Abstract
RNA interference (RNAi) mechanisms are conserved and consist of an interrelated network of activities that not only respond to exogenous dsRNA, but also perform endogenous functions required in the fine tuning of gene expression and in maintaining genome integrity. Not surprisingly, RNAi functions have widespread influences on cellular function and organismal development. Previously, we observed a reduced capacity to mount an RNAi response in nine Caenorhabditis elegans mutants that are defective in ABC transporter genes (ABCRNAi mutants). Here, we report an exhaustive study of mutants, collectively defective in 49 different ABC transporter genes, that allowed for the categorization of one additional transporter into the ABCRNAi gene class. Genetic complementation tests reveal functions for ABCRNAi transporters in the mut-7/rde-2 branch of the RNAi pathway. These second-site noncomplementation interactions suggest that ABCRNAi proteins and MUT-7/RDE-2 function together in parallel pathways and/or as multiprotein complexes. Like mut-7 and rde-2, some ABCRNAi mutants display transposon silencing defects. Finally, our analyses reveal a genetic interaction network of ABCRNAi gene function with respect to this part of the RNAi pathway. From our results, we speculate that the coordinated activities of ABCRNAi transporters, through their effects on endogenous RNAi-related mechanisms, ultimately affect chromosome function and integrity.
WHILE the exploitation of cellular RNA interference (RNAi) mechanisms to target specific sequences for silencing has facilitated analysis of gene function, we have only an incomplete description of the endogenous functions that RNAi mechanisms afford to cells. In Caenorhabditis elegans, the products of the mut-7 and rde-2 genes are required not only for a fully effective RNAi response to dsRNA, but also for endogenous processes that ultimately affect chromosome and micro-RNA functions. In particular, mut-7 and rde-2 mutants display (a) an increased rate of nondisjunction (Him phenotype), (b) aberrant germ-line expression of sequences from a repetitive transgene array in situations where expression is normally silenced, (c) a reduced ability for transgene-induced cosuppression of endogenous genes, (d) a high rate of transposon mobilization from chromosomal sites in the mutant germ line, a tissue that normally represses mobilization of transposons, and (e) defects in the function of endogenous noncoding RNAs (Tabara et al. 1999; Dernburg et al. 2000; Tops et al. 2005; Lee et al. 2006). The mut-7 gene encodes a protein with an exonuclease domain (Ketting et al. 1999). rde-2 encodes a protein with no discernible motifs and little similarity to proteins outside nematodes (Tops et al. 2005). Previous reports indicate both a cytoplasmic and a nuclear localization for MUT-7, whereas RDE-2 is apparently localized exclusively to the cytoplasm where it and MUT-7 reside within a larger protein complex (Tops et al. 2005). Precise roles for MUT-7 and RDE-2 in RNAi and endogenous processes have not been established.
We previously reported a role for C. elegans ATP-binding cassette (ABC) proteins in RNAi, with emphasis on the analysis of one member of this family, haf-6. We also identified an additional eight transporter genes that are similarly required for RNAi: abt-1, pgp-4, pgp-11, haf-2, mrp-1, pmp-1, wht-1(C05D10.3), and wht-3(C16C10.12) (Sundaram et al. 2006). These initial assays were limited in number and were restricted to observations of RNAi activities in germ line and muscle. However, the genome of C. elegans harbors 61 different ABC transporter genes that are collectively expressed in a wide array of tissues (Zhao et al. 2004). Thus, we considered that the narrow scope of our initial set of assays might have limited our ability to identify the complete set of ABCRNAi genes—in particular, those ABC transporter mutants that might have somatic RNAi defects in nonmuscle tissue.
Here we set out to identify additional C. elegans ABC transporters that could be classified as ABCRNAi genes by a more thorough assessment of RNAi activities in all available ABC transporter mutant strains. Of the 61 ABC transporter genes, mutations in 49 different genes are represented. We tested each mutant strain for RNAi defects using additional assays, and we performed complementation testing between nonallelic mutants. Our complementation testing utilized ABC transporter mutants and mutants defective for mut-7 or rde-2. Because deletion alleles were used in each of the complementation tests, any noncomplementation interactions that are observed likely represent cases of combined haplo-insufficiency. Second-site noncomplementation (SSNC) is often an indicator of physical interactions between proteins (Stearns and Botstein 1988; Campbell et al. 1995; Haarer et al. 2007); in other cases the interactions reveal functional requirements from multiple pathways that contribute to a given process (Yook et al. 2001; Jaspersen et al. 2004). In either case, a noncomplementation result implies a common function or influence on a particular process (Hawley and Gilliland 2006). Our results allowed us to classify an additional gene, haf-9, as an ABCRNAi gene. We also reveal common and distinct features related to RNAi defects for each of the mutant strains.
ABC transporters are classified into subfamilies on the basis of relative homologies, as well as the number and positioning of conserved transmembrane and ABC domains. The ABCRNAi subset of transporter genes consists of diverse subfamilies (A, B, C, D, and G), and both full transporter and half molecules are represented in this group. A genetic interaction with mut-7 and rde-2 is a unifying feature of this diverse group of genes; thus, the ABCRNAi subset can be defined on the basis of function, rather than subfamily classification. The genetic interactions with mut-7 and rde-2, as well as other common phenotypes, point to an ability for ABCRNAi transporters to influence RNAi mechanisms that ultimately affect chromosome function and integrity.
MATERIALS AND METHODS
Strains used in complementation tests with haf-6(ne335):
XX47 [(ne309)], WM27 [rde-1(ne219) V], XX52 [rsd-2(ne319) IV], WM30 [mut-2(ne298) I], PD8356 [rde-1(ne300) V], XX51 [sid-1(ne318)], NL917 [mut-7(pk204) III], NL3531 [rde-2(pk1657) I], XX50 [sid-1(ne316)], NL1820 [mut-7(pk720) III], WM29 [rde-2(ne221) I], IG10 [tol-1(nr2033) I], PD5085 [ppw-1(pk2505)], PD8221 [rde-4(ne299) III], CB4043 [smg-2 (e2008) I; him-5(e1490) V].
