Abstract
Reciprocal crosses between species can yield hybrids with different viabilities. The high frequency of this asymmetric hybrid viability (“Darwin's corollary”) places it alongside Haldane's rule and the “large-X effect” as a general feature of postmating reproductive isolation. Recent theory suggests that reciprocal cross asymmetries can arise from stochastic substitutions in uniparentally inherited loci such as mitochondrial genomes, although large systematic differences in mitochondrial substitution rates can also contribute to asymmetries. Although the magnitude of asymmetry will be relatively insensitive to unequal rates of mitochondrial evolution in diverging species, we show here that rate asymmetries can have a large effect on the direction of viability asymmetries. In reciprocal crosses between species, the maternal parent with faster mitochondrial evolution will tend to produce less viable F1 hybrids owing to an increased probability of mito-nuclear incompatibilities. We test this prediction using data on reciprocal hybrid viability and molecular evolution rates from a clade of freshwater fishes, Centrarchidae. As predicted, species with accelerated mitochondrial evolution tend to be the worse maternal parent for F1 hybrids, providing the first comparative evidence for a systematic basis to Darwin's corollary. This result is consistent with the hypothesis that mito-nuclear incompatibilities can play an important role in reproductive isolation. Such asymmetrical reproductive isolation may help explain the asymmetrical mitochondrial introgression observed between many hybridizing species. However, as with any comparative study, we cannot rule out the possibility that our results arise from a mutual correlation with a third variable such as body size.
WHEN species hybridize, their progeny often exhibit some degree of infertility or inviability. This hybrid breakdown prevents introgression between the parental species and so can play an important role in speciation. Therefore, a major focus of evolutionary genetics research has been to understand the genetic basis of reduced hybrid fitness that is largely independent of the environment in which hybrids are reared, i.e., intrinsic postzygotic isolation (Orr and Presgraves 2000; Coyne and Orr 2004). Research on the genetics of intrinsic postzygotic isolation has led to four interrelated generalizations.
First and foremost, there is now a broad consensus that intrinsically low hybrid fitness (not attributable to ecological interactions) is often the result of deleterious epistatic interactions between genes from divergent parental genomes (Coyne and Orr 2004; Noor and Feder 2006). These “Dobzhansky–Muller incompatibilities” (DMIs) can arise when an ancestral population (with genotype AABB) diverges into two isolated populations that fix different derived alleles (aaBB and AAbb, respectively). The derived alleles a and b must be compatible with the ancestral alleles A and B, or else the new mutations would not have been able to pass through a heterozygous state to become fixed. However, there is no guarantee that the derived alleles are compatible with each other, so there is a chance that in hybrids (AaBb) the derived alleles a and b interact (or fail to interact) in a way that reduces hybrid fitness (Dobzhansky 1934; Muller 1939; Orr 1993, 1995; Turelli and Orr 1995, 2000; Orr and Turelli 2001). Several incompatibility loci have been identified (Ting et al. 1998; Barbash et al. 2003; Presgraves 2003; Presgraves et al. 2003), although very few pairwise interactions have been fully described (Schartl et al. 1999; Brideau et al. 2006)
A second generalization, Haldane's rule, states that when one sex is inviable or infertile in hybrids, it is the heterogametic sex (e.g., XY or ZW) (Haldane 1922). This rule holds for the vast majority of taxa surveyed to date (Laurie 1997; Coyne and Orr 2004). It is widely accepted that this pattern arises for hybrid viability because DMIs tend to be recessive: incompatibilities between autosomal alleles a and b will generally be masked by the successful (dominant) interaction between the compatible ancestral alleles A and B. However, incompatibilities involving an X-linked locus will be hemizygous in the XY sex and hence can reduce fitness even if they are recessive (Muller 1942; Turelli 1998). These hemizygous incompatibilities also contribute to a third generalization, the large-X effect (or “Coyne's rule”), which observes that X chromosomes make a disproportionate contribution to heterogametic inviability and/or sterility (Coyne and Orr 1989a, 2004; Turelli and Orr 2000).
The past few years have seen a revival of interest in a fourth trend, “isolation asymmetry,” in which reciprocal hybrid crosses yield different degrees of hybrid inviability or sterility (Tiffin et al. 2001; Bolnick and Near 2005; Turelli and Moyle 2007). These asymmetries have been documented in plants, fungi, insects, and vertebrates (Sturtevant 1920; Muller 1942; Thornton 1955; Oliver 1978; Harrison 1983; Wu and Davis 1983; Rakocinski 1984; Coyne and Orr 1989b; Gallant and Fairbain 1997; Presgraves and Orr 1998; Navajas et al. 2000; Tiffin et al. 2001; Willett and Burton 2001; Presgraves 2002; Dettman et al. 2003). In 14 diverse genera of angiosperms, Tiffin et al. (2001) found asymmetric postmating isolation in 35–45% of species pairs. In a freshwater fish family, Centrarchidae, 18 of 20 species pairs with reciprocal cross viability data exhibited asymmetries (Bolnick and Near 2005). For instance, crosses between the congeners Lepomis gulosus and L. macrochirus yield average hybrid survival rates of 77% (L. gulosus dam) and 35% (L. macrochirus dam). This F1 hybrid asymmetry has recently been dubbed “Darwin's corollary to Haldane's rule” by Turelli and Moyle (2007), in recognition of Darwin's own detailed attention to the pattern (Darwin 1859, Chap. 8).
Asymmetric hybrid incompatibilities cannot be explained by traditional autosome–autosome DMIs. This is because F1 hybrid autosomal genotypes will be the same regardless of cross direction (AaBb); note that this symmetry does not hold for subsequent backcross progeny (Welch 2004) or in cases in which taxa differ in autosomal gene content (for simplicity, we ignore this complication below). F1 asymmetries can be explained by DMIs involving uniparentally inherited genetic factors (Turelli and Moyle 2007). Four major classes of asymmetrical DMIs have been discussed. First, Haldane's rule may be asymmetrical because one species' X chromosome may have different numbers or magnitudes of incompatibilities than the other species' X. That is, hybrids with genotype X1YAa need not have equal fitness to X2YAa hybrids (alternatively, the sex chromosomes can interact asymmetrically). However, these asymmetries will be observed only in the heterogametic sex. Second, asymmetries may arise from DMIs between mitochondrially encoded genes and nuclear genes (Kenyon and Moraes Carlos 1997; Rand et al. 2001, 2004; Rawson and Burton 2002; Sackton et al. 2003; Burton et al. 2006; Harrison and Burton 2006). Incompatibilities involving genes in cytoplasmic organelles may be asymmetric because, as with Haldane's rule, one species' mitochondria (or chloroplasts) may contribute to more or stronger DMIs (i.e., the fitness of M1Aa need not equal the fitness of M2Aa). We refer to this category of DMI as a “mito-nuclear incompatibility,” to distinguish it from the third possible cause of asymmetry, arising from maternal–zygotic incompatibilities. Maternal–zygotic incompatibilities can occur when maternally encoded transcription factors in the oocyte fail to properly regulate expression of paternally derived genes (Whitt et al. 1977; Philipp et al. 1983; Turelli and Moyle 2007). In this case the DMI is not between genes within the hybrid, but between the hybrid and maternal genomes. A fourth category of asymmetry arises from gametophyte–sporophyte interactions in plants (Tiffin et al. 2001; Turelli and Moyle 2007).
Turelli and Moyle (2007) modeled the evolution of reciprocal hybrid asymmetry. In their analysis, they made a crucial distinction between stochastic and deterministic contributions to asymmetry. To understand this difference, consider mito-nuclear DMIs. First, let us assume that both species are equally likely to substitute novel mitochondrial haplotypes and that the mitochondrial differences are equally likely to generate an incompatibility with the other species' nuclear genome. Even so, the stochastic nature of substitutions and DMI occurrence makes it very likely that one hybridization direction accumulates more potent mito-nuclear DMIs than the other, yielding asymmetric hybrid viability. In contrast to this purely stochastic explanation, it is possible that there is a systematic bias as to which species develops more mitochondria-based DMIs. This systematic bias arises when there is an asymmetry in the relative substitution rates of the two populations: whichever species has a relatively faster rate of mitochondrial evolution will carry more substitutions and hence have a greater capacity for incompatibilities with the other species' nuclear genome. By “relatively faster,” we mean that the mitochondrial genome experiences an accelerated substitution rate relative to its nuclear genome substitution rate (Turelli and Moyle 2007). If instead both parts of the genome accelerate equally, one species' mitochondria has more opportunities to pick up incompatibilities with the other species' nuclear genome, but the reciprocal is also true so no systematic asymmetry ensues.
