Abstract
Infectious pancreatic necrosis (IPN) is a viral disease currently presenting a major problem in the production of Atlantic salmon (Salmon salar). IPN can cause significant mortality to salmon fry within freshwater hatcheries and to smolts following transfer to seawater, although challenged populations show clear genetic variation in resistance. To determine whether this genetic variation includes loci of major effect, a genomewide quantitative trait loci (QTL) scan was performed within 10 full-sib families that had received a natural seawater IPN challenge. To utilize the large difference between Atlantic salmon male and female recombination rates, a two-stage mapping strategy was employed. Initially, a sire-based QTL analysis was used to detect linkage groups with significant effects on IPN resistance, using two to three microsatellite markers per linkage group. A dam-based analysis with additional markers was then used to confirm and position any detected QTL. Two genomewide significant QTL and one suggestive QTL were detected in the genome scan. The most significant QTL was mapped to linkage group 21 and was significant at the genomewide level in both the sire and the dam-based analyses. The identified QTL can be applied in marker-assisted selection programs to improve the resistance of salmon to IPN and reduce disease-related mortality.
INFECTIOUS pancreatic necrosis (IPN) is a viral disease that currently presents a major problem in the production of Atlantic salmon, and other salmonid species, in many countries. This highly contagious disease has the unusual characteristic of affecting farmed salmon during two specific windows of the life cycle (Roberts and Pearson 2005). In the freshwater phase of the salmon life cycle, IPN outbreaks in fry have been observed for several decades, with up to 70% mortality. In the marine environments, the emergence of problematic IPN outbreaks (up to 40% mortality) is more recent, coinciding with the dramatic expansion of salmon aquaculture (Roberts and Pearson 2005). The causative agent for IPN is a double-stranded, nonmembraned RNA birnavirus, of which several subtypes have been characterized (Roberts and Pearson 2005). Levels of mortality during an IPN outbreak are determined by numerous factors, although it is increasingly clear that a strong genetic component to IPN resistance exists in salmon (Guy et al. 2006).
The elucidation of the molecular genetic basis of economically important traits in all salmonid species is complicated by the structure and properties of their genome. The salmonid genome is thought to have undergone a relatively recent duplication event (25–100 million years ago) and is currently evolving toward a fully diploid state (Allendorf and Thorgaard 1984; Allendorf and Danzmann 1997). Salmonid fish exhibit several remnants of their tetraploid ancestry, including some exchange of chromatid segments between ancestrally homeologous chromosome arms following the formation of multivalent structures at meiosis (Wright et al. 1983). These structures are also thought to constrain recombination, particularly toward centromeric regions of the chromosome (Allendorf and Danzmann 1997; Sakamoto et al. 2000). Interestingly, these phenomena are unique to male meiosis, and as a result there are striking sex-specific differences in linkage maps within salmonid species, with estimations of female:male map distance ratios of 3.25 in rainbow trout (Oncorhynchus mykiss) (Sakamoto et al. 2000), 3.92 (Gilbey et al. 2004) or 8.26 (Moen et al. 2004a) in Atlantic salmon, and 2.64 for brown trout (Salmo trutta) (Gharbi et al. 2006). The implications of these recombination differences for mapping quantitative trait loci (QTL) include the increased power of QTL detection, combined with the reduced ability to position the QTL in males (Hayes et al. 2006).
Several Atlantic salmon genetic linkage maps currently exist (e.g., Gilbey et al. 2004; Moen et al. 2004a). The most detailed map currently available is the composite map of Høyheim and Danzmann (http://grasp.mbb.sfu.ca/GRASPlinkage.html), which combines molecular marker and mapping data from the Genomic Research on Atlantic Salmon Project (GRASP) (http://grasp.mbb.sfu.ca/), the Salmon Genome Project (SGP) (http://www.salmongenome.no/cgi-bin/sgp.cgi), and the European Union collaborative SALMAP project (http://www.salmongenome.no/cgi-bin/sgp.cgi). These maps have facilitated the detection of QTL in salmon affecting body weight and condition factor (Reid et al. 2005), while molecular markers have also been linked to disease resistance against infectious salmon anemia (ISA) (Grimholt et al. 2003; Moen et al. 2004b, 2007), furunculosis (Grimholt et al. 2003), and the ectoparasite Gyrodactylus salaris (Gilbey et al. 2006). Furthermore, in the closely related rainbow trout, two QTL have been identified with a highly significant effect on freshwater IPN resistance in a backcross family (Ozaki et al. 2001). These IPN-resistance QTL in trout explained between 27 and 34% of the phenotypic variance, demonstrating that major loci affecting resistance to IPN can be segregating in salmonid species.