Mutants used in phenotypic analyses:
VC1046 [abce-1(gk481)], VC712 [mrp-4(ok1095) X], VC561 [abcf-1(ok830) V], GH534 [mrp-4(cd8) X], VC544 [abcf-2(ok771) III], RB1070 [mrp-6(ok1027) X] RB1645 [abch-1(ok2034) II], RB1269 [mrp-8(ok1360) III], VC572 [abcx-1(ok865) II], NL132 [pgp-1(pk17) IV], FX703 [abt-1(tm703) IV], VC134 [pgp-2(gk114) I], RB842 [abt-2((ok669) I], NL131 [pgp-3(pk18) X], FX502 [abt-2(tm502) I], VC21 [pgp-4(gk16) X], FX1050 [abt-4(tm1050) V], RB959 [pgp-5(ok856) X], RB817 [abt-4(ok633) V], FX663 [pgp-5(tm663) X], RB1765 [abt-5(ok2268) I], RB1047 [pgp-6 (ok994) X], RB1151 [cft-1(ok1180) V], VC286 [pgp-7(ok528)/szT1;+/szT1 I], MT4983 [ced-7(n1996) III], FX830 [pgp-9(tm830) V], MT4984 [ced-7(n1997 III], FX996 [pgp-10(tm996) X], MT8886 [ced-7(n2024) III], RB1045 [pgp-10(ok991) X], FX843 [haf-1(tm843) IV], FX333 [pgp-11(tm333) II], RB867 [haf-1(ok705) IV], VC26 [pgp-12(gk19) X], VC16 [haf-2(gk13) II], RB894 [pgp-13(ok747) X], VC1186 [haf-3(gk549) V], RB1041 [pgp-15(ok987) X], RB1107 [haf-3(ok1086) V], RB908 [pmp-1(ok773) II], VC449 [haf-4(gk240) I], RB1108[pmp-3(ok1087) V], RB1080 [haf-4(ok1042) I], FX968 [pmp-3(tm968) V], VC252 [haf-5(gk155) III], RB675 [pmp-4(ok396) IV], VC287 [haf-5(gk161) III], VC189 [pmp-4(ok396) IV], XX193 [haf-6(ne335) I; him-5(e1490) V], XX194 [haf-6(ne335) I; ccIs8160(rpL28∷GFP); him-5(e1490) V], XX195 [haf-6(ne335) I; cIs8160(rpL28∷GFP)], XX364 [haf-6(ne335) I], FX688 [wht-1(tm688) III], VC118 [haf-7(gk46) V], RB1007 [wht-3(ok927) III], VC15 [haf-8(gk12) IV], RB1033[wht-3(ok962) III], VC32 [haf-9(gk23) I], FX714 [wht-3(tm714) III], NL147 [mrp-1(pk89) X], RB1058 [wht-4(ok1007) II], NL147[mrp-1(ut153) X], RB932 [wht-5(ok806) IV], RB1713 [mrp-2(ok2157) X], VC629 [wht-6(ok882) III], RB1028 [mrp-3(ok955) X], RB933 [wht-7(ok812) III], VC692 [wht-9(ok1044) III].
Other strains, outcrossing, and DNA sequence verification:
To help prevent or monitor for accidental introduction of RNAi-related mutations that may be present in the background of wild-type N2 populations, we maintain wild-type strains as subtypes. These subtypes have passed our tests for brood size and sensitivity to pop-1 and unc-22 foods and lack temperature-sensitive Sterility or Him phenotypes. Many of the phenotypes here were obtained using deletion alleles, and in most cases recapitulating the effects of the mutation using RNAi is not a comparable methodology. For example, in our complementation tests, the animals tested are trans-heterozygotes and RNAi would likely induce a null phenotype; therefore, the null mutations and RNAi would produce dissimilar genetic conditions. We repeated experiments using more than one mutant subtype; these were generated by outcrossing the mutant strains to different N2 subtypes. Each N2 subtype, and the corresponding mutant subtype, would be expected to harbor different sets of background mutations.
XX1181 [mut-7(pk204) III; dpy-4(e1166) unc-17(e245) IV] was generated from crosses of PD2027 [dpy-4(e1166) unc-17(e245) IV] and NL917 [mut-7(pk204) III]. XX1187 [rde-2(ne221) I; dpy-4(e1166) unc-17(e245) IV] was generated from crosses of PD2027 [dpy-4(e1166) unc-17(e245) IV] and WM29 [rde-2(ne221) I]. XX1569 and XX1570 [mut-7(pk204) III] are independent F2 sibling subtypes from a cross of XX1181 and NL917. XX1571-3 [rde-2(ne221) I] subtypes were similarly derived from crosses of XX1187 and WM29.
Strains XX1824–1826 harbor the mut-7(pk201) mutation. XX1824 [mut-7(pk201); yyEx62.2-rde-2 cosmid] was derived from a cross between XX1570 mutants and XX506 transgenic animals. XX1825 [mut-7(pk201); yyEx75.1-haf-6 cDNA (G562R)] was derived from a cross using XX1570 and XX752. XX1826 [mut-7(pk201); yyEx97.4-haf-6 cDNA (wild type)] was derived from a cross using XX1570 and XX1318. Strains XX1827–1829 harbor the rde-2(pk1657) mutation. XX1827 [rde-2(pk1657); yyEx62.2-rde-2 cosmid] was derived from a cross between WM29 mutants and XX506 transgenic animals. XX1828 [rde-2(pk1657); yyEx75.1-haf-6 cDNA (G562R)] was derived from a cross using WM29 and XX752. XX1829 [rde-2(pk1657); yyEx97.4-haf-6 cDNA (wild type)] was derived from a cross using WM29 and XX1318.
NL3643 [unc-22(st136) IV] was outcrossed to wild-type males (XX171, an N2 strain from the lab of Andrew Fire) five times before it was used in additional crosses. The unc-22(st136) allele harbors a transposon insertion, and this outcrossed strain (XX366) has not displayed Mutator activity under any growth condition.
The ABCRNAi mutants identified previously [abt-1(tm703), haf-2(gk13), haf-6(ne335), mrp-1(pk89), pgp-4(gk16), pgp-11(tm333), pmp-1(ok773), wht-1/C05D10.3(tm688), wht-3/C16C10.12(ok962), and the newly identified haf-9(gk23)] were tested for RNAi activity as unoutcrossed lines and again after three rounds of outcrossing to wild-type males (XX171). Outcrossed lines were established from cloned individuals, and extensive PCR analyses were performed on the cloned adult and on the ensuing population to ensure homozygosity for each deletion. Most strains that displayed noncomplementing interactions were subjected to additional rounds of outcrossing using wild-type XX937 males (a selected subtype from an N2 stock obtained from the Caenorhabditis Genetics Center, DR subclone of CB origin) or XX366, a transposon-insertion line that does not display Mutator activity.