Turelli and Moyle's (2007) analyses led them to conclude that “unless relative rate differences are extreme…, stochastic effects are more likely to explain observed levels of postmating asymmetry than systematic interspecific differences in the relative rates of molecular evolution” (Turelli and Moyle 2007, p. 1068). Specifically, they found that levels of asymmetry (i.e., the magnitudes of reciprocal cross differences) are likely to have an appreciable deterministic basis only if one species experiences at least a fourfold acceleration in its mitochondrial evolution relative to the other species, while the nuclear genomes remain at their previous rate. However, Turelli and Moyle (2007, p. 1081) did propose that “if systematic rate differences … do play a significant role in postmating asymmetry, then the direction of asymmetry should be predictable from the relative rates of evolution for the loci that contribute to [unidirectional] DMIs.” Here, we extend the analysis of Turelli and Moyle, focusing on the direction of asymmetries (i.e., which maternal parent is more likely to yield lower hybrid viability). Our analysis shows that moderate differences in rates of mitochondrial evolution can have an appreciable impact on which species is likely to be the “worse” maternal parent in reciprocal crosses. We then test this prediction by comparing the directions of F1 viability asymmetry, and mitochondrial and nuclear substitution rates, in a clade of North American freshwater fishes (Centrarchidae). We find a weak but statistically significant tendency for the species with faster mitochondrial evolution to be the worse maternal parent in reciprocal hybrid crosses. Together, our theory and data suggest that systematic effects involving mito-nuclear interactions may frequently contribute to “Darwin's corollary.”
MODEL
Given that our data on hybrid asymmetry in centrarchids concern embryo-to-larval viability and relative rates of mitochondrial vs. nuclear evolution, we focus on analyzing hybrid inviability and mito-nuclear DMIs. However, we emphasize that many asymmetrical DMIs may involve maternal effects (cf. De Renzis et al. 2007), and there is some evidence for maternal effects contributing to hybrid asymmetry in centrarchids (Whitt et al. 1977; Philipp et al. 1983). Following Turelli and Orr (1995, 2000), the asymmetry analysis of Turelli and Moyle (2007) assumes that individual DMIs contribute additively to a “hybrid breakdown score,” denoted S, and that hybrid viability decreases as S increases up to a threshold value, denoted C, beyond which hybrids become completely inviable. Let Sij denote the breakdown score for hybrids between species i and j that have species i mothers.
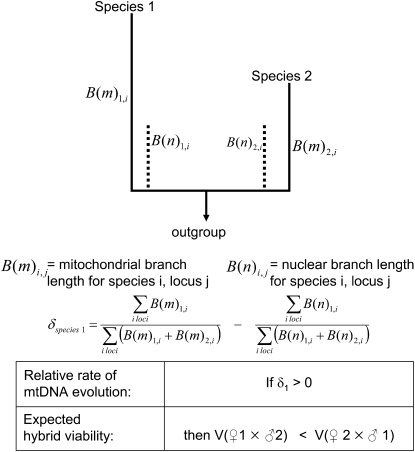
Considering only mito-nuclear DMIs, the existence of a preferred direction for asymmetry is determined by the parameter
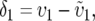 |
(1) |
where υ1 (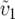 ) is the fraction of the expected mitochondrial (nuclear) substitutions that occur in lineage 1. The parameter δ1 thus represents the degree of asymmetry in rates of mitochondrial evolution, relative to nuclear rates. This quantity can be approximated from estimates of the number of mtDNA and nuclear substitutions occurring in each lineage, using a rooted phylogeny. For instance, in Figure 1 more than half of the mitochondrial substitutions have occurred in the lineage leading to species 1, so υ1 = B(m)1/[B(m)1 + B(m)2] > 0.5, where B(m)i is the mitochondrial branch length from species i to the most recent common ancestor of a given species pair. Turelli and Moyle (2007) showed that when δ1 > 0, corresponding to a higher proportion of mitochondrial vs. nuclear substitutions occurring in lineage 1, the expected hybrid breakdown score is higher for crosses with species 1 as the maternal parent [E(S12) > E(S21)]. This result arises because crosses with taxon 1 mothers will on average experience more mito-nuclear DMIs than do the reciprocal crosses. Rather surprisingly, they found that the level of reciprocal-cross asymmetry, as quantified by the absolute value of the hybrid viability difference between reciprocal crosses, is not appreciably affected by δ1 > 0, unless the relative rate differences are extreme, for instance, δ1 > 0.3. If we assume that both lineages experience equal rates of nuclear substitutions, δ1 = 0.3 corresponds to lineage 1 experiencing on average four times as many mitochondrial substitutions as lineage 2. However, Turelli and Moyle did not investigate how the probability of directional asymmetry, e.g., P(S12 > S21), depends on δ1.
) is the fraction of the expected mitochondrial (nuclear) substitutions that occur in lineage 1. The parameter δ1 thus represents the degree of asymmetry in rates of mitochondrial evolution, relative to nuclear rates. This quantity can be approximated from estimates of the number of mtDNA and nuclear substitutions occurring in each lineage, using a rooted phylogeny. For instance, in Figure 1 more than half of the mitochondrial substitutions have occurred in the lineage leading to species 1, so υ1 = B(m)1/[B(m)1 + B(m)2] > 0.5, where B(m)i is the mitochondrial branch length from species i to the most recent common ancestor of a given species pair. Turelli and Moyle (2007) showed that when δ1 > 0, corresponding to a higher proportion of mitochondrial vs. nuclear substitutions occurring in lineage 1, the expected hybrid breakdown score is higher for crosses with species 1 as the maternal parent [E(S12) > E(S21)]. This result arises because crosses with taxon 1 mothers will on average experience more mito-nuclear DMIs than do the reciprocal crosses. Rather surprisingly, they found that the level of reciprocal-cross asymmetry, as quantified by the absolute value of the hybrid viability difference between reciprocal crosses, is not appreciably affected by δ1 > 0, unless the relative rate differences are extreme, for instance, δ1 > 0.3. If we assume that both lineages experience equal rates of nuclear substitutions, δ1 = 0.3 corresponds to lineage 1 experiencing on average four times as many mitochondrial substitutions as lineage 2. However, Turelli and Moyle did not investigate how the probability of directional asymmetry, e.g., P(S12 > S21), depends on δ1.
Figure 1.—
An illustration of the deterministic hypothesis to explain asymmetric F1 viability. For any two species within a larger phylogeny, one can estimate the branch length from each species back to its common ancestor (arrow indicates outgroup rooting) for both mitochondrial loci (solid lines, m) and nuclear loci (dotted lines, n). In this schematic, species 1 has accelerated mtDNA evolution (relative to nuclear rates), resulting in δ1 > 0. Theory predicts that species 1 thus has more potential for creating genetic incompatibilities in hybrids when it is the maternal parent.
As shown below, the probabilities of directional asymmetry, like the absolute magnitudes of asymmetry, depend on divergence time, t. We measure divergence times in scaled units, τ = t/TC, with TC defined as the harmonic mean of the times until the expected breakdown scores from the reciprocal crosses reach the threshold value C. In principle, three quantities might be used to quantify the probability of directional asymmetry:
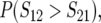 |
(2) |
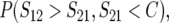 |
(3) |
or
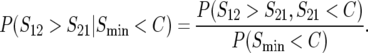 |
(4) |
The first, (2), is simply the probability that the breakdown score for hybrids with taxon 1 mothers is larger than that for hybrids with taxon 2 mothers. Because the expected number of DMIs is proportional to t2 (Orr 1995; Orr and Turelli 2001), differences between the expected values of S12 and S21 will become increasingly large as divergence time (t) increases; and P(S12 > S21) will approach 1 when δ1 > 0 and τ is large. However, this will not be manifest in fitness differences once τ is sufficiently large that both S12 and S21 exceed C, and all hybrids become inviable. This suggests measure (3), which requires that S12 > S21 but S21 < C, so that a fitness difference is observed between the hybrids from reciprocal crosses. However, this quantity must approach 0 as τ increases, because the condition S21 < C will be violated eventually. Hence, we suggest that the time dependence of the probability of directional asymmetry may be most informatively quantified by (4), which conditions on observed fitness asymmetry and reports the probability that the observed asymmetry goes in a specified direction (e.g., lower viability with taxon 1 mothers).
