pod size QTL and a pod dehiscence QTL. Significant epistatic interactions were observed between Pdc3.5.1 on LG5 and Pdc3.4b.1 on LG4b and between Pdc3.5.1 on LG5 and Pdc3.7.1 on LG7.
ECC, SDC, and SDCBM were characterized as qualitative traits (Table 2). Purple epicotyl and black mottle on non-red seed coat from the wild parent were dominant to green epicotyl and red seed coat without black mottle from the cultivated parent in the expression of these traits. The segregation ratios for each trait fitted the expected ratio (3:1). The recessive genes for green epicotyl, red seed coat, and non-black-mottle seed coat from cultivated parent are tentatively named as ecc3.4a.1, sdc3.1.1, and sdcbm3.4a.1, respectively. The ecc3.4a.1 and sdcbm3.4a.1 genes were tightly linked together near SSR marker CEDG185 on LG4b. The gene, sdc3.1.1, for red seed coat was mapped near SSR marker CEDG053 on the LG1.
Distribution of domestication trait-related QTL across the azuki bean genome and linkage groups:
Linkage group 1:
Most QTL are distributed in two regions. In the first region between SSR markers CEDG053 and CEDG001 (interval ∼10 cM), QTL related to seed dormancy (Sdg3.1.1, Sdp3.1.1, and Sdws3.1.1), stem length at the seedling stage (Ecl3.1.1 and St1i3.1.1), and the recessive gene controlling the red seed-coat color (sdc3.1.1) were located. At these loci, alleles from the cultivated parent have the effect of reducing seed dormancy, red seed-coat color, and increasing stem length at the seedling stage. The QTL for organ size of seed (Sdwt3.1.1 etc.), pod (Pdl3.1.1 and Pdw3.1.1) and leaf (Lfpl3.1.1, Lfpw3.1.1, etc.) were found between SSR markers CEDG051 and CEDG140. Alleles from the cultivated parent had the effect of increasing organ size.
Linkage group 4+6:
Most QTL are found in two regions. In the first region near SSR markers CEDG036 and AY1, QTL with a strong effect on flowering time (Fld3.4a.1) and maturity (25Pddm3.4a.1–100Pddm3.4a.1) were found. Alleles from the cultivated parent shorten the days to flowering but increase the period to pod maturity. QTL with a moderate effect on stem thickness, Stt3.4a.1, and total seed number, Sdtn3.4a.1, were also linked. In the second region near SSR marker CEDG037 on the LG6a segment, QTL mainly for seed, pod, and leaf size, and seed productivity-related traits were localized. Most of these QTL near the presumed reciprocal translocation breakpoints showed overdominance.
Linkage group 7:
The QTL for pod dehiscence (Pdt3.7.1), pod size (Pdl3.7.1 and Pdw3.7.1), and pod color (Pdc3.7.1) were found between SSR marker CEDG182 and CEDG174. Alleles from the cultivated parent had the effect of increasing pod size and reducing pod dehiscence and coloration.
Linkage group 9:
The QTL for organ size, growth habit, and yield-related traits were found in two distinct regions. In the first region between markers CEDG024 and E88M40-Codo, QTL for stem twining (Sttw3.9.1), stem-size traits (Stl3.9.1, St7i–St10i3.9.1, and Stt3.9.1), and seed productivity-related traits (Sdtwt3.9.1, Sdtn3.9.1, and Pdtn3.9.1), size of seed (Sd100wt3.9.1 etc.), leaf (Lfml3.9.1, Lfmw3.9.1, etc.), and pod (Pdl3.9.1 and Pdw3.9.1) were clustered. Alleles from the cultivated parent had the effect of reducing stem size, number of pods and seeds, and total seed weight, and increasing seed, leaf, and pod size. QTL for stem length at the seedling stage (Ecl3.1.13.9.1, St1i3.9.1, and St2i3.9.1) were located at the distal end near SSR marker CEDG228. Cultivated-parent alleles have an increasing effect on stem length.
Although it is unknown whether all 162 QTL identified for domestication- and fitness-related traits have independent gene action on each trait, the observed number of QTL was compared with the expected number of QTL calculated on the basis of each linkage group length (Table 4). The χ2-value was 80.2 and this value is significant at the α = 0.001 level, suggesting a departure from random distribution across the azuki bean genome. The number of QTL on LG4b, LG6a, LG9, and LG11 was significantly higher than expected, whereas the number of QTL on linkage groups 3 and 8 was significantly less than the expected number. Furthermore, the number of QTL at each 10-cM interval was counted and compared to the expected values by χ2-tests (Table 5). The χ2-tests indicated a nonrandom distribution of QTL on all linkage groups except for LG2, LG5, LG8, and LG11.
TABLE 4.
Observed and expected numbers of QTL on each linkage group
| No. of QTL
|
||||
|---|---|---|---|---|
| LG | Length (cM) | Detected | Expected | χ2a |
| LG1 | 116.2 | 21 | 26.1 | 1.00 |
| LG2 | 80.2 | 17 | 18.0 | 0.06 |
| LG3 | 73.9 | 4 | 16.6 | 9.57** |
| LG4a | 49.9 | 10 | 11.2 | 0.13 |
| LG4b | 6.6 | 7 | 1.5 | 20.52*** |
| LG5 | 55.6 | 16 | 12.5 | 0.98 |
| LG6a | 36.8 | 17 | 8.3 | 9.22*** |
| LG6b | 3.0 | 0 | 0.7 | 0.67 |
| LG7 | 56.0 | 7 | 12.6 | 2.48 |
| LG8 | 81.5 | 3 | 18.3 | 12.81*** |
| LG9 | 43.3 | 25 | 9.7 | 23.96*** |
| LG10 | 77.9 | 13 | 17.5 | 1.16 |
| LG11 | 40.0 | 22 | 9.0 | 18.83*** |
| Total | 720.9 | 162 | 162 | 80.20*** |
** and *** indicate significance at 1% and 0.1% levels, respectively.
Departure from random distribution across the genome was tested under the null hypothesis in a Poisson goodness of fit.