We sequenced DNA prepared from each of the haf-6, rde-2, and mut-7 noncomplementing strains, and in each mutant we verified that the other two genes were wild type in sequence. DNA sequences were obtained from genomic DNA as well as from RT–PCR products. cDNAs were easily generated, indicating gene expression, and the splicing patterns were as expected. (Both rde-2 mRNA isoforms were observed in all mutant strains.) We sequenced an rde-2 cDNA 1737 bp in length from the following strains: XX194 [haf-6(ne335) I; him-5(e1490) V], NL917 [(mut-7(pk201) III] (260 bp from the middle of this cDNA were not read), and NL1820 [mut-7(pk720) III]. We sequenced a haf-6 cDNA 2000 bp in length from XX1569 [mut-7(pk204) III], XX1571 [rde-2(ne221) I], NL1820 [mut-7(pk720) III], and NL3531 [rde-2(pk1657) I]. A total of 2943 bp of mut-7 cDNA were sequenced in XX194 [haf-6(ne335) I; him-5(e1490) V], XX1571 [rde-2(ne221) I], and NL3531 [rde-2(pk1657) I]. Sequence information from genomic DNA isolated from each mutant strain validated our results derived from cDNAs and provided information in gaps.
Feeding strains and feeding-based assays for RNAi defects:
Feeding plates harboring bacteria engineered to express dsRNA were prepared using the HT115(DE3) host bacteria as described (Timmons et al. 2001; Hull and Timmons 2004; Sundaram et al. 2006). Plasmids transformed into this bacterial strain were derived from L4440 vector (Timmons and Fire 1998). pop-1 food targets the TCF/LEF1 transcription factor and produces sterility in young animals reared on this food; unc-22 food (Timmons and Fire 1998) targets a gene expressed in muscle and induces a Twitching phenotype. Other feeding strains were obtained from the MRC Geneservice RNAi library (Kamath et al. 2003). Male stocks were established for each mutant strain by rearing animals at 20° on feeding strains with plasmids targeting him genes. mut-7 and rde-2 animals are Him, and we observed that mutant males sire progeny only when the males arose by means unrelated to the mut-7/rde-2 defect. When rde-2 or mut-7 males are produced via RNAi phenocopy of Him pathway genes, matings can be successful. RNAi assays were performed as described (Timmons et al. 2001; Hull and Timmons 2004; Sundaram et al. 2006) at 15°, 20°, and 25°.
Generation of transgene lines and transgene-based rescue of RNAi:
Transgene lines were established as complex arrays to better ensure expression in the germ line (Kelly et al. 1997). Plasmid DNA sequences were linearized before injecting. unc-25∷gfp sequences were used as a dominant transformation marker; bacteriophage lambda DNA was used as carrier DNA. Cosmid F21C3 harbors wild-type rde-2 genomic sequence and provides for rescue of the RNAi defects in rde-2 mutants. Plasmid pLT417 harbors let-858 promoter∷let-858 5′-UTR∷haf-6 cDNA∷let-858 3′-UTR sequences. The haf-6 coding region in pLT417 harbors a G562R substitution that affects a conserved amino acid in the ATP-binding cassette domain, and this plasmid provides partial rescue of RNAi defects in haf-6 mutants. Plasmid pLT372 is identical to pLT417 except that the haf-6 cDNA sequence is wild type; this plasmid rescues the RNAi defects in haf-6 mutants (Sundaram et al. 2006). Transgene arrays were generated in wild type and crossed into mutant backgrounds. Strains were verified for homozygosity by RNAi behavior (of nontransgenic siblings), temperature-sensitive sterility phenotypes, PCR, and/or DNA sequencing, as appropriate to the strain. RNAi activity was assessed by placing animals as L2/L3 larvae on pop-1 food. Absence of live progeny is indicative of an intact RNAi response.
Complementation testing:
Conclusions from our complementation tests were based on two or more independent crosses that were tested using different preparations of pop-1 feeding plates. Crosses were performed using one hermaphrodite per mating on standard OP50-seeded plates with no obvious contamination. The presence of 50% males in the progeny indicated a successful cross. RNAi activity was assessed by noting the number of F2 progeny produced per F1 animal placed on pop-1 food. The assays were performed under identical conditions in an incubator set at 20°. We did not assess RNAi activity in trans-heterozygous males. In Figure 2A, mut-7 and rde-2 mutants were used as the hermaphrodite parent with haf-6 males to generate trans-heterozygotes; reciprocal crosses with haf-6 as parental hermaphrodite produced progeny with normal RNAi activity. Complementation testing was performed using all combinations of rde-2(ne221), mut-7(pk204) (both harbor point mutations that result in a premature stop codon and a S812L substitution, respectively), rde-2(pk1657), and mut-7(pk720) (both harbor large deletions). In Tables 2 and 3, conclusions were drawn using data from at least two sets of independent genetic crosses and the progeny from each cross was tested on different preparations of pop-1 food. The F1 cross-progeny were placed as L2/L3 larvae on plates and the number of F2 animals per F1 adult was tabulated. We used the following classification scheme in our assessments: −, 0–5 F2's per F1 trans-heterozygote (indicative of wild-type RNAi activity); w, 6–20 F2's per F1; +, 21–50 F2's per F1; and ++, >50 F2's per F1 (indicative of strong RNAi defects). Trans-heterozygous combinations that displayed − and w results were considered complementary (C); combinations that produced + and ++ results were considered noncomplementary (NC); allele sets that produced + results were considered weak (W); and sets that displayed wide experiment-to-experiment variations (− and ++) were labeled inconsistent (I). Strains that displayed noncomplementation were subjected to three rounds of additional outcrossing and were retested for RNAi defects. A weak result could reflect a maternally supplied protein with a short half-life in the germ line. Alternatively, a weak RNAi defect might indicate that the paternal gene is transcriptionally activated relatively late in the critical period. Such conditions may also contribute to inconsistent results as well.