In F1 hybrids, there is an important difference between mito-nuclear DMIs and DMIs between autosomal loci. Whereas both autosomal loci involved in a DMI are heterozygous for incompatible alleles, mito-nuclear DMIs involve hemizygous loci interacting with heterozygous loci and are expected to have a systematically larger effect. Indeed this distinction underlies Muller's (1942) “dominance theory” for Haldane's rule. The expected ratio of effects of autosomal vs. mito-nuclear DMIs is denoted h0 and, as discussed by Turelli and Orr (2000) and Turelli and Moyle (2007), is expected to be small, on the order of 0.1. As shown by Turelli and Moyle (2007), the quantitative properties of reciprocal-cross asymmetry can be adequately described using a bivariate normal approximation for the reciprocal breakdown scores (S12, S21). For realistic parameter values, the behavior of this distribution depends on h0 only through a composite parameter, denoted η, that quantifies the relative effect of nuclear (symmetrical) vs. mito-nuclear (asymmetrical) DMIs. With η = 0, all DMIs are mito-nuclear and the level of expected asymmetry is greatest. Whereas with η = 10, symmetrical nuclear DMIs contribute on average 10 times as much to the inviability of hybrids as mito-nuclear DMIs, and little asymmetry is expected. Under the bivariate Gaussian approximation, the probabilities in (4) would be calculated by numerical integration using Mathematica 6.0 (Wolfram 2003).
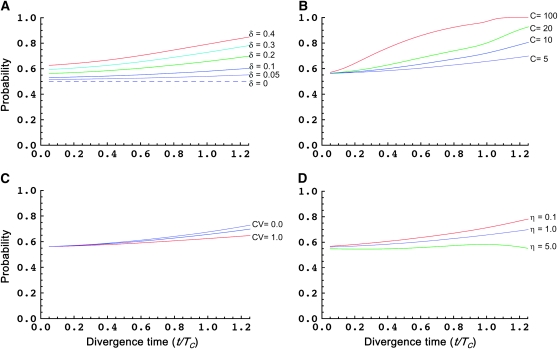
Here we illustrate how these probabilities of directional asymmetry (Equation 4) vary with: divergence time δ1; C, the average number of mito-nuclear DMIs needed to produce complete postmating isolation; CV, the coefficient of variation of DMI effects; and η, which measures the relative effect of symmetrical vs. asymmetrical DMIs. To explain the levels of asymmetry observed in centrarchid fishes by Bolnick and Near (2005), Turelli and Moyle (2007) argued that C and η must both be fairly small, with C on the order of 5 and η on the order of 1. We focus on these values. For additional discussion of the model parameters and their biological interpretation, see Turelli and Moyle (2007).
Figure 2, A–D, shows how the probability of a specified direction of asymmetry, 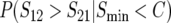 , varies as a function of divergence time (τ = t/TC) while varying the values of δ1 (A), C (B), CV (C), and η (D), around base values of δ1 = 0.2, C = 5, CV = 0.5, and η = 1, while holding h0 = 0.1. Figure 2A shows, as expected, that the probability of directional asymmetry increases with the asymmetry of mitochondrial substitution rates, δ1. The case δ1 = 0 provides a control, showing that the probability of a given direction of asymmetry is 0.5 for all divergence times, as expected. When mitochondrial rates are equal, either species is equally likely to be the maternal parent for the less viable cross direction. As δ1 increases, species 1 exhibits faster mitochondrial evolution and hence is increasingly likely to produce less viable hybrids when acting as the maternal parent. The results with δ1 = 0.3 are notable. This corresponds to mtDNA evolution being four times faster in lineage 1 than in lineage 2, but it produces a very small effect on the expected magnitude of asymmetry (see Figure 6C of Turelli and Moyle 2007). In contrast, there is an appreciable effect on the probability of directional asymmetry, which rises from ∼0.65 to 0.77 as τ increases from 0.6 to 1.2. Over this range of divergence times, the probability that at least one of the reciprocal crosses produces viable hybrids [i.e., the denominator of (4)] decreases from very near 1 to 0.33.
, varies as a function of divergence time (τ = t/TC) while varying the values of δ1 (A), C (B), CV (C), and η (D), around base values of δ1 = 0.2, C = 5, CV = 0.5, and η = 1, while holding h0 = 0.1. Figure 2A shows, as expected, that the probability of directional asymmetry increases with the asymmetry of mitochondrial substitution rates, δ1. The case δ1 = 0 provides a control, showing that the probability of a given direction of asymmetry is 0.5 for all divergence times, as expected. When mitochondrial rates are equal, either species is equally likely to be the maternal parent for the less viable cross direction. As δ1 increases, species 1 exhibits faster mitochondrial evolution and hence is increasingly likely to produce less viable hybrids when acting as the maternal parent. The results with δ1 = 0.3 are notable. This corresponds to mtDNA evolution being four times faster in lineage 1 than in lineage 2, but it produces a very small effect on the expected magnitude of asymmetry (see Figure 6C of Turelli and Moyle 2007). In contrast, there is an appreciable effect on the probability of directional asymmetry, which rises from ∼0.65 to 0.77 as τ increases from 0.6 to 1.2. Over this range of divergence times, the probability that at least one of the reciprocal crosses produces viable hybrids [i.e., the denominator of (4)] decreases from very near 1 to 0.33.
Figure 2.—
The probability that a specified reciprocal-cross direction yields lower hybrid fitness, given that reciprocal-cross asymmetry is observed; i.e., 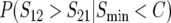 , as a function of scaled divergence time t/TC. (A) The effect of varying the asymmetry in relative rates of mitochondrial evolution, δ1: δ1 = 0 (dashed line), 0.05 (gray–blue), 0.1 (blue), 0.2 (green), 0.3 (light blue), and 0.4 (red), with C = 5, η = 1, h0 = 0.1, and CV = 0.5. (B) The effect of varying the average number of mito-nuclear incompatibilities C at which inviability is complete: C = 5 (gray–blue), 10 (blue), 20 (green), and 100 (red), with δ1 = 0.2, η = 1, h0 = 0.1, and CV = 0.5. (C) The effect of varying the coefficient of variation of DMI effects, CV: CV = 0 (gray–blue), 0.5 (blue), and 1.0 (red), with C = 5, δ1 = 0.2, η = 1, and h0 = 0.1. (D) The effect of varying the relative contribution of symmetrical vs. asymmetrical DMIs η: η = 0.1 (red), 1.0 (gray–blue), and 5.0 (green), with C = 5, δ1 = 0.2, h0 = 0.1, and CV = 0.5.
, as a function of scaled divergence time t/TC. (A) The effect of varying the asymmetry in relative rates of mitochondrial evolution, δ1: δ1 = 0 (dashed line), 0.05 (gray–blue), 0.1 (blue), 0.2 (green), 0.3 (light blue), and 0.4 (red), with C = 5, η = 1, h0 = 0.1, and CV = 0.5. (B) The effect of varying the average number of mito-nuclear incompatibilities C at which inviability is complete: C = 5 (gray–blue), 10 (blue), 20 (green), and 100 (red), with δ1 = 0.2, η = 1, h0 = 0.1, and CV = 0.5. (C) The effect of varying the coefficient of variation of DMI effects, CV: CV = 0 (gray–blue), 0.5 (blue), and 1.0 (red), with C = 5, δ1 = 0.2, η = 1, and h0 = 0.1. (D) The effect of varying the relative contribution of symmetrical vs. asymmetrical DMIs η: η = 0.1 (red), 1.0 (gray–blue), and 5.0 (green), with C = 5, δ1 = 0.2, h0 = 0.1, and CV = 0.5.