TABLE 5.
The number of QTL in each 10-cM interval
| No. of QTL
|
||||
|---|---|---|---|---|
| LG | Detected | Averagea | Rangeb | χ2c |
| LG1 | 21 | 1.75 | 0–6 | 30.37** |
| LG2 | 17 | 1.89 | 0–5 | 14.78 |
| LG3 | 4 | 0.50 | 0–4 | 81.24*** |
| LG4a | 10 | 2.00 | 0–6 | 19.36*** |
| LG4b | 7 | 1.75 | 0–7 | 153.20*** |
| LG5 | 16 | 2.67 | 0–6 | 12.45 |
| LG6a | 17 | 4.25 | 0–17 | 129617.51*** |
| LG6b | 0 | 0.00 | 0 | 2.25 |
| LG7 | 7 | 1.17 | 0–5 | 30.36*** |
| LG8 | 3 | 0.33 | 0–2 | 1.85 |
| LG9 | 25 | 5.00 | 0–12 | 184.41*** |
| LG10 | 13 | 1.63 | 0–6 | 29.78*** |
| LG11 | 22 | 5.50 | 0–9 | 6.01 |
** and *** indicate significance at 1% and 0.1% levels, respectively.
The average QTL density on each linkage group.
The range of the number of QTL per 10-cM interval.
Departure from random distribution of QTL in each 10-cM interval was tested under the null hypothesis in a Poisson goodness of fit.
DISCUSSION
Construction of linkage map:
The linkage map constructed here covers 92.8% of the standard linkage map of azuki bean, has lower marker density, and an uneven marker distribution compared to the standard linkage map (Han et al. 2005). One reason for the differences in the two linkage maps is that the map constructed here was based on two accessions, wild and cultivated azuki bean of Japanese origin, while the standard map was based on accessions that were more genetically diverged—a Japanese cultigen and a wild accession from Nepal (Zong et al. 2003). In the population analyzed here, a large percentage (40%) of SSR polymorphisms show a 2-bp difference between parents and no strong segregation distortion. This compares with the mapping population using an accession from Nepal that had only 12% SSR polymorphism with a 2-bp difference but 63% with a >10-bp difference. This agrees with previous reports of a high degree of genetic similarity between Japanese wild and cultivated azuki bean on the basis of isozyme (Yasuda and Yamaguchi 1996), RAPD (Xu et al. 2000a), and AFLP (Xu et al. 2000b) analyses.
Problems encountered in developing the linkage map here were the lack of polymorphic SSR markers in the middle of LG9 and the identification of a pseudolinkage group LG4+6 (Figure1). In this population only half the number of SSR markers were mapped to LG9 compared to the previously studied mapping population (Isemura et al. 2007). The reason the parents used here are genetically similar on LG9 compared to the other linkage groups is not known. Since many domestication-related QTL are located on LG9, AFLP primer sets were screened enabling some markers to be found and mapped to the middle of LG9. The pseudolinkage group LG4+6 consists of markers from LG4 and LG6 in the previous linkage map of azuki bean (Han et al. 2005; Figure 1, right), rice bean (T. Isemura and A. Kaga, unpublished results) and other related Vigna species (Somta et al. 2006). In a simulation study Livingstone et al. (2000) reported that markers of both chromosomes near reciprocal translocation breakpoints clustered together forming a single pseudolinkage group. Markers in the present linkage map have been identified on separate nonhomologous linkage groups in other mapping populations and are present in two interstitial segments. These data suggest that a reciprocal translocation has occurred between the cultivated and wild accession used in the present study. In a maize interchange heterozygote, cross-shaped pachytene configurations between homologous segments occur (Burnham 1962). This results in three types of meiotic segregants, alternate, adjacent-1, and adjacent-2. The adjacent-2 segregants usually produce nonviable gametes and the genotype resulting from adjacent-2 segregants was not observed in double haploid lines (Osborn et al. 2003). In the present study, a reciprocal translocation would account for the unfertilized ovules in the pod of F1 hybrids and F2 progenies (discussed below). Studies of the cytological configuration, genomic in situ hybridization, and pollen viability will be required to demonstrate reciprocal translocation between LG4 and LG6 and to determine whether such a translocation occurs widely in cultivated or wild azuki bean.
Genomic regions and distribution and characteristics of QTL involved in the domestication process:
Domestication genetic studies in many crops have shown that domestication traits are controlled by several major genes plus some minor genes, and these genes are generally not randomly distributed across the crop genomes (Gepts 2004). The results of this study show that a similar situation exists in azuki bean. A few major genes plus some minor genes control most domestication-related traits studied. For example, two genes control seed-coat color; one QTL of large effect was found for pod shattering, twining habit, and seed dormancy. For FLD and PDDM, one QTL with large effect and 3–4 minor QTL were found. For SD100WT, SDL, and PDL, one major QTL and 2–6 minor QTL were found. These results accord with results of genetic analyses of domestication-related traits from other crops such as common bean (Koinange et al. 1996), pearl millet (Poncet et al. 2000), and rice (Xiong et al. 1999). The distribution of domestication-related QTL across the azuki bean genome is shown by the clustering of QTL on LG1, LG4a, LG6a, LG7, LG9, and LG11 (Figure 3). LG1 is associated with changes in seed coat and plant size (seed-color change, the loss of seed dormancy, and increased organ size of seeds, pods, stems, and leaves). LG4a is associated with changes in phenology and seed productivity (hastened flowering time and reduction in the number of seeds). LG6a is also associated with changes in seed productivity (increased seed size, delayed maturity, and reduction in the number of seeds). LG7 is associated with changes in pods (loss of pod dehiscence and increased pod size). LG9 is associated with changes in plant type, plant size, and seed productivity (loss of the twining, reduction of stem length, and reduction in the number and weight of seeds and pods). These results highlight the importance of major genes in domestication (Poncet et al. 2000; Wang et al. 2005).