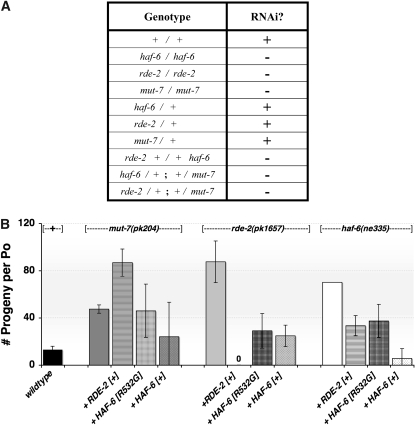
Figure 2.—
haf-6, mut-7, and rde-2 strains constitute a noncomplementing set of mutants with RNAi defects. (A) Animals with the genotype indicated in the left column were placed on pop-1 food as described (materials and methods). The number of progeny produced from each animal was tabulated. + denotes RNAi activity (the average number of progeny produced ranged from 0 to 7 per adult animal in this experiment); − denotes an RNAi defect or lack of response to pop-1 food (an average of 83–154 progeny per animal was observed). mut-7 and rde-2 mutants were the hermaphrodite parent in each of these experiments. Similar noncomplementation results were observed using strains harboring deletions in mut-7 and rde-2 in comparison to strains harboring point mutations. (B) RNAi defects are cross-complemented using transgenes. RNAi activity was assessed using pop-1 food. The animals were homozygous for the mutation at the top of the graph; the x-axis label indicates the additional sequences that were provided by transgenes. Wild-type RDE-2[+] protein was provided by cosmid F21C3. HAF-6[R532G] protein was provided by plasmid pLT417; this plasmid provides incomplete rescue of the RNAi defects in haf-6 mutants. Wild-type HAF-6 protein is expressed from pLT372, which harbors a haf-6 cDNA (Sundaram et al. 2006). Bars represent an average of three experiments; error bars indicate SD.
TABLE 2.
A genetic interaction with mut-7 or rde-2 is not a general feature of ABC transporter mutants
| Complementation with
|
|||
|---|---|---|---|
| Subfamily | Mutation | mut-7 | rde-2 |
| E | abce-1(gk481) | No males | |
| F | abcf-1(ok830) | No males | |
| F | abcf-2 (ok771) | C | C |
| H | abch-1(ok2034) | C | C |
| H | abcx-1(ok865) | C | C |
| A | abt-2(ok669) | C | C |
| abt-2(tm502) | C | C | |
| A | abt-4(ok633) | C | C |
| abt-4(tm1050) | C | C | |
| A | abt-5(ok2268) | C | C |
| A | ced-7(n1996) | C | C |
| ced-7(n1997) | C | C | |
| ced-7(n2094) | C | C | |
| C | cft-1(ok1180) | I | I |
| B | haf-1(ok705) | C | C |
| haf-1(tm843) | C | C | |
| B | haf-3 (ok1086) | C | C |
| haf-3 (gk549) | C | C | |
| B | haf-4(gk240) | I | I |
| haf-4(ok1042) | C | C | |
| B | haf-5(gk161) | C | C |
| haf-5(gk155) | C | C | |
| B | haf-7(gk46) | C | C |
| B | haf-8(gk12) | C | C |
| C | mrp-2(ok2157) | C | C |
| C | mrp-3(ok995) | C | C |
| C | mrp-4 (ok1095) | C | C |
| C | mrp-4 (cd8) | C | C |
| C | mrp-6(ok1027) | C | C |
| C | mrp-8(ok1360) | C | C |
| B | pgp-1(pk17) | C | C |
| B | pgp-2(gk114) | C | C |
| B | pgp-3(pk18) | C | C |
| B | pgp-5(ok856) | C | C |
| pgp-5(tm663) | C | C | |
| B | pgp-6(ok994) | C | C |
| B | pgp-7(ok528) | C | C |
| B | pgp-9(tm830) | C | C |
| B | pgp-10(ok991) | C | C |
| pgp-10(tm996) | C | C | |
| B | pgp-12(gk19) | C | C |
| B | pgp-13(ok747) | C | C |
| B | pgp-15(ok987) | C | C |
| D | pmp-3(ok1087) | C | C |
| pmp-3(tm968) | C | C | |
| D | pmp-4(ok396) | C | C |
| G | wht-3(ok927) | C | C |
| wht-3(tm714) | C | C | |
| G | wht-4(ok1007) | C | C |
| G | wht-5(ok806) | C | C |
| G | wht-6(ok882) | C | C |
| G | wht-7(ok812) | C | C |
| G | wht-9(ok1044) | C | C |
| Other RNAi-related mutants | |||
| K08H10.7 | rde-1(ne219) | C | C |
| T20G5.11 | rde-4(ne299) | C | C |
| C04F5.1 | sid-1(ne318) | C | C |
| F52G2.2 | rsd-2(ne319) | C | C |
| F16D3.2 | rsd-6(pk2011) | C | C |
Complementation tests were performed using mut-7(pk204) or rde-2(ne221) strains as the hermaphrodite parent; however, the reciprocal cross was performed for abce-1 and abcf-1 as we were unable to generate male stocks from these mutants. trans-heterozygous progeny were tested for RNAi activity using pop-1 food. False positives were observed in cft-1 mutants due to contamination on the plates. C, complementing alleles; NC, noncomplementing alleles; W, weakly complementing alleles; I, alleles that inconsistently complement (see materials and methods for a description of the scoring system and supplemental Table 3 at http://www.genetics.org/supplemental/ for data).
TABLE 3.
RNAi defects in cross-progeny
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ♂ | abt-1 | haf-2 | haf-6 | haf-9 | mrp-1 | pgp-4 | pgp-11 | pmp-1 | wht-1 | wht-3 | rde-2 | mut-7 | |
| A | abt-1 | NC | I | C | C | C | C | C | C | C | NC | NC | NC |
| B | haf-2 | C | NC | C | C | C | C | C | C | C | C | NC | NC |
| B | haf-6 | C | C | NC | C | C | I | C | C | C | C | NC | NC |
| B | haf-9 | C | C | C | NA | Ia | C | C | C | C | C | NC | NC |
| C | mrp-1 | C | I | C | C | NC | C | Ia | C | C | Ia | NC | NC |
| B | pgp-4 | C | I | I | C | C | NC | I | I | I | Ia | NCa | NCa |
| B | pgp-11 | C | I | NC | C | NC | NC | NC | C | C | NC | NC | NC |
| D | pmp-1 | C | C | C | C | C | I | C | NC | I | I | NC | NC |
| G | wht-1 | C | C | C | C | C | C | C | C | NC | C | NC | NC |
| G | wht-3 | Ia | NC | C | C | I | C | C | C | C | NC | NC | NC |
| – | rde-2 | I | C | C | C | C | W | C | C | C | I | NC | NC |
| – | mut-7 | I | C | C | C | C | C | C | C | C | W | NC | NC |
All ABC transporter genes in the ABCRNAi category display genetic interactions with mut-7 and rde-2. Reciprocal crosses were performed for each set of mutants; cross-progeny were tested for RNAi activity using pop-1 food. Column headings show the homozygous mutation from the hermaphrodite parent; row headings indicate the parental male genotype; entries in table body show complementation results observed in trans-heterozygous progeny. C, complementing alleles; NC, noncomplementing alleles; W, weakly complementing alleles; I, alleles that inconsistently complement. Animals homozygous for haf-9 are not RNAi defective.