In common with the majority of aquaculture species, the selective breeding of Atlantic salmon is relatively new in comparison to that of terrestrial livestock. Farmed Atlantic salmon typically have a 4-year generation interval and, therefore, are currently just a few generations from their wild ancestors (Muir 2005). The genetic progress made in readily measurable traits (such as growth or sexual maturation) has been rapid and substantial. Where the traits are more difficult or impossible to measure on the selection candidate (such as disease resistance or fillet quality), full-sib phenotype information has been effectively utilized in selection programs (Sonesson 2005). However, for these difficult or expensive to measure traits, marker-assisted selection (MAS) directly on the selection candidate could substantially increase the accuracy of selection in salmon breeding programs (Sonesson 2005). The detection of QTL is an effective starting point for the application of marker-assisted, or gene-assisted, selection. Furthermore, identification of the genes underlying the QTL may lead to fundamental knowledge of genetic regulation of viral disease resistance and of host–virus interactions in fish.
Both experimental tank challenges and large-scale field challenges have resulted in consistent evidence for familial differences in resistance to IPN (Guy et al. 2006; Wetten et al. 2007). Selective breeding for IPN resistance, utilizing challenge information from siblings of commercial broodstock fish, is currently practiced in the breeding programs of major salmon breeding companies. The narrow-sense heritability for resistance to the marine phase of the disease has been estimated from field challenges at ∼0.4, which is higher than is typically associated with disease-resistance traits (Guy et al. 2006). The purpose of the current study is to utilize the extensive data and samples available from IPN field-challenge trials, combined with a salmonid-specific strategy for a genomewide molecular marker scan, to detect QTL affecting resistance to the marine phase of IPN in a commercial Atlantic salmon population.
MATERIALS AND METHODS
Animals:
The fish chosen for genotyping and analysis were from a cohort of ∼200 families that were siblings to the broodstock fish of Landcatch Natural Selection. These fish were spawned in 1999 in Ormsary, United Kingdom, and subsequently incubated and hatched into individual family tanks in March 2000, before being moved to larger mixed tanks in October 2000. The fish all received a routine vaccination against the bacterial disease furunculosis in November 2000. In April 2001, ∼55,000 smolts were transferred from the freshwater tanks to a single seawater site in Shetland, United Kingdom, where the IPN virus was known to be endemic. IPN is known to cause mortalities typically between 2 and 3 months post-transfer to seawater (Roberts and Pearson 2005). Therefore, dead fish were collected from the water between 5 and 12 weeks following transfer and veterinary inspection confirmed that deaths during this period were due to IPN. Approximately 16,000 mortalities were attributed to IPN, representing an overall mortality rate of ∼30%. Of these, ∼5000 dead fish and 5000 surviving fish, chosen at random, were sampled and genotyped to assign to family (see Genotyping below). Ten full-sib families were chosen from this cohort on the basis of having a large number of fish with DNA available, with the average full-sib family size being 58 (ranging from 51 to 70), giving a total sample size of 584 offspring with approximately equal numbers of mortalities and survivors. Large families were chosen to increase the statistical power for within-family linkage analysis, while the strategy of having close to equal numbers of mortalities and survivors was used to increase the chance that any IPN resistance QTL would be segregating within the chosen families.
Genotyping:
DNA was extracted from all fin clips using a Biosprint DNA kit (QIAGEN, Crawley, UK) following the manufacturer's protocol and fish were assigned to family by DNA profiling, using the 10-marker assignment multiplex system described in Guy et al. (2006). The genome scan was accomplished using microsatellite markers informed by the composite linkage map available from the GRASP website (http://grasp.mbb.sfu.ca/GRASPlinkage.html; hereafter referred to as the “composite linkage map”). Each new marker was optimized for ABI 377-mediated fluorescent detection by screening at annealing temperatures from 47° to 67°, using a Mastercycler gradient thermal cycler (Eppendorf, Hamburg, Germany) to determine the optimal annealing temperature for each marker. Markers with compatible PCR parameters and size ranges were combined into PCR multiplexes. Primer details and GenBank accession numbers, where available, for the microsatellite loci are given in supplemental Table 1 at http://www.genetics.org/supplemental/.
The genotyping strategy for the genome scan was designed to optimize the resources available, accounting for the low recombination rate in males compared to females. Therefore, a two-stage genotyping and analysis strategy was employed, whereby initially two to three markers per linkage group (29 linkage groups in total; Atlantic salmon 2n = 58) were genotyped on all parents and progeny, and QTL effects were assessed using a sire-family-based regression analysis (see QTL mapping below). Additional markers were then genotyped on the linkage groups that showed statistically significant evidence for a QTL (supplemental Table 3 at http://www.genetics.org/supplemental/), and the position of the QTL on the linkage group was estimated using a dam-family-based regression analysis. The genotype data were used to assign offspring to families and were checked for potential Mendelian inheritance errors using the family assignment program (FAP) (Taggart 2007). In the overall data set, including the chosen QTL families, the successful family assignment rate was >99% for survivors and >94% for mortality samples (lower due to some instances of degraded template).