Figure 2B varies C, the average number of mito-nuclear DMIs needed to produce complete inviability, holding δ1 = 0.2, η = 1, h0 = 0.1, and CV = 0.5. For moderate divergence times, corresponding to significant probabilities of hybrid inviability, the probability of directional asymmetry approaches 1 as C becomes large. This reflects the fact that as C increases, the stochastic process of DMI accumulation becomes increasingly deterministic. (As shown by Orr and Turelli 2001, the variance in the number of DMIs is close to the mean; so the coefficient of variation of breakdown scores producing high levels of inviability decreases as C increases.) As shown by Turelli and Moyle (2007), when C is ≥10, we expect much lower magnitudes of asymmetry than observed in the centrarchid data of Bolnick and Near (2005).
Figure 2C varies CV, the coefficient of variation of DMI effects, with δ1 = 0.2, C = 5, h0 = 0.1, and η = 1. It shows that for realistic values of CV (namely, CV < 1), the magnitude of CV has a negligible effect on the probability of directional asymmetry. As expected, as CV increases, the probability of directional asymmetry decreases slightly as this additional source of stochasticity masks the deterministic signal associated with δ1 > 0. Figure 2D varies η, the ratio of the expected contribution of symmetrical vs. asymmetrical DMIs to hybrid inviability, holding C = 5, δ1 = 0.2, h0 = 0.1, and CV = 0.5. With very large η, almost all DMIs are symmetrical and reciprocal-cross asymmetry should be negligible, in contrast to the considerable asymmetry documented by Bolnick and Near (2005). Figure 2D shows that the probability of directional asymmetry, like its expected magnitude, increases with decreasing η.
In conclusion, while asymmetric rates of mitochondrial evolution (δ1 > 0) have a negligible effect on the magnitude of asymmetries in reciprocal F1 crosses (Turelli and Moyle 2007), the direction of asymmetry is sensitive to δ1. We therefore predict that for a given pair of species, the maternal parent species with relatively faster mitochondrial evolution will tend to produce less viable offspring. Below we present a test of this prediction using data from centrarchid fishes. However, it is necessary to clarify the connection between our parameters δ1 and η and our mtDNA data. As noted in the Introduction, mito-nuclear DMIs are not the only potential source of postmating reproductive asymmetry. In particular, the data of Whitt et al. (1977) and Philipp et al. (1983) indicate that maternal–zygotic interactions are also likely to be important. Hence, mito-nuclear interactions probably make up only a portion of the asymmetrical DMIs that contribute to η and the relevant value of δ1 is likely to differ from estimates based on mtDNA alone. To the extent that the other uniparentally inherited factors evolve independently of mtDNA (i.e., do not tend to accelerate with mtDNA acceleration), our mtDNA estimates are likely to correctly estimate the sign of δ1 but overestimate its magnitude. We simply do not have enough data to predict how far above 0.5 the probabilities of directional asymmetry are likely to be. However, our best guesses for the parameters suggest that values >0.6 are unlikely.
METHODS
Hybrid viability data and calculating reciprocal asymmetry:
We compiled published data on hybrid viability among centrarchid species (see Bolnick and Near 2005 for details). Centrarchids are external fertilizers, so it is possible to strip eggs and sperm from mature individuals to generate in vitro hybrids. Viability (Vij) of a particular cross direction (species i and j as dam and sire, respectively) was measured as the percentage of hybrid embryos that hatched, divided by the hatch rate of intraspecific embryos from the same clutch of eggs, to control for egg maturity. Note that Vij can exceed 100% if hybrids exhibit higher hatching rates than embryos from intraspecific crosses. Fertilization rates are consistently high (>90%) even for distantly related species pairs and are not correlated with evolutionary divergence (West and Hester 1966; Merriner 1971), so embryo hatch rates are a good measure of viability.
The compiled data set contains 18 species pairs with reciprocal-cross data, not including 3 reciprocal pairs with zero viability in both directions (Table 1). For each of the 18 pairs, we identified which species was the “worse dam,” i.e., the species i for which Vij < Vji. Note that many of the published reciprocal crosses do not report sufficient information to determine whether reciprocal-cross viabilities are significantly different. However, large clutch sizes (often ≫500) mean that even slight differences in viability are usually statistically significant. All nine reciprocal crosses for which sample sizes were available exhibited significant asymmetries (see Bolnick and Near 2005 for details). Note also that centrarchids do not appear to follow Haldane's rule (Bolnick and Near 2005), consistent with the finding that they have no karyotypically distinct sex chromosomes (Roberts 1964). In an intraspecific cross of L. cyanellus, only 1 of 429 polymorphic amplified fragment length polymorphism (AFLP) markers was fully linked to sex (present in all females and in no males; López-Fernández and Bolnick 2007). Consequently, asymmetric hybrid viabilities are unlikely to have arisen via Haldane's rule and are more likely to reflect mito-nuclear or maternal–zygotic incompatibilities (Whitt et al. 1977; Philipp et al. 1983).
TABLE 1.
Data on reciprocal crosses among centrarchid species
| Species pair
|
Hybrid viability (% of control cross)
|
Relative viability difference | Worse dam | Rate asymmetry δi
|
Species with δi > 0 | Worse dam: faster mtDNA? | Smaller species
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Node | A | B | A × B | B × A | A | B | Species | Faster mtDNA? | Worse dam? | ||||
| A | A. rupestris | L. gulosus | 2 | 5.5 | −0.636 | A | 0.002 | −0.002 | A | Yes | B | No | No |
| A | A. rupestris | L. macrochirus | 26 | 5 | 0.808 | B | −0.294 | 0.294 | B | Yes | B | Yes | Yes |
| B | A. rupestris | P. nigromaculatus | 41 | 86.5 | −0.526 | A | 0.029 | −0.029 | A | Yes | A | Yes | Yes |
| I | L. cyanellus | L. gulosus | 99 | 107 | −0.075 | A | 0.012 | −0.012 | A | Yes | A | Yes | Yes |
| H | L. cyanellus | L. macrochirus | 105 | 90 | 0.143 | B | 0.045 | −0.045 | A | No | Equal | NA | NA |
| G | L. cyanellus | L. microlophus | 118 | 107 | 0.093 | B | −0.104 | 0.104 | B | Yes | B | Yes | Yes |
| D | L. cyanellus | M. salmoides | 11 | 80 | −0.863 | A | 0.062 | −0.062 | A | Yes | A | Yes | Yes |
| H | L. gulosus | L. macrochirus | 77 | 35 | 0.545 | B | 0.045 | −0.045 | A | No | A | Yes | No |
| G | L. gulosus | L. microlophus | 107 | 82 | 0.234 | B | 0.224 | −0.224 | A | No | B | No | Yes |
| D | L. gulosus | M. salmoides | 75 | 103 | −0.272 | A | 0.062 | −0.062 | A | Yes | A | Yes | Yes |
| A | L. gulosus | P. nigromaculatus | 1.7 | 2.3 | −0.261 | A | −0.155 | 0.155 | B | No | A | No | Yes |
| G | L. macrochirus | L. microlophus | 77 | 102 | −0.245 | A | −0.144 | 0.144 | B | No | B | Yes | No |
| D | L. macrochirus | M. salmoides | 7 | 70 | −0.900 | A | 0.324 | −0.324 | A | Yes | A | Yes | Yes |
| A | L. macrochirus | P. nigromaculatus | 13 | 44 | −0.705 | A | 0.290 | −0.290 | A | Yes | A | Yes | Yes |
| D | L. microlophus | M. salmoides | 0 | 43 | −1.0 | A | 0.399 | −0.399 | A | Yes | A | Yes | Yes |
| E | M. dolomieu | M. salmoides | 82 | 102 | −0.196 | A | 0.058 | −0.058 | A | Yes | A | Yes | Yes |
| F | M. floridanus | M. salmoides | 109 | 91 | 0.165 | B | −0.121 | 0.121 | B | Yes | A | No | No |
| C | P. annularis | P. nigromaculatus | 98 | 77 | 0.214 | B | −0.003 | 0.003 | B | Yes | B | Yes | Yes |
Node identities correspond to Figure 3. For each species pair we arbitrarily assign a species A and B, which we use to specify the viabilities of the respective reciprocal crosses (female parent listed first). Viabilities are calculated by dividing the percentage of hybrid embryos that survive to larval stage by the percentage of intraspecific control cross embryos that survive. Relative viability difference is calculated as the difference in viabilities, standardized by the higher value. The “worse dam” is the species providing the eggs for the lower viability reciprocal cross. Relative mitochondrial rates, δi, for both species are calculated from branch length data in supplemental Table 1 of the supplemental Appendix (http://www.genetics.org/supplemental/). The species with the higher value exhibits relatively faster mtDNA evolution. Under the heading “Smaller species” we identify the species with the smaller maximum body size and state whether this coincides with having faster-evolving mtDNA or being the worse dam.