Noncomplementation was more obvious after further outcrosses of stocks (see Table 2 legend and materials and methods for description of scoring and supplemental Table 2 for data).
RESULTS
Forty of 49 C.elegans ABC transporter mutants tested are not RNAi defective:
The C. elegans genome harbors 61 ABC transporter genes. Previously, we identified nine ABC transporter mutants that are defective in RNAi (Sundaram et al. 2006). In these earlier experiments, bacterial strains were used as a dsRNA delivery vehicle to deliver pop-1 dsRNA or unc-22 dsRNA to animals. When ingested by C. elegans, these bacterial strains can induce RNAi responses. We refer to this methodology as a “feeding” protocol and to the specific reagents as “pop-1 food” or “unc-22 food.” The pop-1 gene encodes a Wnt-responsive Tcf/Lef-related protein with an HMG box and is required for germ-line development; the unc-22 gene encodes a large extracellular matrix protein required for muscle function. Ingestion of pop-1 food elicits sterility in wild-type animals, and unc-22 food elicits a twitching phenocopy. In contrast, RNAi-defective mutants that ingest either of these foods produce progeny and have normal movements.
In earlier experiments, most of the ABC transporter mutants we tested displayed normal RNAi activities, except for the nine mutants we now refer to as ABCRNAi mutants. The fact that only about one-sixth of the ABC transporter genes, and not all ABC transporter genes, are required for efficient RNAi leads one to speculate that there is some degree of specificity with regard to how ABC transporters might affect RNAi function. The goal of the experiments reported here is to address the question of specificity by determining if additional ABC transporters are required for efficient RNAi, functions that may have been missed in previous examinations. Because the RNAi defects in ABC transporter mutants were revealed using feeding protocols, and because RNAi feeding protocols have some degree of experiment-to-experiment variability, we initially retested all available ABC transporter mutants. We first simply repeated our previous observations using pop-1 and unc-22 foods. All tests were again conducted at three different temperatures (15°, 20°, and 25°) to uncover potential heat or cold sensitivities, and the experiments were performed on the original strains as well as strains that had been subjected to additional outcrossings in our lab (see materials and methods). The results obtained did not differ from our previous reports (Sundaram et al. 2006 and data not shown).
We next utilized a more comprehensive set of feeding strains, targeting a wider array of genes expressed mostly in the soma (Table 1). Again, this more comprehensive set of assays did not reveal additional ABCRNAi mutants; RNAi activity was observed in all of the strains except those already designated as ABCRNAi (data not shown). Because all the ABCRNAi transporter mutants display RNAi defects when bacterial feeding strains are used to target genes expressed in the germ line, this broader set of data allows us to speculate that ABCRNAi transporter functions are particularly required in the germ line.
TABLE 1.
ABC transporter mutants defective in 40 different genes are not defective in RNAi
| Gene targeted | Expected expression pattern | Observed RNAi phenocopy |
|---|---|---|
| acn-1 | Hypodermis, seam cells, excretory gland | Sterile and Unc, the few surviving progeny die as embryos or larvae |
| (Brooks et al. 2003; Frand et al. 2005) | ||
| bar-1 | Vulval precursor cells, seam cells, somatic gonad, P12 | Ruptured vulva, egg-laying defective, Protruding vulva |
| (Eisenmann et al. 1998; Natarajan et al. 2004) | ||
| bli-3 | Peripheral to muscle bundles, hypodermal cells | Sterile, treated animals and surviving progeny display blistered cuticles |
| (Fraser et al. 2000; Frand et al. 2005) | ||
| ceh-13 | A, D, E, MS lineages | Small, Unc, larval arrest |
| (Wittmann et al. 1997; Brunschwig et al. 1999; Streit et al. 2002) | ||
| cks-1(dom-6) | Germ line, embryo | Embryonic lethality |
| (Polinko and Strome 2000) | ||
| pop-1 | Germ line, embryo, vulval precursor cells, Q neuroblasts, hypodermis | Sterility |
| (Lin et al. 1995, 1998; Siegfried et al. 2004; Deshpande et al. 2005) | ||
| elt-2 | Intestine | Early larval arrest |
| (Fukushige et al. 1998, 1999) | ||
| F38E11.5 | Somatic gonad, embryo | Sick, slow growing, and sterile |
| (Kamath et al. 2003; Simmer et al. 2003; Rual et al. 2004) | ||
| peb-1 | Pharynx, vulva, hindgut | Unc, sterile |
| (Thatcher et al. 2001) | ||
| sma-1 | Hypodermal cells, pharynx, excretory cell, intestine, some neurons | Slow growth, small, Dumpy |
| (McKeown et al. 1998; Praitis et al. 2005) | ||
| unc-15 | Various muscles | Paralysis |
| (Rual et al. 2004; Deshpande et al. 2005) | ||
| unc-22 | Muscle | Twitching |
| (Moerman et al. 1988) | ||
| unc-112 | Various muscles | Paralysis |
| (Rogalski et al. 2000; Edens et al. 2001) | ||
| wnk-1 | Excretory canal, hypodermis | Egg-laying abnormal |
| (Kamath et al. 2003; Simmer et al. 2003) |
The reagents utilized for RNAi assays and how RNAi defects were monitored are described. The RNAi target is listed in the first column. Bacteria were used as the dsRNA delivery vehicle. The collected information regarding expression patterns is listed in the second column. The RNAi phenocopies we observed in wild-type animals are listed in the third column. Experiments were performed on strains defective for 49 different ABC transporter genes. Tests for RNAi activity were performed at 15°, 20°, and 25°.
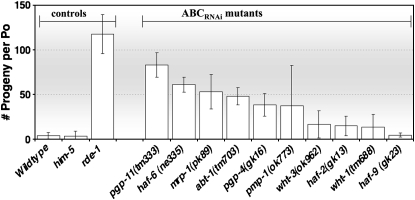
We directly compared the relative expressivity of the RNAi defects in the ABCRNAi strains by assessing each of their RNAi activities simultaneously, using an identical preparation of pop-1 food. Some ABCRNAi strains display strong RNAi defects, while in other strains the defects are rather weak (Figure 1).