On LG1 and LG9, QTL for various fitness- and domestication-related traits are clustered in a narrow region (Figure 3). These clusters may be due to pleiotropy or close linkage of QTL. Single mutations can have a pleiotropic effect on various unrelated organs. In maize, change in inflorescence sex and number and length of the internode in lateral branches and inflorescences are distributed within a narrow genomic region (Doebley et al. 1995). Change in these traits is due to the pleiotropic effect of tb1 based on the complementation test for tb1. In Arabidopsis, the testa color mutant gene transparent testa (tt) causes a reduction in seed weight (Debeaujon et al. 2000). Similarly testa color mutant gene black seed (bks) in tomato decreases seed weight and increases fruit pH (Downie et al. 2003). Thus the QTL distribution within a narrow region in this study may be explained by the pleiotropic effect of a single mutation. Further studies are required to determine whether pleiotropy and/or genetic linkage are occurring in azuki bean.
In this study, 28 of the quantitative traits measured were the same as those in the previously reported azuki bean mapping population that was based on accessions that were more genetically diverged—a Japanese cultigen and a wild accession from Nepal (Isemura et al. 2007). It was expected that there would be a high degree of similarity between the results of previous and present studies. Of QTL or genes for the common traits, on the basis of approximate position in the linkage group ∼40% are considered likely to be the same (Table 3, supplemental data 6 at http://www.genetics.org/supplemental/). Hence a large number of QTL found here were not found in the previous study.
The seeds of both the cultivated and wild parents used in this study were 40–50% larger than those of parents in the previous study (Isemura et al. 2007). This may explain, in part, the differences in number and effect of QTL between the populations studied in relation to seed size and germination. A number of QTL associated with seed and pod size were located at the breakpoint of the presumed reciprocal translocation between chromosomes 4 and 6. These QTL exhibited overdominance. Seed fertility related to the reciprocal translocation can explain the expression of the QTL at this location (see below). No seed-related QTL were found on LG6 in the previous study.
Here two QTL for seed-coat permeability were found on LG1 and LG6a compared to five QTL detected on LG1, 4, and 9 in the previous study. One QTL on LG1 appears to be common in both studies. The numerous genetic differences between this and the previous study reflect evolutionary divergence of germplasm from different locations and selection history of both wild and cultivated azuki bean. The gene pool of azuki bean therefore has abundant genetic variation for exploitation in breeding.
Seed dormancy and germination:
Seed dormancy and the control of germination are traits that enable plants to survive in natural conditions where the seed environment can change rapidly. Seed dormancy is determined by multiple factors, usually divided into physical and physiological factors, and their interactions (Baskin and Baskin 2004). Seed dormancy prevents uniform germination and reduction of seed dormancy is an important step in the domestication process. Wild azuki bean seeds have dormancy. In cultivated azuki bean, seed dormancy is not completely lost since transient escaped cultivars have been found in Japan so, weak dormancy enables them to over winter.
Physical dormancy is caused by water impermeable layers of palisade cells in the seed coat that control water movement (Finch-Savage and Leubner-Metzger 2006). In azuki bean, water is taken up through a specific tissue, the lens (strophiolar cleft), adjacent to the hilum and from there, water is distributed through the seed coat to inner tissues (Kikuchi et al. 2006). In other Asian Vigna, the wild species V. minima, V. dalzelliana, and V. calcarata auct. pl. have water-impermeable seed coats whereas cultivated V. umbellata does not exhibit seed dormancy. Loose lens structure in the cultivated Vigna species is an important physical difference from wild Vigna species (Gopinathan and Babu 1985).
To determine the genetic factors that, in turn, determine physical dormancy in azuki bean, SDP was evaluated in laboratory conditions and two QTL, Sdp3.1.1 and Sdp3.6a.1, were identified on LG1 and LG6a, respectively. Sdp3.1.1 is tightly linked with the seed-coat color gene, sdc3.1.1. The same relationship has previously been observed in the cross between the genetically diverged Japanese cultigen and the wild accession from Nepal (Isemura et al. 2007). When the genotype is the cultivated parent homozygote at sdc3.1.1 and Sdp3.1.1 the seeds have a red seed coat and increased water permeability. Thus, red seeds tend to imbibe water more rapidly than tan seeds in laboratory conditions. No QTL for SDP was found close to the other seed color gene on LG4b, sdcbm3.4.1, controlling the black mottle on seed coat.
A close relationship between seed-coat color and water permeability of the seed coat has also been reported in Arabidopsis (Debeaujon et al. 2000) and tomato (Downie et al. 2003). In Arabidopsis the tt mutants with light testa color exhibit higher water permeability compared with the wild type. Similarly in tomato, water permeability of mutant bks was lower than that of the wild type with pale brown seed color. In Arabidopsis mutants, defects in flavonoid pigments in cell layers of the seed coat are responsible for seed color change and thin seed coat. On the other hand, in the tomato mutant, the biosynthesis and accumulation of a large quantity of melanin pigment in cell layers of the seed coat is responsible for seed color change and toughens the seed coat. Further studies are needed to determine the relationship between variation in the lens structure and the uptake of water in azuki bean and whether there is a pleiotropic effect from red seed coat.