Phylogenetic analyses and branch length estimation:
We sequenced five loci, totaling 5533 bp, from at least one individual of all 32 described centrarchid species (Near et al. 2004, 2005). The loci include multiple mitochondrial genes (ND2, 16S, and a set of tRNAs) and four nuclear genes (rhodopsin, TMO4C4, and introns from calmodulin and S7 ribosomal protein). We obtained a consensus phylogeny using a partitioned Bayesian analysis (see Near et al. 2005 for details). Similar phylogenies were found using maximum-likelihood and parsimony methods (Near et al. 2004, 2005). Phylogenies estimated from individual loci were often not as well resolved as those resulting from analysis of the combined data set; however, there was little phylogenetic incongruence among the trees estimated from each locus. The one exception concerned Micropterus treculi, which mtDNA and nuclear gene phylogenies place in very different locations within the Micropterus clade. We omitted this species from the present molecular analyses, but its inclusion has no effect on our results.
We obtained branch lengths for each locus by estimating their substitution rates on a constrained topology, the Bayesian consensus tree (Figure 3). The optimal likelihood model was obtained for each nuclear locus and mitochondrial gene using ModelTest 3.0 (Posada and Crandall 1998), and branch lengths (the expected number of substitutions per nucleotide site) were estimated using these specific maximum-likelihood models in PAUP* 4.0b10 (Swofford 2002). For each species pair, these branch lengths represent Bij, the number of substitutions from each species i to the most recent common ancestor of the pair, for locus j (Figure 1). Whichever species has a larger Bij has a faster rate of molecular evolution at that locus. We summed the branch lengths across all mitochondrial loci to obtain a total mitochondrial branch length B(m)i for each member of each species pair. Similarly, we summed across nuclear loci to obtain total nuclear branch lengths B(n)i. Next, we calculated the rate asymmetry for mitochondrial (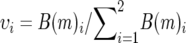 ) and nuclear (
) and nuclear (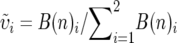 ) loci. Finally, we calculated the relative mitochondrial rate asymmetry δi (Equation 1, Figure 1) for each species in each of the 18 species pairs. In a given species pair, the species with the positive δi is inferred to have accelerated mitochondrial evolution, and the other species of the pair will have a negative δi of equal magnitude. Note that the theoretical predictions concern overall substitution rates, rather than rates of nonsynonymous substitutions per se. Furthermore, we do not necessarily expect that the particular genes used here are the causal agents in asymmetric incompatibilities. We therefore focus on overall substitution rates, rather than nonsynonymous substitutions.
) loci. Finally, we calculated the relative mitochondrial rate asymmetry δi (Equation 1, Figure 1) for each species in each of the 18 species pairs. In a given species pair, the species with the positive δi is inferred to have accelerated mitochondrial evolution, and the other species of the pair will have a negative δi of equal magnitude. Note that the theoretical predictions concern overall substitution rates, rather than rates of nonsynonymous substitutions per se. Furthermore, we do not necessarily expect that the particular genes used here are the causal agents in asymmetric incompatibilities. We therefore focus on overall substitution rates, rather than nonsynonymous substitutions.
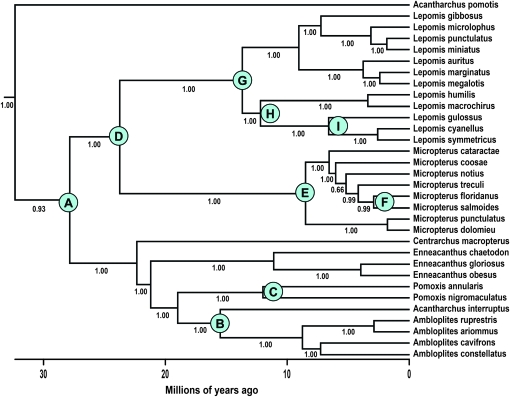
Figure 3.—
A phylogenetic hypothesis for the Centrarchidae, based on DNA sequences from three mtDNA gene regions and four nuclear genes. Bayesian posterior probabilities are provided for all nodes. See Near et al. (2005) for details.
To assess the robustness of our results, we used a second approach to calculating δi. We calculated υi and 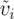 separately for each locus, then averaged across loci to obtain a mean mitochondrial and mean nuclear υi and
separately for each locus, then averaged across loci to obtain a mean mitochondrial and mean nuclear υi and 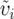 , and took the difference to obtain δi. Our results were qualitatively identical with this method, so we focus on the first formulation, which more closely approximates the theoretical values. Using either approach, we acknowledge that this estimate is based on a very limited sample of genes. This limitation should introduce additional error into our analysis and reduce the likelihood of detecting an effect if one existed, so our empirical tests should tend to be conservative.
, and took the difference to obtain δi. Our results were qualitatively identical with this method, so we focus on the first formulation, which more closely approximates the theoretical values. Using either approach, we acknowledge that this estimate is based on a very limited sample of genes. This limitation should introduce additional error into our analysis and reduce the likelihood of detecting an effect if one existed, so our empirical tests should tend to be conservative.
Testing for systematic causes of asymmetric reciprocal F1 inviability:
On the basis of our model, we predicted that systematic asymmetries might arise because, for a given species pair, the maternal parent with relatively faster mitochondrial evolution (positive δi) should produce less viable hybrids (Figure 1). We tested this directional prediction with a one-tailed sign test. We counted the number of cases (X) out of N reciprocal crosses where the species with δi > 0 was also the worse dam (Vij < Vji), converted this into a fraction (X/N), and tested the null hypothesis that X/N = 0.5 using a binomial (sign) test. However, the 18 species pairs are not phylogenetically independent. We therefore applied a version of Coyne and Orr's (1989b) node-based phylogenetic correction. Each species pair was assigned a score of 1 if the species with a positive δi also was the worse dam and 0 if the theoretical expectation was not met. We then averaged the scores across all species pairs that diverged at particular node, leaving us with nine node-averaged scores. This averaging used the node-weighted approach of Fitzpatrick (2002), although results were equivalent when we used the nonweighted approach of Coyne and Orr (1989b). A t-test then evaluated the null hypothesis that scores averaged 0.5, against a directional alternative hypothesis that scores tend to exceed 0.5.
Another way to test for the same trend is to regress viability asymmetry against mitochondrial rate asymmetry. If the species with faster-evolving mitochondria is the worse maternal parent, we also expect that the rate asymmetry measure is negatively correlated with relative viability. We measured relative viability for species 1 as
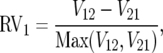 |
(5) |
where Vij is the viability of hybrids with species i and j as the maternal and paternal parents, respectively. Arbitrarily focusing on one species in each pair, δi (faster mitochondrial evolution) should be negatively correlated with relative viability RVi. Regressing RVi against δi is redundant with the sign test (above), but illustrates the robustness of our results to another statistical method. To apply phylogenetic corrections, we calculated the mean δi and RVi for each node and repeated the regression. In calculating mean δi and RVi, we randomly selected one of the sister clades to be the source of the focal species for each species pair.
While we expected that mitochondrial rate asymmetry affects the direction of viability asymmetry, Turelli and Moyle (2007) predicted that the magnitude of viability asymmetry should be relatively insensitive to δi. To test the latter relationship, we calculated the absolute magnitude of rate and viability asymmetries for each species pair. The magnitude of asymmetry was calculated by taking the absolute value of δi and RVi. We regressed |RVi| against |δi|, using nodes as the level of replication (averaging multiple species pairs at a given node). No significant relationship is expected.