Figure 1.—
The RNAi defects are unequal in ABCRNAi mutants. ABCRNAi mutants were reared on pop-1 food and the number of progeny produced by each strain was tabulated. Bars represent the average of values obtained from three plates, each with five Po animals. The experiment utilized identical conditions for each plate. Error bars denote SD. (Some pmp-1 parental animals were lost from one plate, hence the larger error.)
haf-6, mut-7, and rde-2 alleles are noncomplementing:
Previous reports have documented SSNC interactions between alleles of rde-2 and mut-7(ne311), a mut-7 allele with a missense mutation in the presumptive nuclease domain (Tops et al. 2005). During the course of our genetic analyses of haf-6(ne335) mutants, we observed SSNC between alleles of rde-2 and haf-6: doubly heterozygous haf-6(ne335) +/+ rde-2(ne221) animals were RNAi defective (Figure 2A). Using feeding-based assays to score for RNAi defects, we also observed SSNC using rde-2(ne221) and a different allele of mut-7–mut-7(pk204) (Figure 2A). This allele harbors a nonsense codon and disrupts the ability of MUT-7 to bind RDE-2 (Tops et al. 2005). We then extended the syllogism by testing whether the RNAi defects in mut-7 could be complemented by mutations in haf-6, and, indeed, we observed noncomplementation between mut-7(pk204) and haf-6(ne335) (Figure 2A). Similar noncomplementation results were obtained using mutants harboring deletions in rde-2 and mut-7, rde-2(pk1657), and mut-7(pk720). Thus rde-2, mut-7, and haf-6 constitute a noncomplementing group of alleles with respect to RNAi activity.
RNAi defects in haf-6 mutants are dosage dependent with respect to the amount of dsRNA introduced (Sundaram et al. 2006), and we considered that the SSNC results may indicate dosage sensitivity with respect to HAF-6, MUT-7, and RDE-2 proteins as well. This hypothesis is supported by the observation that the RNAi defects in mut-7 and rde-2 mutants can be partially rescued by extra copies of haf-6(+) DNA (Figure 2B). We also observed an ability of rde-2 sequences to partially rescue the RNAi defects in haf-6 mutants. Wild-type rde-2 sequences do not provide rescue of RNAi defects in mut-7(pk204) (Figure 2B); however, these mutants harbor a version of MUT-7 that cannot bind RDE-2. Thus, rescue of RNAi defects in mut-7(pk204) animals by overexpression of HAF-6 may occur independently of an RDE-2/MUT-7 protein complex.
All ABCRNAi mutants display genetic interactions with mut-7 and rde-2 mutants:
We reasoned that we might use complementation testing with mut-7 or rde-2 alleles as an assay to help reveal additional ABC transporter genes that are required for RNAi. We therefore extended our complementation testing to all available ABC transporter mutant strains. The vast majority of the ABC transporter alleles in these experiments harbor deletions; whereas both deletions and point mutations in mut-7 and rde-2 alleles are available and were used in our assays. The mut-7 point mutations displayed similar genetic behavior to the deletion strains, and this was the case for rde-2 as well. Because the results we obtained using mut-7 strains were similar to those obtained using rde-2 mutants, we refer to the use of either of these two mutant strains, their activities, and their genetic behaviors as mut-7/rde-2.
The RNAi defects in mut-7/rde-2 mutants were complemented by all the ABC transporter mutants that display wild-type RNAi activity, with the exception of haf-9 (Tables 2 and 3 and supplemental Tables 1 and 2 at http://www.genetics.org/supplemental/). Therefore, we now include haf-9 in our list of ABCRNAi genes, despite the fact that homozygous mutants with a single-gene defect in haf-9 are wild type for RNAi activity. Our collective results imply that the SSNC assays are a more sensitive indicator for RNAi function than feeding-based RNAi assays. This additional set of data points to specificity with regard to the ability of ABC transporters to influence RNAi mechanisms, as >75% of the 49 ABC transporter mutants tested complement the RNAi defects in mut-7/rde-2. In addition, we hypothesize that the RNAi defects in ABC transporter mutants are specific to the mut-7/rde-2 branch of the RNAi pathway, as a number of strains with defects in other RNAi pathways, such as the RNA-induced silencing complex (RISC) pathway or pathways that facilitate dsRNA trafficking, complemented the RNAi defects in mut-7 and rde-2 (Table 2 and supplemental Table 1). Thus not only are the SSNC tests sensitive assays for the detection of RNAi-related functions for ABC transporters, but also the assays help narrow the ABCRNAi functions to a particular branch of the RNAi network (Lee et al. 2006; Yigit et al. 2006).
Long-lasting parent-of-origin effects influence whether RNAi phenotypes are observed in trans-heterozygous progeny:
In the sections above, we demonstrated SSNC interactions between ABCRNAi and mut-7/rde-2 mutants: trans-heterozygous progeny displayed RNAi defects. We also observed a parent-of-origin effect in those SSNC interactions. In complementation tests using mut-7/rde-2 as the parental hermaphrodite and ABCRNAi mutants as parental males, RNAi defects were observed in trans-heterozygous progeny (Table 3 and supplemental Table 2). However, in most reciprocal crosses utilizing mut-7 or rde-2 as the parental male, the resulting trans-heterozygous progeny displayed a wild-type level of RNAi activity (Table 3 and supplemental Table 2). In control experiments, a parent-of-origin effect is not revealed in simple crosses utilizing wild-type males and hermaphrodites that are homozygous for mut-7 (or rde-2): wild-type levels of RNAi activity are displayed in both sets of genetically identical progeny produced from reciprocal crosses. Thus, a dependency on maternal MUT-7/RDE-2 is revealed only when the animal is also heterozygous for an ABCRNAi transporter gene.
The RNAi defects in trans-heterozygotes are surprisingly long lasting. Animals were placed on feeding plates as young larvae, and RNAi activities were assessed later in development, in the germ line of the ensuing adult. mut-7 and rde-2 mutations are recessive, and RNAi activity can be provided from paternal sources in animals that are singly heterozygous for mut-7 or rde-2. However, in trans-heterozygous (mut-7/rde-2)/+; +/ABCRNAi progeny that originate from oocytes lacking maternal MUT-7 or RDE-2, paternal mut-7/rde-2 does not provide rescue of RNAi defects. In these trans-heterozygous progeny, the maternal load of any ABCRNAi transporters should be inherited at wild-type levels. How the genetic condition of heterozygosity for an ABCRNAi gene prevents paternal rescue of mut-7/rde-2 is a question that remains to be answered.