For the SDP QTL on LG6a, the allele from the cultivated parent had a weak but dominant inhibitory effect on water uptake. This might reflect the fact that the cultivated parent has not completely lost seed dormancy. Among cultivated legumes azuki bean seeds have a relatively poor ability to take up water, and hard seed coat sometimes causes trouble for processors of the sweet azuki bean paste “Ann” because some seeds do not soften on boiling (Ozawa 1978). Decline in seed moisture content in azuki bean during maturation and preharvest appears to be important in determining cooking efficiency (Hsieh et al. 1992). On LG6a, QTL with strong effects on days to pod maturity, Pddm3.6a.1, and seed water content, Sdwc3.6a.1, were identified in a similar region. At both QTL, the alleles from the cultivated parent have a positive effect, increasing both seed water content and the time to maturity. These QTL had the same dominant effect on trait expression as the seed-coat permeability QTL. Thus, alleles from the cultivated parent related to pod and seed maturation could indirectly increase hard seed number and this could potentially be an advantage for the survival of hybrid progenies in natural habitats.
Seed germination was also evaluated in the field in two different seasons, summer and winter. Two QTL, Sdg3.1.1 and Sdws3.1.1, for SDG and SDWS were reproducibly identified near the gene for seed-coat color (sdc3.1.1) and QTL for seed-coat permeability (Sdp3.1.1) on LG1. The lower PEV for the two germination-related QTL than Sdp3.1.1 reflects field environmental heterogeneity. The direction of allele effects at both germination QTL can be understood because the allele from the cultivated parent at Sdg3.1.1 promoted germination rate as well as seed-coat permeability (Sdp3.1.1) while the allele from the cultivated parent at Sdws3.1.1 for seed winter survival reduced the rate of seed survival during winter. These results suggest that a single major QTL, possibly the gene for seed-coat color, is the gene responsible for controlling physical seed dormancy. With respect to seed survival in the soil over winter the cultivated-parent allele would be a disadvantage to hybrid progenies in natural habitats.
Wild azuki bean is a summer annual; pods disperse seeds in autumn and seeds remain in the soil over winter until some germinate the following spring. The winter seed survival field experiment (SDWS) reflects better post dispersal conditions than the summer evaluation. The soil temperature for SDWS was low (average 2.3° for January and February). Most of the seeds that imbibed soil water failed to germinate at the low temperature and decayed. Generally, seeds from F2 plants revealed low seed survival (average 20.4%) in the winter field experiment even though average seed-coat permeability was 41.8%. Thus the physical dormancy alone might not completely explain seed dormancy. The seed survival from winter to spring and subsequent germination may rely on competition (or balance) between physiological activity associated with seed dormancy and germination and microorganism activity associated with seed decay. Microorganisms interact on weed seed viability and cause depletion of seed bank (Kremer 1993). The lens (Gopinathan and Babu 1985) and hilum (authors' observation) are thought to be a target digested by microorganisms or softened by wet and dry cycles. In the winter experimental field, fluctuation of the soil conditions where the seeds were buried was affected not only by wet and dry cycles but also by temperature variation (lowest −3° and highest 21°, average 4.9°) until first germination was observed. Cold stratification is not considered a major factor for breaking dormancy because mechanical scarification of post-harvested wild azuki bean seed coat can break seed-coat dormancy completely. However, quick germination in response to higher temperatures might enable seeds to escape microorganism degradation. Unexpectedly, the first germination in spring was observed in the hybrid progenies, followed by the cultivated parent and then the wild parent. The SDWSGD was controlled by a major QTL, Sddgws3.2.1, on LG2 (PEV 35.3%) close to flowering and maturity QTL. The effect of the cultivated-parent allele accelerates germination timing and is dominant to the wild-parent allele. Similarly, the germination in summer was controlled by a major QTL, Sddg3.9.1, on LG9 (PEV 25.8%). The cultivated-parent allele promoting quick germination is dominant to the wild-parent allele. Thus alleles from the cultivated parent on Sddg3.9.1 could be advantageous for early germination.
Seed productivity:
Cultivated and wild azuki bean and their hybrid progenies were evaluated for their phenotypes in field conditions to determine components of fitness. The number of seeds produced by a plant is a major determinant for survival under natural conditions. In legumes such as cowpea (Peken and Artk 2004), common bean (Raffi and Nath 2004), and soybean (Iqbal et al. 2003), the number of pods and seeds per plant, number of seeds per pod, and pod length have a positive effect on seed productivity. This is also the case with cultivated and wild azuki bean. In this study seed size had a negative effect on total seed number (r = −0.36 to −0.47). Similarly a significant and negative correlation (r = −0.34) was observed between seed size and total seed number in common bean (Raffi and Nath 2004). In the present study, SDTN of the cultivated parent was 10% less than the wild parent, whereas in hybrid progenies with alleles from the cultivated parent, seed productivity varied widely. Some hybrid plants produced as many seeds as the wild parent. Total seed number in the hybrid progenies showed significant and positive correlations with PDTN, SDNPPD, and ST8–10I (supplemental data 4). However, total seed number showed significant and negative correlations with seed- and leaf-size-related traits.