Comparative studies such as this one are fundamentally limited in their ability to infer causation, relying on correlations across species. When correlations between two variables do occur, they might be explained by mutual correlation with a third variable. Previous analyses suggested a possible association between asymmetric viability and adult body size, in which the smaller species tends to be the worse dam (Bolnick et al. 2006). To evaluate whether body size might represent a confounding variable, we conducted two phylogenetically corrected sign tests (t-tests on averaged 0/1 scores for each node), to test (1) whether the smaller species is the worse dam and (2) whether the smaller species tends to have faster mitochondrial evolution.
RESULTS
A sign test confirmed that species with faster-evolving mitochondria tend to be the worse dam, producing F1 hybrids with lower viability than the reciprocal cross with a slow-mitochondria female parent (Table 1). Of 18 species pairs, 13 pairs matched our expectation, with a one-tailed binomial probability P = 0.0481 under the null hypothesis of no association between mtDNA rate and asymmetry. A one-tailed test is appropriate because we are evaluating an a priori directional hypothesis. The phylogenetically corrected t-test also confirmed that node averages of the sign test scores consistently exceeded the null expectation of 0.5 (mean node averages = 0.79, t9 = 2.32, P = 0.024). Both the sign test and the phylogenetically corrected t-test are consistent with our model showing that that variation in mitochondrial evolution rates generates systematic differences in reciprocal F1 hybrid viabilities. The theory also emphasized that asymmetric viability should result from asymmetric mitochondrial rates relative to nuclear rates (δ), not mitochondrial asymmetry alone (ν) (Turelli and Moyle 2007). Consistent with this expectation, we found no association between the direction of viability asymmetry and ν: the species with the larger ν was also the worse dam in only 7 of 18 cases.
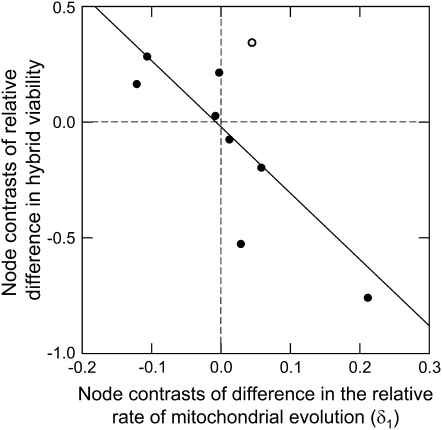
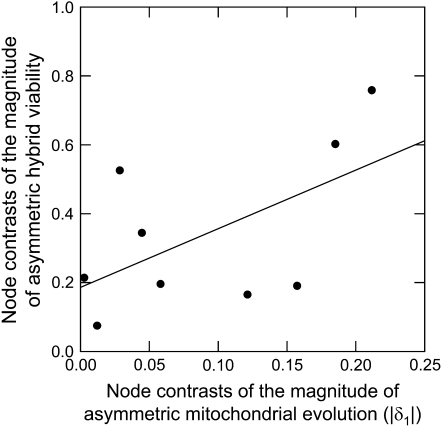
A quantitative comparison of viability and molecular evolution rate asymmetries is also consistent with a deterministic contribution to Darwin's corollary. There was a significant negative correlation between the relative viability difference and the difference in relative mtDNA rates (F1,16 = 10.11, P = 0.006, r2 = 0.387). The regression with node averages also supported a negative correlation (Figure 4, F1,7 = 8.392, P = 0.023, r2 = 0.545). This trend confirms the sign test results because it incorporates the direction of the asymmetries. That is, when species A has the faster evolution rate (positive rate difference), it is also the worse dam (negative relative viability). In contrast to results for the direction of asymmetry, we found no significant correlation between mitochondrial rate asymmetry and the magnitude of viability asymmetry (Figure 5, regression of node averages: F1,7 = 3.45, P = 0.106, r2 = 0.33), although the trend was positive.
Figure 4.—
The quantitative relationship between the mitochondrial substitution rate of asymmetry δ1 and the degree of asymmetry in F1 viability, using node averages. Both relative rate and relative asymmetry are calculated, selecting species 1 for each pair from an arbitrary clade subtending each node, allowing both positive and negative values of each variable. This regression is partially redundant with the sign test described in the text, as it tests for effect direction: this is illustrated by dividing the figure into quadrants (dotted lines). The one node that runs counter to our expectation is indicated by an open rather than a solid circle.
Figure 5.—
The quantitative relationship between the magnitude (rather than direction) of mitochondrial rate asymmetry δ1 and the magnitude of viability asymmetry, using node averages.
The association between mitochondrial rates and viability asymmetry is admittedly a correlation, rather than evidence of causation. Highlighting this fact, we also found that the worse maternal parent tended to have the smaller body size in a given pair (sign test on raw data, 13/17 cases, P = 0.049; t-test on node averages, mean score = 0.796, t7 = 2.53, P = 0.017; note that one species pair was omitted due to identical maximum body sizes). The smaller species also has a marginally nonsignificant tendency to exhibit accelerated mitochondrial evolution (sign test on raw data, 13/17 cases, P = 0.049; t-test on node averages, mean score = 0.712, t7 = 1.51, P = 0.085).
DISCUSSION
Asymmetric viability of reciprocal hybrid crosses is likely to arise from DMIs between uniparentally inherited factors and autosomal biparentally inherited loci (Turelli and Moyle 2007). Uniparentally inherited factors include hemizygous sex chromosomes, mitochondria, and cytoplasm. These confer asymmetries because when one species' mitochondria have a deleterious interaction with an autosomal locus from the other species, there is no reason why the second species' mitochondria must also confer incompatibility, or if it does there is no reason why their effects need be the same. Consequently, Coyne and Orr (2004, p. 310) stated that “asymmetry provides prima facie evidence for the role of the cytoplasm in postzygotic isolation.”
Substitutions generating these uniparental DMIs (UDMIs) are expected to occur stochastically in both parental species. Consequently, the direction of the asymmetry (which reciprocal cross is less viable) may be entirely random. However, there may also be a deterministic contribution if one species is more likely to acquire a UDMI, for instance, if it exhibits accelerated mitochondrial evolution and so is likely to harbor more substitutions. Our theoretical results predict that the direction of F1 viability asymmetries depends on relative rates of mitochondrial evolution in the parental taxa. Our empirical results are consistent with this prediction: hybrid viability tends to be lower when the species with accelerated mtDNA evolution is the maternal parent. This trend was confirmed by both a sign test and linear regression, with both raw and phylogenetically corrected data. However, we find no evidence that the magnitude of asymmetry is related to the difference in evolutionary rates, consistent with previous theoretical predictions (Turelli and Moyle 2007). The agreement between our theory and empirical results suggests that mito-nuclear incompatibilities contribute to postzygotic reproductive isolation in centrarchids, as has been previously demonstrated in other taxa [e.g., Drosophila (Rand et al. 2001; Sackton et al. 2003) and intertidal copepods (Burton et al. 2006)]. In particular, mito-nuclear incompatibilities may help explain the widespread asymmetrical viabilities of reciprocal crosses.
We emphasize two caveats regarding our results. First, the node-averaging approach to reducing phylogenetic nonindependence does not guarantee full independence of the data. For example, node averages may not be independent if the same substitution(s) are involved in hybrid inviability between L. microlophus and L. cyanellus (Figure 3, node G) and between L. microlophus and M. salmoides (Figure 3, node D). At present there is no evidence that the same substitutions are involved in incompatibility across different nodes, and the epistatic nature of DMIs makes this appear unlikely. Other more formal approaches to phylogenetic correction are not applicable here because they require branch lengths (e.g., Bolnick and Near 2005), which are an explanatory variable in our study, or they are not equipped to handle characters that are properties of pairs of taxa rather than individual taxa [e.g., independent contrasts (Felsenstein 1985)].
A second caveat is that our results are correlative, as in any comparative analysis. We are not able to conclusively establish causation, because we are not able to rule out the possibility that the association we have found arises from mutual dependence on an unidentified third variable. For instance, it is conceivable that both asymmetric viability and asymmetric mitochondrial branch lengths are a result of differences in body size. We previously noted that for a given species pair, the smaller species tends to be the worse maternal parent (Bolnick and Near 2005; Bolnick et al. 2006). In the present study, we confirmed this trend with a phylogenetically corrected analysis and also found that small body size exhibits a marginally significant tendency to be associated with accelerated mitochondrial evolution. We are not, at present, able to rigorously tease apart the directions of causation in this triangle of correlations. However, we emphasize that theory provided an a priori prediction that mitochondrial rate asymmetries should give rise to viability asymmetry, whereas there was no prediction regarding body-size effects.