Some combinations of ABCRNAi mutations also fail to complement:
We extended our analyses to see if pairs of different ABCRNAi mutants would complement one another (Table 3). We anticipated that we might gain information regarding redundancy with respect to ABC transporter functions in RNAi or results that might indicate heterodimerization partners for the half transporters. We observed a few pairs of noncomplementing ABCRNAi transporter alleles and several instances where the SSNC results were parent-of-origin dependent (Table 3). In general, most combinations of ABC transporter mutants were complementary, highlighting the specificity of these interactions and ruling out the possibility of a common background mutation in each of the strains.
Most of our noncomplementation experiments utilized pop-1 food. We considered that the RNAi defects we observed in trans-heterozygous animals might reflect an ability of this genetic condition to influence the pop-1 pathway. To address this question, we retested noncomplementing pairs of alleles using cks-1 food and again observed noncomplementation. cks-1 is a conserved cell-cycle regulator required for cyclin-dependent kinase activity and proper cell division (Polinko and Strome 2000; Shirayama et al. 2006), confirming that the defect in trans-heterozygotes resides in the RNAi pathway and not in the pop-1 pathway (supplemental Table 2).
Conditional phenotypes are observed in ABCRNAi mutants:
The SSNC interactions we uncovered led us to investigate whether the ABCRNAi genes, like mut-7 or rde-2, are required for endogenous RNAi-associated processes. We first looked to see if ABCRNAi mutants shared common phenotypes with mut-7 and rde-2. Unlike mut-7 and rde-2, the ABCRNAi mutants do not have a high rate of chromosome nondisjunction, as they do not display a high incidence of males (Him) phenotype under any of the environmental conditions we tested, including elevated temperature, the presence of exogenous dsRNAs or heavy metal cations, or starvation. However, we did observe additional phenotypes in some ABCRNAi transporter mutants that are indicative of defects in RNAi mechanisms. We report three main observations.
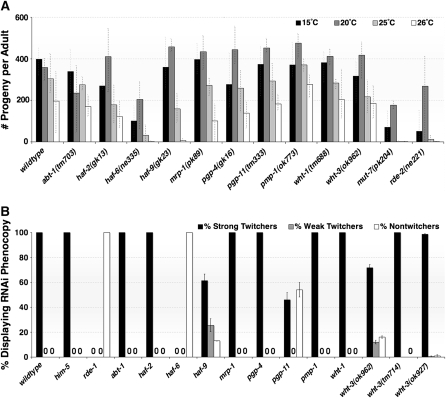
First, the brood sizes are reduced in some mutants reared under conditions of elevated temperature (Figure 3A). While most mutants have normal fertility, haf-6 and haf-9 mutants produce fewer progeny at elevated temperatures. mut-7 and rde-2 mutants also have reductions in brood sizes, and it is widely appreciated that both rde-2 and mut-7 stocks will eventually stop producing progeny when reared at 25° for several generations.
Figure 3.—
ABCRNAi mutants display additional temperature-sensitive phenotypes. (A) The brood sizes from each strain were assessed at different temperatures under otherwise identical conditions. Progeny were counted and removed from plates daily for 5 days (15° and 20° experiments) or 3 days (25°). Bars represent results from duplicate experiments. Error bars denote SD. (B) Animals with the indicated genotypes were placed on unc-22 food and Twitching phenocopies were assessed at 15°, 20°, and 25°. No twitching was observed in rde-1 or in sid-1mutants at any temperature, as expected. Twitching was observed in all other strains reared at 15° and 20°; the results from the 25° experiment are displayed here. Animals from each plate were classified as nontwitchers, weak twitchers (good movement with twitching), or strong twitchers (paralysis and twitching). Bars represent the average of three experiments; error bars denote SD.
Second, the RNAi defects in haf-6(ne335), mut-7(pk204), and rde-2(ne221) mutants are temperature sensitive for one target, unc-22, when the feeding method is used to deliver dsRNA (Figure 3B). No temperature sensitivity is observed when unc-22 dsRNA is introduced by soaking, injection, or transgene expression of unc-22 sequences in muscle (data not shown). To date, unc-22 is the only sequence for which we have observed a method- and temperature-sensitive RNAi defect. This phenotype is not a general consequence of defects in RNAi and is not observed in RISC-defective animals or in animals defective for systemic RNAi due to loss of sid-1 (data not shown). However, we have observed similar temperature-sensitive RNAi defects from unc-22 food in rsd-2 and rsd-6 mutants (W. Han, unpublished results). We hypothesize that this temperature- and method-of-delivery-dependent RNAi phenotype is a hallmark for a subset of RNAi-defective mutants that includes rde-2, mut-7, haf-6, rsd-2, and rsd-6.
Finally, mut-7 and rde-2 mutants display an increased rate of transposon mobilization (Mutator activity), and some ABCRNAi mutants display temperature-sensitive Mutator activity as well (Table 4). Strains with Mutator activity include haf-6(ne335), haf-9(gk23), wht-1(tm688), and mrp-1(pk89). While mut-7 and rde-2 animals display Mutator activity at all temperatures, all the ABCRNAi mutants are temperature sensitive for this phenotype. We hypothesize that this collective set of temperature-sensitive phenotypes in haf-6 and other ABCRNAi mutants reflects endogenous roles for ABCRNAi transporters in cellular processes that also require the function of MUT-7/RDE-2, roles that ultimately affect chromosome integrity.
TABLE 4.
Mutants have increased rates of transposon mobilization
| % revertants
|
|||
|---|---|---|---|
| Strain | 15° | 20° | 25° |
| unc-22(st136) | 0 | 0 | 0 |
| (0/33,000) | (0/32,000) | (0/41,000) | |
| rde-2(ne221); unc-22(st136) | 0.08 | 0.40 | |
| (10/12,750) | (90/22,200) | ||
| mut-7(ne221); unc-22(st136) | 3.74 | ||
| (127/3,400) | |||
| abt-1(tm0703); unc-22(st136) | 0 | 0 | 0 |
| (0/26,000) | (0/24,500) | (0/38,000) | |
| haf-2(gk13); unc-22(st136) | 0 | 0 | 0 |
| (0/25,500) | (0/27,220) | (0/38,000) | |
| haf-6(ne335); unc-22(st136) | 0 | 0 | 0.09 |
| (0/21,150) | (0/21,150) | (17/18,000) | |
| haf-9 (gk23); unc-22(st136) | 0 | 0 | 0.04 |
| (0/27,300) | (0/25,500) | (15/41,200) | |
| mrp-1(pk89); unc-22(st136) | 0 | 0 | 0.02 |
| (0/26,000) | (0/26,050) | (8/41,000) | |
| pgp-4(gk16); unc-22(st136) | 0 | 0 | 0 |
| (0/24,850) | (0/25,100) | (0/38,000) | |
| pgp-11(tm33); unc-22(st136) | 0 | 0 | 0 |
| (0/28,500) | (0/28,000) | (0/38,000) | |
| pmp-1(ok773); unc-22(st136) | 0 | 0 | 0 |
| (0/26,900) | (0/28,700) | (0/38,000) | |
| wht-1(tm688); unc-22(st136) | 0 | 0 | 0.06 |
| (0/28,300) | (0/22,000) | (26/42,000) | |
| wht-3(ok962); unc-22(st136) | 0 | 0 | 0 |
| (0/26,800) | (0/25,000) | (0/38,000) | |
| wht-3(ok927); unc-22(st136) | 0 | 0 | |
| (0/13,500) | (0/11,800) | ||
Strains are marked with an unc-22(st136) allele, which harbors a Tc1 transposable element. Germ-line excisions were assessed by noting the number of nontwitching revertants that appeared in the progeny of homozygous mutants. Revertants were set aside and a 3:1 phenotypic ratio was observed in the progeny from each, as expected; PCR was performed on some revertants and the amplified unc-22 sequence was the size expected for wild type.