Linkage map:
Markers were chosen for the genome scan on the basis of their position in the composite linkage map (http://grasp.mbb.sfu.ca/GRASPlinkage.html) to achieve wide coverage within and across linkage groups. The linkage between all markers was initially evaluated using the “twopoint” option in Crimap version 2.4 (Green et al. 1990). A LOD score of >3 was considered as significant linkage between markers. Markers showing significant linkage were then grouped and, for linkage groups with more than two markers, the most likely marker order was evaluated using the “build” and “flipsn” options. The genetic distance between the markers was then calculated using the “fixed” option, with the large difference in recombination patterns between the sexes requiring the calculation of sex-specific map distances. The second stage of genotyping involved adding additional markers to the linkage groups and families showing evidence for a QTL from stage one, with the marker order and positions being reevaluated using the methods described above. In this stage, the “chrompic” option was also used, to highlight putative phase errors, which were then removed from the analysis.
It is important to note that the numbers assigned to Atlantic salmon linkage groups are not fully consistent between the existing linkage maps. In this study, the linkage groups are numbered consecutively from 1 to 29. However, in the composite linkage map (http://grasp.mbb.sfu.ca/GRASPlinkage.html) the linkage groups are numbered from 1 to 32, but with the omission of nos. 26, 27, and 29.
QTL mapping:
The phenotype data were binary (1, died; 0, survived) and, under the assumption that underlying IPN resistance is a continuous variable, the phenotypic expression of which is dependent on a critical threshold, it is appropriate to apply the same QTL-mapping methods as used for quantitative traits (Visscher et al. 1996a; Zhang et al. 2004). For the genome scan, a two-stage linear-regression approach (Knott et al. 1996) was used for QTL detection, using the web-based software package “QTL Express” (Seaton et al. 2002). Briefly, the conditional probability of inheriting a particular haplotype from the sire or dam was inferred from the marker genotypes in all offspring, at 1-cM intervals. Subsequently, the phenotypic value (i.e., died or survived) was regressed on the probability that a particular haplotype allele was inherited from the sire or the dam. The large full-sib families available facilitated the choice of either sire or dam as the mapping parent within families. However, the detection of QTL in the initial genome scan was based on the sire analysis, due to the low recombination giving greater power to detect QTL using few markers per linkage group (Hayes et al. 2006).
Where significant evidence for a QTL was detected in the sire-based scan, additional markers were genotyped in the QTL-segregating families, and the QTL analysis described above was repeated using a dam-family-based analysis. An estimation of the percentage of within-family variance explained (PVE) by the QTL was calculated according to the formula of Knott et al. (1996). In the sire-based analysis, the formula of h2QTL = 4[1 − (MSEfull/MSEreduced)] was used, where MSEfull is the mean squared error of the model including the QTL, and MSEreduced is the mean squared error of the model fitting only a family mean. Additionally, an estimate of PVE was obtained from both the sire and the dam analysis in stage 1, such that h2QTL = 2{[1 − (MSEfull/MSEreduced)Sire] + [1 − (MSEfull/MSEreduced)Dam]}.
Significance thresholds and confidence intervals:
The appropriate significance thresholds for the study were determined empirically by a permutation analysis (Churchill and Doerge 1994). The chromosomewide thresholds were calculated for each linkage group, using 10,000 permutations. With 29 linkage groups, we expect ∼1.45 false positives per genome scan. The genomewide thresholds (the level at which 1 false positive is expected in 20 genome scans; Lander and Kruglyak 1995) were calculated by applying a Bonferroni correction to account for the analysis of 29 independent linkage groups, as described in Knott et al. (1998). Significance thresholds were initially calculated for the genome scan and then recalculated with the additional markers used to position the QTL on the significant linkage groups. In the analysis of QTL position, confidence intervals were calculated using a bootstrapping approach (Visscher et al. 1996b), whereby the top and bottom 2.5% of resampled position estimates define the 95% confidence interval for the QTL. For significant QTL, parents were judged to be segregating (i.e., heterozygous for alternative QTL alleles) on the basis of the t-test of the estimated allelic effect of that parent. For this test, where the overall QTL effect had already been declared as significant, the nominal 5% significance threshold was used.