To elucidate the relationship among traits, path analysis was applied. On the basis of path analysis, ∼90% of the variation in total seed number can be directly explained by seed number per pod and total pod number (Figure 2). Variation in seed number per pod can be explained by 100-seed weight and pod length. The significant positive correlation between pod length and number of seeds per pod in the F2 population was unexpected since wild azuki bean has short pods but many seeds, whereas cultivated azuki bean has long pods but fewer seeds (Table 2). On the basis of the parental phenotype it was expected that as the pod length increased, seed number per pod would decrease. So we assume that pod length depends on mature seed number in the pod, and pods elongate in proportion to the number of developing fertilized ovules. Fertility (seeds developed compared to number of ovules) of F2 plants with shorter pods was lower than that in plants with longer pods. Therefore, an increase of unfertilized ovules leads to a decrease in pod length. The reciprocal translocation between parents would account for the unfertilized ovules in the pod of F1 hybrids and F2 plants. Among three types of meiotic segregants, alternate, adjacent-1, and adjacent-2 of interchanged hybrid between such parents, the adjacent-2 segregants usually produce nonviable gametes, and high sterility has been observed on the hybrid and the progenies with interchange heterozygote in maize (Burnham 1962). These accord well with the identification of QTL, Sdnppd3.6a.1 and Pdl3.6a.1, for seed number per pod and pod length, respectively, near the presumed translocation breakpoint on LG6a. Only when the genotype was in the heterozygous state, unlike the parents, the QTL showed a strong negative effect: a decrease in seed number per pod and pod length.
On the other hand, pod total number increases when upper internode length and branch numbers increase. Both longer upper internode and increased branch number are derived from the wild parent. Increased leaf size derived from the cultivated parent reduces the total pod number. Therefore cultivated alleles for larger seeds and leaves indirectly reduce total seed number via reduction of seed number per pod and total pod number. These results may reflect physiological constraints on the size of organs for the production and assimilation of photosynthate and trade-off in the distribution of photosynthate among various reproductive organs (Stearns 1992).
QTL for total seed number were found on LG4a, LG6a, LG9, and LG10. These QTL were generally close to those for pod number, seed number per pod, and organ size (seed, pod, and leaf). The wild-parent allele at these QTL increase total seed number, pod number, seeds per pod, and decrease seed and pod size, while the allele from the cultivated parent has the reverse effects. The close association of these traits can be explained in part by allometry, the differential growth rates of the parts as a result of a trade-off relationship between number of organs and organ size (Sinnott 1960). In addition, genetic linkage or pleiotropy may be involved. The presumed reciprocal translocation also might affect seed size and needs investigation in crosses involving other azuki bean accessions. A QTL (Sdtn3.6a.1) for total seed number at the distal end on LG6a has a very low additive effect but high dominant effect. Dominant effects were found in the QTL for seed weight (Sd100wt3.6a.1). This helps explain the allometric relationship between organ number and size where reduction in seed number corresponds to an increase in seed size.
Besides the effect of organ size, path analysis suggests that stem characteristics have a large effect on seed number. Alleles derived from the cultivated parent decrease the length of upper internodes and diminish stem twining, resulting in shorter plant height. As the stem becomes shorter the number of nodes decreases and flowers are borne closer to the ground. A consequence of this might be reduced seed and pod number (Figure 2). The loss of twining would be a disadvantage for supporting growing plants in natural habitats where competition for light with other species is important.
The QTL for total seed and pod number with positive effect from the wild parent on LG4a were close to a QTL, Fld3.4a.1, for flowering time (PEV 43.7%) with negative effect from the cultivated parent. Although total biomass was not measured in this study, an allele for shorter time to flowering from the cultivated parent means a reduced vegetative growth period and suggests reduced total biomass, leading to lower seed production. These results suggest that azuki bean genes for increased organ (sink) size, especially seeds, and shorter growth habit are disadvantageous to plants under natural habitat with respect to seed productivity.
Fitness of hybrid progenies:
Analysis of plant habit and its genetic foundation in relation to fitness of hybrid derivatives between cultivated and wild azuki bean offers several insights into the dynamics of the azuki bean crop complex. Generally crop genes are not believed to have a selective advantage in natural conditions and are expected to disappear, at least in the homozygous state, from wild populations. The data presented here suggest that genes from cultivated azuki bean reduce seed production and dormancy of wild/cultivated hybrid derivatives. Even if introgression occurs from cultivated azuki bean into wild populations, hybrid derivatives carrying disadvantageous crop genes would not become established in the wild azuki bean habitat. However, cultivated, wild, and weedy populations of azuki bean in Japan constitute a crop complex (Vaughan et al. 2004). In addition, populations consisting of a mixture of wild and weedy phenotypes, referred to as complex populations, are reported throughout Japan (Tomooka et al. 2002) and the microhabitat structure of these complex populations in Tottori prefecture have been well-characterized (Kaga et al. 2004). The hybrid progenies from the cross studied here have survived three years in experimental seminatural conditions (A. Kaga, unpublished results). It is not known how hybrid weedy forms evolve after introgression and become established.
Genetic control for fitness-related traits occur in complex regulatory networks (Figure 3). The QTL for most traits are observed separately on several linkage groups while two-linked QTL on the same linkage group are rare. This is especially noteworthy where we infer the combination of QTL and predict the phenotype of hybrid progenies on the basis of an independent assortment of chromosomes. Since the chromosome number of azuki bean is 11, the possible combination in the case of two QTL without crossing over is 211 (2048 combinations). This suggests that many combinations with or without disadvantageous or advantageous alleles from cultivated azuki bean at various QTL will produce various phenotypes in the progenies. Since natural selection occurs on phenotypes, hybrids with various proportions of advantageous and disadvantageous alleles from cultivated azuki bean would be targets of selection depending on the proportion of these alleles. Due to the limitations of the experimental design, particularly population size due to using a wild plant, we need further studies to predict fitness performance in overlooked progenies having different gene combinations. We intend to construct a computer simulation model in relation to population dynamics based on information of fitness-related QTL such as rate of winter-survived seeds and total seed number, the genotypes of hybrid progenies, and life history characteristics. At the same time, development of recombinant inbred lines from the population in the present study is ongoing and the fitness characteristics will be evaluated in various habitats. By these approaches the probability of hybrid progeny persisting in competition with wild plants under various environmental conditions can be assessed and will enable improved understanding of how wild populations evolve into crop complexes.