One possible explanation for the body-size effect is that asymmetric hybrid viability could reflect a combination of mito-nuclear and maternal–zygotic incompatibilities [or genomic imprinting (Haig 2004)]. Although our data are consistent with a systematic effect driving mito-nuclear UDMIs, this does not preclude other forms of incompatibilities acting simultaneously. Allozyme expression studies in centrarchids have previously implicated cyto-nuclear incompatibilities as a cause for asymmetry (Whitt et al. 1977; Philipp et al. 1983). When two species were crossed experimentally, F1 hybrids exhibited delayed onset of expression for a number of allozymes. When the maternally and paternally derived allozymes were electrophoretically distinguishable, it was frequently found that the maternal alleles were expressed normally but the paternal alleles were delayed, even in reciprocal crosses (Whitt et al. 1977). The simplest explanation is that maternally encoded transcription factors in the oocyte cytoplasm were successfully regulating embryonic expression of maternal alleles, but failing to regulate paternal alleles. The magnitude of delayed onset was correlated with the degree of hybrid inviability (Philipp et al. 1983). Such cytoplasmic–nuclear incompatibilities could, in principle, be exacerbated by body size differences between species. However, the most obvious reason for such an effect does not appear to hold: body size is not correlated with egg size (Bolnick et al. 2006), and egg size differences are not correlated with hybrid viability (Merriner 1971). However, evolutionary shifts in development rate and other life-history characteristics may be associated with evolutionary shifts in either maternally encoded cytoplasmic transcription factors or metabolic function, which could result in UDMIs.
The dominance theory for Haldane's rule also relies on UDMIs, because it arises from incompatibilities between a hemizygous and therefore uniparentally inherited factor (e.g., the X chromosome in males) and autosomal loci. Asymmetric instances of Haldane's rule are widespread (Laurie 1997; Presgraves and Orr 1998), but these asymmetries are restricted to the heterozygous sex. There are several reasons why we believe Haldane's rule does not contribute to asymmetric viability in centrarchids. First, there is no published evidence that reciprocal hybrid crosses produce different sex ratios or that asymmetric viabilities are sex limited (Bolnick and Near 2005). Second, although biased sex ratios are observed in some centrarchid hybrids, they cannot be explained by inviability of one sex, because hybrid hatching success remains high (>90%) in many crosses with strongly biased sex ratios (Bolnick and Near 2005). We hypothesize that sex-ratio bias may reflect a breakdown in sex determination rather than Haldane's rule (Bolnick and Near 2005). Third, there is no evidence that the centrarchid genome includes a large hemizygous region linked to sex. Karyotypic studies have identified no heteromorphic sex chromosomes (Roberts 1964). A genomic scan using AFLPs found very low levels of sex linkage (López-Fernández and Bolnick 2007). If indeed the sex-linked region of the genome is small, there may be little opportunity for hemizygous sex-linked loci to generate DMIs and asymmetric incompatibility (Turelli and Begun 1997) [but the “effective” size of the sex-linked region may be magnified by genetic conflicts over sex ratio (Tao et al. 2007a,b)].
In conclusion, our results provide the first tentative evidence that asymmetric reproductive isolation is at least partly due to systematic biases in molecular evolutionary rates. This suggests that mito-nuclear incompatibilities may have a general role in reproductive isolation among centrarchid species, as asymmetries have been found in nearly all species pairs for which reciprocal-cross data are available. This general pattern of asymmetrical F1 viabilities, termed Darwin's corollary by Turelli and Moyle (2007), remains relatively poorly understood. Elaborating on these mechanisms may help illuminate the mechanisms of speciation. Asymmetric hybrid viabilities may also have important consequences for modern taxa, because hybrid viabilities may influence the direction of introgression during hybridization. For example, asymmetric mito-nuclear incompatibilities might contribute to the widespread tendency for mitochondrial DNA to introgress asymmetrically between hybridizing populations (Wilson and Bernatchez 1998; Chan and Levin 2005).
Acknowledgments
We thank D. Rand and L. Moyle for comments. This research was supported by National Science Foundation grants IBN-0076436 to P.C.W. and DEB-0412802 to D.I.B., the University of Texas at Austin, the University of California at Davis, and Yale University.
References
- Barbash, D. A., D. F. Siino, A. M. Tarone and J. Roote, 2003. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc. Natl. Acad. Sci. USA 100 5302–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolnick, D. I., and T. J. Near, 2005. Tempo of hybrid inviability in sunfish (Centrarchidae). Evolution 59 1754–1767. [PubMed] [Google Scholar]
- Bolnick, D. I., T. J. Near and P. C. Wainwright, 2006. Body size divergence promotes post-zygotic reproductive isolation in centrarchids. Evol. Ecol. Res. 8 903–913. [Google Scholar]
- Brideau, N. J., H. A. Flores, J. Wang, S. Maheshwari, X. Wang et al., 2006. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314 1292–1295. [DOI] [PubMed] [Google Scholar]
- Burton, R. S., C. K. Ellison and J. S. Harrison, 2006. The sorry state of F2 hybrids: consequences of rapid mitochondrial DNA evolution in allopatric populations. Am. Nat. 168 S14–S24. [DOI] [PubMed] [Google Scholar]
- Chan, K. M. A., and S. A. Levin, 2005. Leaky prezygotic isolation and porous genomes: rapid introgression of maternally inherited DNA. Evolution 59 720–729. [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 1989. a Two rules of speciation, pp. 180–207 in Speciation and its Consequences, edited by D. Otte and J. A. Endler. Sinauer Associates, Sunderland, MA.
- Coyne, J. A., and H. A. Orr, 1989. b Patterns of speciation in Drosophila. Evolution 43 362–381. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 2004. Speciation. Sinauer Associates, Sunderland, MA.
- Darwin, C., 1859. On the Origin of Species. Harvard University Press, Cambridge, MA.