RESULTS
Linkage map:
The mapping of QTL affecting resistance to IPN was achieved by analyzing 10 full-sib families with intermediate mortality levels from a natural IPN field challenge. Initially, by tracking the inheritance of microsatellite marker alleles from parents to offspring, the linkage groups and genetic distance between markers were defined (supplemental Table 1). The linkage groups calculated were consistent with the composite linkage map (http://grasp.mbb.sfu.ca/GRASPlinkage.html). The expected pattern of reduced male recombination was evident in the linkage maps, with the total map distance for males being 253 cM compared to 1209 cM for females. Therefore, the female:male ratio was 4.77:1. This ratio is comparable with other Atlantic salmon linkage maps (Gilbey et al. 2004; Moen et al. 2004a; http://grasp.mbb.sfu.ca/GRASPlinkage.html).
Sire-based QTL analysis:
The relevant significance thresholds for the first-stage sire-based QTL genome scan were calculated using a permutation analysis. The genomewide significance threshold was F = 3.4, with the chromosomewide thresholds ranging from F = 1.8 to 2.0.
The sire-based genome scan of the 29 Atlantic salmon linkage groups revealed two genomewide significant QTL on linkage groups 21 (LG 21) and 26 (LG 26) and a suggestive QTL on LG 19 (Table 1). For the most significant QTL, on LG 21, four sires showed evidence for segregation, and the mean additive effect on mortality in those sires was 0.41 (SE 0.13). Furthermore, the evidence for this QTL also reached genomewide significance in a dam-based analysis, with three dams showing statistically significant evidence for QTL segregation (Table 2). The percentage of within-family variation (across all 10 families) explained by the LG 21 QTL is 24.6%, when estimated using the sire-based analysis only, and 20.9% when estimated using both sire and dam analyses. Within the four segregating families, the estimated percentage of within-family variation explained is 79%. There was a single family (n = 57) where both sire and dam were segregating for the LG 21 QTL, and there was a difference in mortality rate of 75% between the alternative QTL homozygotes (as predicted from flanking marker genotypes; Table 3).
TABLE 1.
Details of the significant and suggestive QTL from the genome scan (stage 1), showing the significance levels and estimated percentage of within-family variation explained, and the QTL positioning using additional markers for segregating dams (stage 2), showing the significance level, position, and confidence intervals
| Linkage group | Sire F ratio | Significance level | Dam F ratio | Estimated PVE (%) |
|---|---|---|---|---|
| Stage 1: genome scan | ||||
| 21 | 4.77 | Genomewide | 3.57 | 24.6 |
| 26 | 3.74 | Genomewide | 2.54 | 18.2 |
| 19 | 2.32 | Chromosomewide | 1.45 | 8.9 |
| Linkage group | Dam F ratio | Significance level | QTL position (cM) | 95% confidence interval (cM) |
| Stage 2: QTL positioning | ||||
| 21 | 14.40 | Genomewide | 69 | 63–73 |
| 26 | 4.96 | Chromosomewide | 55 | 3–72 |
| 19 | 4.27 | Chromosomewide | 52 | 27–52 |
TABLE 2.
The QTL effect on mortality and associated absolute T values in segregating individual parents for the significant and suggestive QTL
| LG | Sire | QTL effect estimate (standard error) | Absolute T value |
|---|---|---|---|
| Stage 1: genome scan | |||
| 21 | L3M3076 | 0.57 (0.13) | 4.5 |
| L2M0167 | 0.50 (0.12) | 4.1 | |
| L2M0243 | 0.26 (0.12) | 2.1 | |
| L3M3080 | 0.26 (0.13) | 2.0 | |
| 26 | L2M0167 | 0.52 (0.14) | 3.6 |
| L3M3051 | 0.40 (0.15) | 2.7 | |
| L3M3069 | 0.60 (0.25) | 2.4 | |
| L2M0149 | 0.33 (0.14) | 2.4 | |
| 19 | L3M3076 | 0.39 (0.15) | 2.6 |
| L2M0082 | 0.40 (0.17) | 2.3 | |
| LG | Dam | QTL effect estimate (standard error) | Absolute T value |
| Stage 2: QTL positioning | |||
| 21 | L2F0056 | 0.73 (0.14) | 5.3 |
| L2F0067 | 0.35 (0.12) | 2.8 | |
| L2F0133 | 0.32 (0.13) | 2.6 | |
| 26 | L2F0034 | 0.68 (0.20) | 3.4 |
| L2F0622 | 0.47 (0.22) | 2.1 | |
| 19 | L2F0622 | 0.55 (0.16) | 3.4 |
TABLE 3.
The differences in IPN mortality levels between alternative LG 21 QTL genotypes, within a single full-sib family where both parents were segregating for the QTL
| Sire QTL haplotypea
|
||
|---|---|---|
| Resistant | Susceptible | |
| Dam QTL haplotypea | ||
| Resistant | 9% mortality (n = 11) | 62% mortality (n = 13) |
| Susceptible | 22% mortality (n = 9) | 84% mortality (n = 19) |
The haplotypes were determined by the segregation of alleles at the markers BHMS217 and Rsa476 to offspring.