This study on azuki bean has significance not only in relation to the dynamics of the crop complexes but also in relation to gene introgression. In much of Japan wild azuki bean and wild soybean are common and are sometimes sympatric. The frequency of introgression from cultivated to wild azuki bean is greater than from cultivated to wild soybean (Kaga et al. 2005). These differences may reflect differential gene control for fitness of hybrid progeny in the azuki bean complex compared to the soybean complex. Here a QTL, Sdp3.1.1, for seed-coat permeability with a relatively large effect was identified on LG1 near the locus controlling seed-coat color. Such a close relationship between seed germination-related QTL and seed-coat color locus is also reported in the progenies between soybean and wild soybean (Keim et al. 1990; Sakamoto et al. 2004). Yellow seed-coat color, which is predominant in soybean, is mainly controlled by dominant suppressor gene I due to gene silencing (Todd and Vodkin 1993). Yellow seeds imbibe water more rapidly than colored seeds and exhibit a higher exudation of sugars that affects seedling emergence (Zheng and Watanabe 2000). Gene introgression from currently grown soybean cultivars with yellow seed coat to wild soybean might affect the fitness of hybrid progenies by reducing hard seed coat and subsequently the number of progenies. In contrast, in azuki bean the seed color change from tan to red seed coat (red is the most common in Japanese cultivated azuki beans) is controlled by a recessive gene. The direction of the seed color mutation in currently grown varieties of azuki bean is the reverse of soybean. Therefore, seeds from hybrid progenies resulting from gene introgression from cultivated to wild azuki bean might have a greater chance of survival than between cultivated and wild soybean. Tan-colored weedy azuki bean can commonly be found in Japan.
In conclusion, this study has identified the genomic regions where QTL for 46 traits related to domestication in azuki bean are found. Although the distribution of the QTL followed a common pattern of being distributed in clusters on a limited number of linkage groups, a large number were positioned in proximity to the breakpoint of a presumed translocation. The results provide details of the genetic relationship of traits related to seed characteristics that explain why azuki bean forms a crop-wild-weedy complex in Japan. The results also reveal that domestication of azuki has involved a trade-off between yield and seed size with fewer but longer pods and fewer but larger seeds on plants with shorter stature in cultivated azuki bean being at the expense of overall seed yield.
Acknowledgments
The authors thank the following staff of the National Institute of Agrobiological Sciences for technical support: T. Nobori, N. Karino, S. Hirashima, K. Sugimoto, H. Uchiyama, T. Taguchi, and H. Tomiyama. In addition, they thank A. Korol, Institute of Genetics, University of Haifa, Israel, for advice regarding QTL mapping. This study was conducted while T.I. was a fellow of the Japan Society for Promotion of Science. Support from Global Environment Research Fund of the Japanese Ministry of the Environment to A.K. is acknowledged.
References
- Baskin, J. M., and C. C. Baskin, 2004. A classification system for seed dormancy. Seed Sci. Res. 14 1–16. [Google Scholar]
- Benjamini, Y., and Y. Hogberg, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57 289–300. [Google Scholar]
- Blair, M. W., F. Pedraza, H. F. Buendia, E. Gaitan-Solis, S. E. Beebe et al., 2003. Development of a genome-wide anchored microsatellite map for common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 107 1362–1374. [DOI] [PubMed] [Google Scholar]
- Burnham, C. R., 1962. Discussions in cytogenetics, pp. 66–116 in Interchanges. Burgess Publishing, Minneapolis.
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, G. W., 2006. East Asian plant domestication, pp. 77–95 in Archaeology of Asia, edited by M. T. Stark. Blackwell Publishing, Oxford.
- Debeaujon, I., K. M. Leon-Kloosterziel and M. Koornneef, 2000. Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol. 122 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., A. Stec and C. Gustus, 1995. Teosinte branched 1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie, A. B., D. Zhang, L. M. A. Dirk, R. R. Thacker, J. A. Pfeiffer et al., 2003. Communication between the maternal testa and the embryo and/or endosperm affect testa attributes in tomato. Plant Physiol. 133 145–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant, J. D., B. W. Gardunia, K. D. Livingstone, M. R. Stevens and E. N. Jellen, 2006. An algorithm for analyzing linkages affected by heterozygous translocations: QuadMap. J. Hered. 97 62–66. [DOI] [PubMed] [Google Scholar]
- Ellstrand, N. C., H. C. Prentice and J. F. Hancock, 1999. Gene flow and introgression from domesticated plants into their wild relatives. Annu. Rev. Ecol. Syst. 30 539–563. [Google Scholar]
- Finch-Savage, W. E., and G. Leubner-Metzger, 2006. Seed dormancy and the control of germination. New Phytol. 171 501–523. [DOI] [PubMed] [Google Scholar]
- Gaitan-Solis, E., M. C. Duque, K. J. Edwards and J. Tohme, 2002. Microsatellite repeats in common bean (Phaseolus vulgaris): isolation, characterization, and cross-species amplification in Phaseolus ssp. Crop Sci. 42 2128–2136. [Google Scholar]
- Gepts, P., 2004. Crop domestication as a long-term selection experiment. Plant Breed. Rev. 24(2): 1–44. [Google Scholar]
- Gopinathan, M. C., and C. R. Babu, 1985. Structural diversity and its adaptive significance in seeds of Vigna minima (Roxb.) Ohwi & Ohashi and allies (Leguminosae-Papilionoideae). Ann. Bot. 56 723–732. [Google Scholar]
- Gressel, J. (editor), 2005. Crop Ferality and Volunteerism. Taylor & Francis, Boca Raton, FL.