- De Renzis, S., O. Elemento, S. Tavazoie and E. F. Wieschaus, 2007. Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 5 1036–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettman, J. R., D. J. Jacobson, E. Turner, A. Pringle and W. J. Taylor, 2003. Reproductive isolation and phylogenetic divergence in Neurospora: comparing methods of species recognition in a model eukaryote. Evolution 57 2721–2741. [DOI] [PubMed] [Google Scholar]
- Dobzhansky, T., 1934. Studies on hybrid sterility. I. Spermatogenesis in pure and hybrid Drosophila pseudoobscura. Proc. Natl. Acad. Sci. USA 19 397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J., 1985. Phylogenies and the comparative method. Am. Nat. 125 1–15. [Google Scholar]
- Fitzpatrick, B. M., 2002. Molecular correlates of reproductive isolation. Evolution 56 191–198. [DOI] [PubMed] [Google Scholar]
- Gallant, S. L., and D. J. Fairbain, 1997. Patterns of postmating reproductive isolation in a newly discovered species pair, Aquarius remigis and A. remigoides (Hemiptera; Gerridae). Heredity 78 571–577. [DOI] [PubMed] [Google Scholar]
- Haig, D., 2004. Genomic imprinting and kinship: How good is the evidence? Annu. Rev. Genet. 38 553–585. [DOI] [PubMed] [Google Scholar]
- Haldane, J. B. S., 1922. Sex ratio and unisexual sterility in animal hybrids. J. Genet. 12 101–109. [Google Scholar]
- Harrison, R. G., 1983. Barriers to gene exchange between closely related cricket species. I. Laboratory hybridization studies. Evolution 37 245–251. [DOI] [PubMed] [Google Scholar]
- Harrison, J. S., and R. S. Burton, 2006. Tracing hybrid incompatibilities to single amino acid substitutions. Mol. Biol. Evol. 23 559–564. [DOI] [PubMed] [Google Scholar]
- Kenyon, L., and T. Moraes Carlos, 1997. Expanding the functional human mitochondrial DNA database by the establishment of primate xenomitochondrial cybrids. Proc. Natl. Acad. Sci. USA 94 9131–9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie, C. C., 1997. The weaker sex is heterogametic: 75 years of Haldane's rule. Genetics 147 937–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Fernández, H., and D. I. Bolnick, 2007. What causes partial F1 hybrid viability? Incomplete penetrance versus genetic variation. PLoS One 2 e1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merriner, J. V., 1971. Development of intergeneric centrarchid hybrid embryos. Trans. Am. Fish. Soc. 100 611–618. [Google Scholar]
- Muller, H. J., 1939. Reversibility in evolution considered from the standpoint of genetics. Biol. Rev. Camb. Philos. Soc. 14 261–280. [Google Scholar]
- Muller, H. J., 1942. Isolating mechanisms, evolution and temperature. Biol. Symp. 6 71–125. [Google Scholar]
- Navajas, M., A. Tsagkarov, J. Lagnel and M. J. Perrot-Minnot, 2000. Genetic differentiation in Tetranychus urticae (Acari: Tetranychidae): Polymorphism, host races or sibling species? Exp. Appl. Acarol. 24 365–376. [DOI] [PubMed] [Google Scholar]
- Near, T. J., D. I. Bolnick and P. C. Wainwright, 2004. Investigating phylogenetic relationships of the Centrarchidae (Actinopterygii: Perciformes) using DNA sequences from mitochondrial and nuclear genes. Mol. Phylogenet. Evol. 32 344–357. [DOI] [PubMed] [Google Scholar]
- Near, T. J., D. I. Bolnick and P. C. Wainwright, 2005. Fossil calibrations and molecular divergence time estimates in centrarchid fishes (Teleostei: Centrarchidae). Evolution 59 1768–1782. [PubMed] [Google Scholar]
- Noor, M. A. F., and J. L. Feder, 2006. Speciation genetics: evolving approaches. Nat. Rev. Genet. 7 851–861. [DOI] [PubMed] [Google Scholar]
- Oliver, C. G., 1978. Experimental hybridization between the nymphalid butterflies Phyciodes tharos and P. campestris montana. Evolution 32 594–601. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 1993. A mathematical model of Haldane's rule. Evolution 47 1606–1611. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 1995. The population genetics of speciation: the evolution of hybrid incompatibilities. Genetics 139 1805–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., and D. C. Presgraves, 2000. Speciation by postzygotic isolation: forces, genes, and molecules. BioEssays 22 1085–1094. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., and M. Turelli, 2001. The evolution of postzygotic isolation: accumulating Dobzhansky-Muller incompatibilities. Evolution 55 1085–1094. [DOI] [PubMed] [Google Scholar]
- Philipp, D. P., H. R. Parker and G. S. Whitt, 1983. Evolution of gene regulation: isozymic analysis of patterns of gene expression during hybrid fish development. Genet. Evol. 10 193–237. [PubMed] [Google Scholar]
- Posada, D., and K. A. Crandall, 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14 817–818. [DOI] [PubMed] [Google Scholar]
- Presgraves, D. C., 2002. Patterns of postzygotic isolation in Lepidoptera. Evolution 56 1168–1183. [DOI] [PubMed] [Google Scholar]
- Presgraves, D. C., 2003. A fine-scale genetic analysis of hybrid incompatibilities in Drosophila. Genetics 163 955–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves, D. C., and H. A. Orr, 1998. Haldane's rule in taxa lacking hemizygous X. Science 282 952–954. [DOI] [PubMed] [Google Scholar]
- Presgraves, D. C., L. Balagopalan, S. M. Abmayr and H. A. Orr, 2003. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature 423 715–719. [DOI] [PubMed] [Google Scholar]
- Rakocinski, C. F., 1984. Aspects of reproductive isolation between Campostoma oligolepis and Campostoma anomalum pullum (Cypriniformes: Cyprinidae) in northern Illinois. Am. Midland Nat. 112 138–145. [Google Scholar]
- Rand, D. M., A. G. Clark and L. M. Kann, 2001. Sexually antagonistic cytonuclear fitness interactions in Drosophila melanogaster. Genetics 159 173–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand, D. M., R. A. Haney and A. J. Fry, 2004. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol. Evol. 19 645–653. [DOI] [PubMed] [Google Scholar]
- Rawson, P. D., and R. S. Burton, 2002. Functional coadaptation between cytochrome c and cytochrome c oxidase within allopatric populations of a marine copepod. Proc. Natl. Acad. Sci. USA 99 12955–12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, F. L., 1964. A chromosome study of twenty species of Centrarchidae. J. Morphol. 115 401–418. [DOI] [PubMed] [Google Scholar]
- Sackton, T. B., R. A. Haney and D. M. Rand, 2003. Cytonuclear coadaptation in Drosophila: disruption of cytochrome c oxidase activity in backcross genotypes. Evolution 57 2315–2325. [DOI] [PubMed] [Google Scholar]
- Schartl, M., U. Hornung, H. Gutbrod, J.-N. Volff and J. Whitbrodt, 1999. Melanoma loss-of-function mutants in xiphophorus caused by Xmrk-oncogene deletion and gene disruption by a transposable element. Genetics 153 1385–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant, A. H., 1920. Genetic studies on Drosophila simulans. I. Introduction. Hybrids with Drosophila melanogaster. Genetics 5 488–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D. L., 2002. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. Sinauer, Sunderland, MA.
- Tao, Y., J. P. Masly, L. Araripe, Y. Ke and D. L. Hartl, 2007. a A sex-ratio meiotic drive system in Drosophila simulans. I. An autosomal suppressor. PLoS Biol. 5 e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Y., L. Araripe, S. B. Kingan, Y. Ke, H. Xiao et al., 2007. b A sex-ratio meiotic drive system in Drosophila simulans. II. An X-linked distorter. PLoS Biol. 5 e293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton, W. A., 1955. Interspecific hybridization in Bufo woodhousei and Bufo valliceps. Evolution 9 455–468. [Google Scholar]
- Tiffin, P., M. S. Olson and L. C. Moyle, 2001. Asymmetrical crossing barriers in angiosperms. Proc. R. Soc. Lond. 268 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting, C.-T., S.-C. Tsaur, M.-L. Wu and C. I. Wu, 1998. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science 282 1501–1504. [DOI] [PubMed] [Google Scholar]
- Turelli, M., 1998. The causes of Haldane's rule. Science 282 889–891. [DOI] [PubMed] [Google Scholar]
- Turelli, M., and D. J. Begun, 1997. Haldane's rule and X-chromosome size in Drosophila. Genetics 147 1799–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli, M., and L. C. Moyle, 2007. Asymmetric postmating isolation: Darwin's corollary to Haldane's rule. Genetics 176 1059–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli, M., and H. A. Orr, 1995. The dominance theory of Haldane's rule. Genetics 140 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli, M., and H. A. Orr, 2000. Dominance, epistasis, and the genetics of postzygotic isolation. Genetics 154 1663–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, J. J., 2004. Accumulating Dobzhansky-Muller incompatibilities: reconciling theory and data. Evolution 58 1145–1156. [DOI] [PubMed] [Google Scholar]
- West, J. L., and F. E. Hester, 1966. Intergeneric hybridization of centrarchids. Trans. Am. Fish. Soc. 95 280–288. [Google Scholar]
- Whitt, G. S., D. P. Philipp and W. F. Childers, 1977. Aberrant gene expression during the development of hybrid sunfishes (Perciformes, Teleosti). Differentiation 9 97–109. [DOI] [PubMed] [Google Scholar]
- Willett, C. S., and R. S. Burton, 2001. Viability of cytochrome C genotypes depends on cytoplasmic backgrounds in Tigriopus californicus. Evolution 55 1592–1599. [DOI] [PubMed] [Google Scholar]
- Wilson, C. C., and L. Bernatchez, 1998. The ghost of hybrids past: fixation of arctic charr (Salvelinus alpinus) mitochondrial DNA in an introgressed population of lake trout (S. namaycush). Mol. Ecol. 7 127–132. [Google Scholar]
- Wolfram, S., 2003. The Mathematica Book, Ed. 5. Wolfram Media, Champaign, IL.
- Wu, C.-I., and A. W. Davis, 1983. Evolution of post-mating reproductive isolation - the composite nature of Haldane's rule and its genetic bases. Am. Nat. 142 187–212. [DOI] [PubMed] [Google Scholar]