The LG 26 QTL also surpassed the genomewide threshold for significance in the sire-based analysis. The mean effect of the QTL on mortality in the four segregating sires was 0.46 (SE 0.17), and the estimated PVE was 18.2% (Tables 1 and 2). The LG 19 QTL reached the chromosomewide level of significance, with two segregating sires with an average effect on mortality of 0.40 (SE 0.17).
Dam-based QTL analysis:
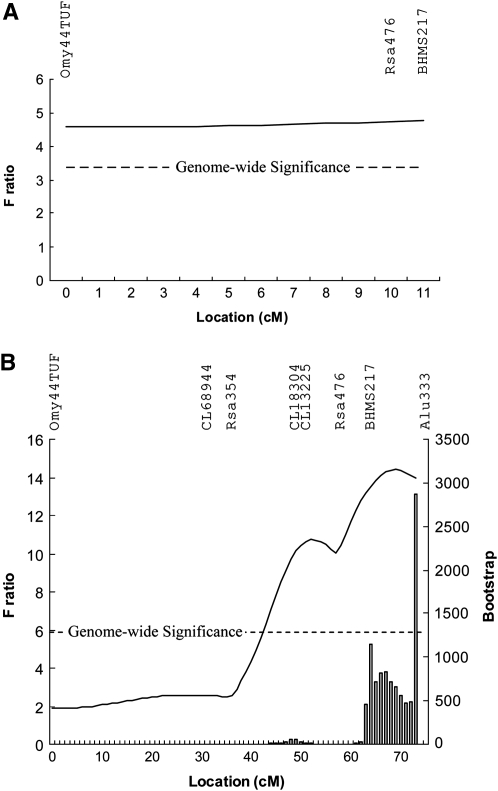
In the second stage of genotyping and analysis, to position the QTL on the linkage group, additional markers from LG 21 were genotyped in the families where the data suggested that the dam was segregating for the QTL. Five additional microsatellite markers were added to LG 21 (supplemental Table 2 at http://www.genetics.org/supplemental/), and the linkage mapping and QTL analysis were rerun to reveal the best-estimated position of the QTL at 69 cM (Figure 1). The F ratio associated with the QTL is substantially higher than that in the sire-family-based analysis, although this can be partially explained by the reduced number of families included in this analysis. The genomewide threshold for significance was F = 6.2, while the chromosomewide threshold was F = 3.6. The 95% confidence interval associated with this QTL, estimated using bootstrapping, was ∼10 cM (from 63 to 73 cM), although the possibility that the QTL may be distal to the end of the linkage map cannot be excluded.
Figure 1.—
The likelihood profile for linkage group 21 following the sire-based genome scan (A) and the dam-based QTL positioning with additional markers (B).
An additional three markers were added to position the suggestive QTL on LG 26, and two markers were added to position the suggestive QTL on LG 19 (although one of these was poorly informative and removed from the analysis). With the additional markers on LG 26, it is clear that there is a large gap between BHMS437 and the rest of the linkage group (supplemental Table 3), and the QTL was positioned within the interval (55 cM). The additional markers on LG 19 (supplemental Table 4 at http://www.genetics.org/supplemental/) assisted with positioning the QTL at 52 cM, although the majority of evidence for this QTL stems from a single segregating dam (Table 2).
DISCUSSION
This study is the first to report the detection and positioning of major loci affecting resistance to IPN in Atlantic salmon. Significant evidence for QTL segregation within the 10 chosen families was found in the sire-based analysis on LGs 21, 26, and 19. In the dam-based analysis, the LG 21 QTL was positioned with a reasonable degree of accuracy (C.I. ∼ 10 cM) given the limited number of molecular markers available in Atlantic salmon compared to many model organism and agricultural species. The size of effect of this QTL, and its potential for utility in marker-assisted selection, was highlighted in a single family where both parents were segregating, with the difference in mortality between the predicted alternative QTL homozygotes being ∼75%.
In the first-stage sire analysis, the QTL on LG 21 was estimated to explain ∼25% of the observed within-family variance in the overall data set, while the QTL on LGs 26 and 19 were estimated to explain ∼18 and 9% of the variance, respectively. The corresponding percentages, when calculated using data from both sire and dam analyses in stage 1, were ∼21% for LG 21, 14% for LG 26, and 6% for LG 19. These percentages may be overestimates due to the intermediate mortality in the families chosen, the binary nature of the data, and the tendency for QTL studies to overestimate the size of effect of significant QTL. Nonetheless, the evidence does suggest that the identified QTL are significant contributors to the within-family variation in IPN mortality in the studied salmon population.