- Guerra-Sanz, J. M., 2004. New SSR markers of Phaseolus vulgaris from sequence databases. Plant Breed. 123 87–89. [Google Scholar]
- Han, O. K., A. Kaga, T. Isemura, X. W. Wang, N. Tomooka et al., 2005. A genetic linkage map for azuki bean. [Vigna angularis (Willd.) Ohwi & Ohashi] Theor. Appl. Genet. 111 1278–1287. [DOI] [PubMed] [Google Scholar]
- Harlan, J. R., 1966. Plant introductions and biosystematics, pp. 55–83 in Plant Breeding, edited by K. J. Frey. Iowa State University Press, Ames, IA.
- Hawkes, J. G., 1983. The Diversity of Crop Plants. Harvard University Press, Cambridge, MA.
- Hsieh, H. M, Y. Pomeranz and B. G. Swanson, 1992. Composition, cooking time, and maturation of azuki (Vigna angularis) and common bean (Phaseolus vulgaris). Cereal Chem. 69 244–248. [Google Scholar]
- Iqbal, S., T. Mahmood, T. A. Ali, M. Anwar and M. Sarwar, 2003. Path coefficient analysis in different genotypes of soybean (Glycine max (L) Merril). Pakistan J. Biol. Sci. 6 1085–1087. [Google Scholar]
- Isemura, T., A. Kaga, S. Konishi, T. Ando, N. Tomooka et al., 2007. Genome dissection of traits related to domestication in azuki bean (Vigna angularis) and their comparison with other warm season legumes. Ann. Bot. 100 1053–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, R.C., J. M. Van Ooijen, P. Stam, C. Lister and C. Dean, 1995. Genotype-by-environment interaction in genetic mapping of multiple quantitative trait loci. Theor. Appl. Genet. 91 33–37. [DOI] [PubMed] [Google Scholar]
- Kaga, A., O. K. Han, S. Hirashima, P. Saravankumar, H. M. P. S. Kumari et al., 2004. Collecting and monitoring of the azuki bean (Vigna angularis) complex populations in Tottori prefecture, Japan. Annual report on exploration and introduction of plant genetic resources. Natl. Inst. Agrobiol. Sci. 20 61–74 (in Japanese with an English summary). [Google Scholar]
- Kaga, A., N. Tomooka, U. Phuntsho, Y. Kuroda, N. Kobayashi et al., 2005. Exploration and collection for hybrid derivatives between wild and cultivated soybean: preliminary survey in Akita and Hiroshima prefectures, Japan. Annual report on exploration and introduction of Plant Genetic Resources. Natl. Inst. Agrobiol. Sci. 21 59–71 (in Japanese with an English summary). [Google Scholar]
- Kao, C. H., Z. B. Zeng and R. D. Teasdale, 1999. Multiple interval mapping for quantitative trait loci. Genetics 152 1203–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim, P., B. W. Diers and R. C. Shoemaker, 1990. Genetic analysis of soybean hard seededness with molecular markers. Theor. Appl. Genet. 79 465–469. [DOI] [PubMed] [Google Scholar]
- Kikuchi, K., M. Koizumi, N. Ishida and H. Kano, 2006. Water uptake by dry beans observed by micro-magnetic resonance imaging. Ann. Bot. 98 545–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koinange, E. M. K., S. P. Singh and P. Gepts, 1996. Genetic control of the domestication syndrome in common bean. Crop Sci. 36 1037–1045. [Google Scholar]
- Korol, A. B., Y. I. Ronin, E. Nevo and P. M. Hayes, 1998. Multi-interval mapping of correlated trait complexes. Heredity 80 273–284. [Google Scholar]
- Kosambi, D. D., 1944. The estimation of map distances from recombination values. Ann. Eugen. 12 172–175. [Google Scholar]
- Kremer, R. J., 1993. Management of weed seed banks with microorganisms. Ecol. Appl. 3 42–52. [DOI] [PubMed] [Google Scholar]
- Li, C.D., C. A. Fatokun, B. Ubi, B. B. Singh and G. J. Scoles, 2001. Determining genetic similarities and relationships among cowpea breeding lines and cultivars by microsatellite markers. Crop Sci. 41 189–197. [Google Scholar]
- Livingstone, K. D., G. Churchill and M. K. Jahn, 2000. Linkage mapping in populations with karyotype rearrangements. J. Hered. 91 423–428. [DOI] [PubMed] [Google Scholar]
- Maeda, K., 1987. Legumes and Humans: A 10,000 Year History. Kokonshin, Tokyo (in Japanese).