The dam-based second-stage analysis with additional markers provided highly significant evidence to confirm the large QTL on LG 21. However, when additional markers were added to LGs 26 and 19 in the dam-based analysis, the evidence for a QTL reached only the suggestive level. It is interesting to note that LGs 26 and 19 share several duplicated marker loci in the composite linkage map (http://grasp.mbb.sfu.ca/GRASPlinkage.html). This strongly suggests that parts of these two linkage groups are ancestrally homeologous and raises the possibility that the two QTL detected may represent two functional paralogs of the same gene(s). For example, the growth hormone gene has two functional paralogs (GH1 and GH2) in salmonids that are both inherited in a diploid fashion (McKay et al. 2004), although the locations of these genes on the salmon linkage map are unknown.
The Atlantic salmon genome shows a great deal of homology to other salmonids, including rainbow trout, Arctic charr, and brown trout (Danzmann et al. 2005; Gharbi et al. 2006). The two previously identified QTL affecting IPN resistance in rainbow trout (RT) map to RT LG 3 (IPN R/S 1) and RT LG 22 (IPN R/S 2) (Ozaki et al. 2001, 2005), for which there has not been any clearly identified homology to the Atlantic salmon QTL linkage groups identified here (Danzmann et al. 2005). However, the locus OmyRGT44TUF maps to the opposite end of RT LG 22 to the IPN R/S 2 QTL (Ozaki et al. 2001, 2005), and OmyRGT44TUF also maps to the opposite end of Atlantic salmon (AS) LG 21 to the QTL identified in the current study. This raises the prospect that these QTL may be due to the effect of the same gene in the two species, although further evidence of homology between RT LG 22 and AS LG 21 would be required to assess this possibility. The markers closest to the RT IPN-resistance QTL were tested in our populations, but they proved uninformative.
The salmonid QTL screening strategy of utilizing the low male recombination to detect QTL in an initial scan has clear advantages in terms of minimizing the required genotyping resources and increasing the experimental power due to the use of fewer independent tests. In addition, the two-stage analysis provides a QTL detection stage (sire based) and a confirmation and positioning stage (dam based). However, one potential drawback of the approach is that the lower male recombination is not universal across the genome, and toward the telomeres of the chromosomes the sex-specific recombination ratios may actually reverse (Sakamoto et al. 2000; Gharbi et al. 2006). As a result, the initial QTL screening using sire-based segregation of just two or three markers may fail to detect QTL residing toward the telomeres of the chromosomes. However, data from the current Atlantic salmon linkage maps suggest that the majority of available markers are inherited with very tight linkage and often almost as a single unit (Gilbey et al. 2004; Moen et al. 2004a; http://grasp.mbb.sfu.ca/GRASPlinkage.html). Therefore, either the increased male recombination toward the telomeres is less pronounced in Atlantic salmon than in other salmonid species or there are simply few markers available for Atlantic salmon telomere regions. In either case, the two-stage QTL-mapping strategy utilized in the current study would appear to be an effective use of the available markers.
Resistance to IPN is a key target trait for breeders of Atlantic salmon and a trait where MAS may have crucial advantages over traditional selection based on sib-challenge trials (Sonesson 2005). The IPN resistance QTL can be applied immediately in within-family MAS in commercial breeding programs, provided that IPN resistance data have been collected from sibs of the selection candidates, thus enabling the phase relationship between the marker alleles and the QTL alleles to be established. However, to improve the utility of the QTL in MAS, and to move toward the identification of positional candidate genes, fine mapping of the QTL to a smaller region of the chromosome is necessary. Unfortunately, the number of available markers currently limits the possibility for fine mapping QTL in Atlantic salmon; therefore the development of additional markers in the QTL regions is an important future target. Comparative mapping and exploitation of the extensive physical genetic map available (Ng et al. 2005) may facilitate the identification of further markers or candidate genes in the QTL regions for Atlantic salmon. These additional markers, in combination with the analysis of additional IPN-challenged populations, will be critical to fine mapping the QTL.
The identified QTL may represent the physiological effect of variation in genes critical to the prevention of, or response to, IPN infection in the marine environment. Whether these genes play a similar role in the resistance of salmon fry to freshwater IPN is unknown, although there is a strong genetic correlation between fresh and marine water IPN resistance (Wetten et al. 2007). Experimental infection of Atlantic salmon with the IPN virus has revealed that the interferon pathways are paramount in the host response, and this is particularly evident through the upregulation of interferon-induced Mx gene expression (e.g., Lockhart et al. 2007). Investigation of the physiological/immune response to infection by fish with alternative QTL genotypes may give valuable insight into the genes and pathways responsible for the QTL effects. The integration of such functional studies with the aforementioned fine-mapping approach may be an effective route toward the identification of the genes underlying the major IPN resistance QTL in Atlantic salmon.
Acknowledgments
The authors gratefully acknowledge William Davidson (Simon Fraser University, Canada), Bjorn Høyheim (Norwegian College of Veterinary Medicine, Norway) and Karim Gharbi (University of Glasgow, UK) for assisting with the provision of microsatellite markers for this project. We acknowledge funding from the British Biotechnology and Biological Sciences Research Council and the European Animal Disease Network of Excellence for Animal Health and Food Safety.
References
- Allendorf, F. W., and R. G. Danzmann, 1997. Secondary tetrasomic segregation of MDH-B and preferential pairing of homeologues in rainbow trout. Genetics 145 1083–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allendorf, F. W., and G. R. Thorgaard, 1984. Tetraploidy and evolution of salmonid fishes, pp. 1–53 in Evolutionary Genetics of Fishes, edited by B. J. Turner. Plenum Press, New York.
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danzmann, R. G., M. Cairney, W. S. Davidson, M. M. Ferguson, K. Gharbi et al., 2005. A comparative analysis of the rainbow trout genome with 2 other species of fish (Arctic charr and Atlantic salmon) within the tetraploid derivative Salmonidae family (subfamily: Salmoninae). Genome 48 1037–1051. [DOI] [PubMed] [Google Scholar]
- Gharbi, K., A. Gautier, R. G. Danzmann, S. Gharbi, T. Sakamoto et al., 2006. A linkage map for brown trout (Salmo trutta): chromosome homeologies and comparative genome organization with other salmonid fish. Genetics 172 2405–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbey, J., E. Verspoor, A. McLay and D. Houlihan, 2004. A microsatellite linkage map for Atlantic salmon (Salmo salar). Anim. Genet. 35 98–105. [DOI] [PubMed] [Google Scholar]
- Gilbey, J., E. Verspoor, T. A. Mo, E. Sterud, K. Olstad et al., 2006. Identification of genetic markers associated with Gyrodactylus salaris resistance in Atlantic salmon Salmo salar. Dis. Aquat. Organ. 71 119–129. [DOI] [PubMed] [Google Scholar]
- Green, P., K. Falls and S. Crooks, 1990. Documentation for Crimap, Version 2.4. Washington University School of Medicine, St. Louis.
- Grimholt, U., S. Larsen, R. Nordmo, P. Midtlyng, S. Kjoeglum et al., 2003. MHC polymorphism and disease resistance in Atlantic salmon (Salmo salar); facing pathogens with single expressed major histocompatibility class I and class II loci. Immunogenetics 55 210–219. [DOI] [PubMed] [Google Scholar]
- Guy, D. R., S. C. Bishop, S. Brotherstone, A. Hamilton, R. J. Roberts et al., 2006. Analysis of the incidence of infectious pancreatic necrosis mortality in pedigreed Atlantic salmon, Salmo salar L., populations. J. Fish Dis. 29 637–647. [DOI] [PubMed] [Google Scholar]
- Hayes, B. J., A. Gjuvsland and S. Omholt, 2006. Power of QTL mapping experiments in commercial Atlantic salmon populations, exploiting linkage and linkage disequilibrium and effect of limited recombination in males. Heredity 97 19–26. [DOI] [PubMed] [Google Scholar]
- Knott, S. A., J. M. Elsen and C. S. Haley, 1996. Methods for multiple-marker mapping of quantitative trait loci in half-sib populations. Theor. Appl. Genet. 93 71–80. [DOI] [PubMed] [Google Scholar]
- Knott, S. A., L. Marklund, C. S. Haley, K. Andersson, W. Davies et al., 1998. Multiple marker mapping of quantitative trait loci in a cross between outbred wild boar and large white pigs. Genetics 149 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E., and L. Kruglyak, 1995. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat. Genet. 11 241–247. [DOI] [PubMed] [Google Scholar]
- Lockhart, K., A. J. McBeath, B. Collet, M. Snow and A. E. Ellis, 2007. Expression of Mx mRNA following infection with IPNV is greater in IPN-susceptible Atlantic salmon post-smolts than in IPN-resistant Atlantic salmon parr. Fish Shellfish Immunol. 22 151–156. [DOI] [PubMed] [Google Scholar]
- McKay, S. J., J. Trautner, M. J. Smith, B. F. Koop and R. H. Devlin, 2004. Evolution of duplicated growth hormone genes in autotetraploid salmonid fishes. Genome 47 714–723. [DOI] [PubMed] [Google Scholar]
- Moen, T., B. Hoyheim, H. Munck and L. Gomez-Raya, 2004. a A linkage map of Atlantic salmon (Salmo salar) reveals an uncommonly large difference in recombination rate between the sexes. Anim. Genet. 35 81–92. [DOI] [PubMed] [Google Scholar]