- Osborn, T.C., D. V. Butrulle, A. G. Sharpe, K. J. Pickering, I. A. P. Parkin et al., 2003. Detection and effects of a homeologous reciprocal transposition in Brassica napus. Genetics 165 1569–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa, Y., 1978. On the viability of stored adzuki bean (Phaseolus angularis) and its hardness after cooking. Rept. Natl. Food Res. Inst. 33 37–40 (in Japanese). [Google Scholar]
- Peken, E., and C. Artk, 2004. Comparison of some cowpea (Vigna unguiculata L. Walp) genotypes from Turkey for seed yield and yield related characters. J. Agron. 3 137–140. [Google Scholar]
- Peng, J., Y. Ronin, T. Fahima, M. Roder, Y. Li et al., 2003. Domestication quantitative trait loci in Triticum dicoccoides, the progenitor of wheat. Proc. Natl. Acad. Sci. USA 100 2489–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncet, V., F. Lamy, K. M. Devos, M. D. Gale, A. Sarr et al., 2000. Genetic control of domestication traits in pearl millet (Pennisetum glaucum L., Poaceae). Theor. Appl. Genet. 100 147–159. [DOI] [PubMed] [Google Scholar]
- Raffi, S. A., and U. K. Nath, 2004. Variability, heritability, genetic advance and relationships of yield and yield contributing characters in dry bean (Phaseolus vulgaris L.). J. Biol. Sci. 4 157–159. [Google Scholar]
- Ronin, Y., A. Korol and E. Nevo, 1999. Single- and multiple-trait analysis of linked quantitative trait loci: some asymptotic analytical approximations. Genetics 151 387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, S., J. Abe, A. Kanazawa and Y. Shimamoto, 2004. Marker-assisted analysis for soybean hard seededness with isozyme and simple sequence repeat loci. Breed. Sci. 54 133–139. [Google Scholar]
- Sinnott, E. S., 1960. Plant Morphogenesis. McGraw-Hill, New York.
- Somta, P., A. Kaga, N. Tomooka, K. Kashiwaba, T. Isemura et al., 2006. Development of an interspecific Vigna linkage map between Vigna umbellata (Thunb.) Ohwi & Ohashi and V. nakashimae (Ohwi) Ohwi & Ohashi and its use in analysis of bruchid resistance and comparative genomics. Plant Breed. 125 77–84. [Google Scholar]
- Stearns, S. C., 1992. The Evolution of Life Histories. Oxford University Press, Oxford.
- Todd, J. J., and L. O. Vodkin, 1993. Pigmented soybean (Glycine max) seed coats accumulate proanthocyanidins during development. Plant Physiol. 102 663–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomooka, N., D. A. Vaughan, H. Moss and N. Maxted, 2002. The Asian Vigna: Genus Vigna Subgenus Ceratotropis Genetic Resources. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- Van Ooijen, J. W., and R. E. Voorrips, 2001. JoinMap 3.0: software for the calculation of genetic linkage maps. Plant Research International, Wageningen, The Netherlands.
- Vaughan, D. A., N. Tomooka and A. Kaga, 2004. Azuki bean, pp. 341–353 in Genetic Resources, Chromosome Engineering, and Crop Improvement. Grain Legumes, edited by R. J. Singh and P. P. Jauhar. CRC Press, Boca Raton, FL.
- Vos, P., R. Hogers, M. Bleceker, M. Reijans, T. van de Lee et al., 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acid Res. 23 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. W., A. Kaga, N. Tomooka and D. A. Vaughan, 2004. The development of SSR markers by a new method in plants and their application to gene flow studies in azuki bean. [Vigna angularis (Willd.) Ohwi and Ohashi] Theor. Appl. Genet. 109 352–360. [DOI] [PubMed] [Google Scholar]
- Wang, H., T. Nussbaum-Wagler, B. Li, Q. Zhao, Y. Vigouroux et al., 2005. The origin of the naked grains of maize. Nature 436 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L. Z., K. D. Liu, X. K. Dai, C. G. Xu and Q. Zhang, 1999. Identification of genetic factors controlling domestication-related traits of rice using an F2 population of a cross between Oryza sativa and O. rufipogon. Theor. Appl. Genet. 98 243–251. [Google Scholar]
- Xu, R. Q., N. Tomooka, D. A. Vaughan and K. Doi, 2000. a The Vigna angularis complex: genetic variation and relationships revealed by RAPD analysis, and their implications for in situ conservation and domestication. Genet. Resour. Crop Evol. 47 123–134. [Google Scholar]
- Xu, R. Q., N. Tomooka and D. A. Vaughan, 2000. b AFLP markers for characterizing the azuki bean complex. Crop Sci. 40 808–815. [Google Scholar]
- Yamaguchi, H., 1992. Wild and weed azuki beans in Japan. Econ. Bot. 46 384–394. [Google Scholar]
- Yamamoto, Y., C. M. Sano, Y. Tatsumi and H. Sano, 2006. Field analysis of horizontal gene flow among Vigna angularis complex plants. Plant Breed. 125 156–160. [Google Scholar]
- Yano, A., K. Yasuda and H. Yamaguchi, 2004. A test for molecular identification of Japanese archaeological beans and phylogenetic relationship of wild and cultivated species of subgenus Ceratotropis (genus Vigna, Papilionaceae) using sequence variation in two non-coding regions of the trnL and trnF genes. Econ. Bot. 58(Suppl): S135–S146. [Google Scholar]
- Yasuda, K., and H. Yamaguchi, 1996. Phylogenetic analysis of the subgenus Ceratotropis (genus Vigna) and an assumption of the progenitor of azuki bean using isozyme variation. Breed. Sci. 46 337–342. [Google Scholar]
- Yu, K., S. J. Park, V. Poysa and P. Gepts, 2000. Integration of simple sequence repeat (SSR) markers into a molecular linkage map of common bean (Phaseolus vulgaris L.). J. Hered. 91 429–434. [DOI] [PubMed] [Google Scholar]
- Zheng, S. H., and R. Watanabe, 2000. Relationship between sugar exudation from imbibing seeds and seedling emergence in soybean. Jpn. J. Crop Sci. 69 520–524. [Google Scholar]
- Zong, X. X., A. Kaga, N. Tomooka, X. W. Wang, O. K. Han et al., 2003. The genetic diversity of the Vigna angularis complex in Asia. Genome 46 647–658. [DOI] [PubMed] [Google Scholar]