- Moen, T., K. T. Fjalestad, H. Munck and L. Gomez-Raya, 2004. b A multistage testing strategy for detection of quantitative trait loci affecting disease resistance in Atlantic salmon. Genetics 167 851–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moen, T., A. K. Sonesson, B. Hayes, S. Lien, H. Munck et al., 2007. Mapping of a quantitative trait locus for resistance against infectious salmon anemia in Atlantic salmon (Salmo salar): comparing survival analysis with analysis on affected/resistant data. BMC Genet. 8 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir, J., 2005. Managing to harvest? Perspectives on the potential of aquaculture. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360 191–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, S. H., C. G. Artieri, I. E. Bosdet, R. Chiu, R. G. Danzmann et al., 2005. A physical map of the genome of Atlantic salmon, Salmo salar. Genomics 86 396–404. [DOI] [PubMed] [Google Scholar]
- Ozaki, A., T. Sakamoto, S. Khoo, K. Nakamura, M. R. Coimbra et al., 2001. Quantitative trait loci (QTLs) associated with resistance/susceptibility to infectious pancreatic necrosis virus (IPNV) in rainbow trout (Oncorhynchus mykiss). Mol. Genet. Genomics 265 23–31. [DOI] [PubMed] [Google Scholar]
- Ozaki, A., O. Masanori, S. Khoo, E. Ohara, K. Fuji et al., 2005. Quantitative trait loci (QTL) analysis and marker-assisted breeding for economical important traits in aquaculture. Proceeding of the 34th UJNR Panel Meeting ‘Aquaculture and Stock Enhancement of Finfish.’ http://www.lib.noaa.gov/japan/aquaculture/presentation_slides/34th/ozaki_ujnr_2005.pdf.
- Reid, D. P., A. Szanto, B. Glebe, R. G. Danzmann and M. M. Ferguson, 2005. QTL for body weight and condition factor in Atlantic salmon (Salmo salar): comparative analysis with rainbow trout (Oncorhynchus mykiss) and Arctic charr (Salvelinus alpinus). Heredity 94 166–172. [DOI] [PubMed] [Google Scholar]
- Roberts, R. J., and M. D. Pearson, 2005. Infectious pancreatic necrosis in Atlantic salmon, Salmo salar L. J. Fish Dis. 28 383–390. [DOI] [PubMed] [Google Scholar]
- Sakamoto, T., R. G. Danzmann, K. Gharbi, P. Howard, A. Ozaki et al., 2000. A microsatellite linkage map of rainbow trout (Oncorhynchus mykiss) characterized by large sex-specific differences in recombination rates. Genetics 155 1331–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaton, G., C. S. Haley, S. A. Knott, M. Kearsey and P. M. Visscher, 2002. QTL Express: mapping quantitative trait loci in simple and complex pedigrees. Bioinformatics 18 339–340. [DOI] [PubMed] [Google Scholar]
- Sonesson, A. K., 2005. Possibilities for marker-assisted selection in aquaculture breeding schemes, pp 309–328 in Marker-Assisted Selection: Current Status and Future Application in Crops, Livestock, Forestry and Fish, edited by E. P. Guimarães, J. Ruane, B. D. Scherf, A. Sonnino and J. D. Dargie. FAO, Rome.
- Taggart, J. B., 2007. FAP: an exclusion-based parental assignment program with enhanced predictive functions. Mol. Ecol. Notes 7 412–415. [Google Scholar]
- Visscher, P. M., C. S. Haley and S. A. Knott, 1996. a Mapping QTLs for binary traits in backcross and F2 populations. Genet. Res. 68 55–63. [Google Scholar]
- Visscher, P. M., R. Thompson and C. S. Haley, 1996. b Confidence intervals in QTL mapping by bootstrapping. Genetics 143 1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetten, M., T. Aasmundstad, S. Kjøglum and A. Storset, 2007. Genetic analysis of resistance to infectious pancreatic necrosis (Salmo salar L.). Aquaculture 272 111–117. [Google Scholar]
- Wright, Jr., J. E., K. Johnson, A. Hollister and B. May, 1983. Meiotic models to explain classical linkage, pseudolinkage, and chromosome pairing in tetraploid derivative salmonid genomes. Isozymes Curr. Top. Biol. Med. Res. 10 239–260. [PubMed] [Google Scholar]
- Zhang, C., D. J. De Koning, J. Hernández-Sánchez, C. S. Haley, J. L. Williams et al., 2004. Mapping of multiple quantitative trait loci affecting bovine spongiform encephalopathy. Genetics 167 1863–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]