DISCUSSION
A subclass of C. elegans ABCRNAi transporters can be defined on the basis of function:
This study rigorously distinguishes ABCRNAi transporters from the remaining ABC transporter genes and points to roles for ABCRNAi transporters in silencing events that affect chromosome function. The C. elegans genome encodes ∼61 ABC transporter proteins that collectively reside on most intracellular membranes. Because ABC transporter-dependent movement of substrates is essential for subcellular organelle biogenesis and function, and because substrates can be transported between organelles (Nunes et al. 2005; Schaheen et al. 2006; Schroeder et al. 2007), it is conceivable that alterations in ABC transporter function might simply affect cell physiology in a more general manner, with indirect effects on RNAi function. If we assume this possibility is correct, then we would expect to observe RNAi defects in multiple tissues in most of the ABC transporter mutants, yet this is not what we observed. RNAi defects were observed in a limited subset of the mutants tested.
ABCRNAi transporters have specific effects on RNAi functions:
Evidence of specificity with regard to RNAi phenotypes is provided by the following observations: only the ABCRNAi subset of transporters displays defects in RNAi, with the exception of haf-9 mutants. While this is a functional definition that we applied on the basis of our results, this definition applies only to 30% of the 49 transporter mutants tested. Furthermore, only the ABCRNAi subset of transporter mutants displays SSNC interactions with mut-7/rde-2.
The fact that the RNAi functions of ABCRNAi transporters are required in germ-line tissue provides additional evidence of specificity. Even though several of the ABCRNAi genes are expressed in various somatic tissues (Zhao et al. 2004), all ABCRNAi transporters are required for efficient RNAi in the germ line. Moreover, our extensive set of RNAi assays has not revealed an ABC transporter gene that is required for RNAi solely in somatic cells. mut-7 and rde-2, which interact genetically with ABCRNAi mutants, are also required for efficient RNAi in the germ line, and the corresponding mutants display phenotypes indicative of germ-line defects (meitoic nondisjunction, cosuppression, and high rates of transposon mobilization). By contrast, some ABC transporters are expressed in the germ line, yet are not required for RNAi. For example, ced-7 is required in the germ line for engulfment of apoptotic corpses (Wu and Horvitz 1998), yet ced-7 mutants are wild type with respect to the RNAi activities assessed in this report. Finally, some ABCRNAi mutants display temperature-sensitive phenotypes in the germ line, phenotypes that are similar to those observed in mut-7/rde-2 mutants. These temperature-sensitive phenotypes include Mutator activity, reductions in brood size, and RNAi defects in response to ingestion of unc-22 food. While germ-line tissue is clearly the focus of ABCRNAi function, we cannot rule out additional trafficking roles between germ-line and somatic tissues.
Finally, within the RNAi network of mechanisms (Lee et al. 2006; Yigit et al. 2006), evidence of specificity is observed with respect to ABCRNAi transporter function. This is revealed in SSNC assays using mut-7/rde-2, genes that are required in RNAi-related processes that affect chromosome as well as miRNA functions. By contrast, ABCRNAi transporters do not interact genetically with genes that are required in other RNAi-related processes, including genes that facilitate systemic silencing by dsRNA trafficking (sid-1) or genes that are required in dsRNA processing and RISC-based mechanisms (rde-1 and rde-4) (Tabara et al. 2002; Feinberg and Hunter 2003; Yigit et al. 2006). Additionally, even though ABCRNAi transporter mutants display temperature-sensitive defects in transposon silencing, they do not interact genetically with other mutants that display temperature-sensitive transposon silencing defects (rsd-6) (Han et al., unpublished results). Similarly, while mut-7/rde-2 mutants have increased rates of nondisjunction, ABCRNAi transporter mutants did not interact with other mutants that display dsRNA-inducible nondisjunction phenotypes (rsd-2) (Han et al., unpublished results). It may be the case that SSNC interactions are observed between pairs of genes that normally produce limited product. RSD-2, RSD-6, SID-1, RDE-1, and RDE-4 may not be limiting in quantity, and therefore the corresponding mutants would not be expected to display SSNC with ABCRNAi mutants. On the other hand, Him and Mutator phenotypes can be induced in rsd-2 and rsd-6 mutants (Han et al., unpublished results), which is consistent with the possibility that RSD-2 and RSD-6 proteins are limiting.
The common phenotypes and genetic interactions between ABCRNAi transporters and mut-7/rde-2 imply common functions that affect chromosome function and transcriptional silencing:
In addition to roles in chromosome function and integrity, mut-7 and rde-2 are also required for cosuppression and silencing of transgenes in the germ line (Tabara et al. 1999; Dernburg et al. 2000; Ketting and Plasterk 2000; Kim et al. 2005). Both these phenomena are observed when transgenes are introduced into C. elegans, and both phenomena have similar features that are suggestive of transcriptional-level silencing. Cosuppression refers to an ability of a transgene to silence gene expression from the transgene itself, as well as from endogenous genes with sequence homology to the transgene. While specific sequences may be more prone to cosuppression, the exact nature of the triggering molecule is not known. Silencing of transgene expression is frequently observed for sequences that express in the germ line. Both cosuppression and transgene silencing require chromatin factors and RNAi, and the transgenes that are silenced in the germ line display histone methylation patterns reminiscent of heterochromatin (Kelly et al. 2002; Robert et al. 2005). Both transgene silencing and cosuppression require mut-7 and rde-2, but not rde-1 (Dernburg et al. 2000).
Here, we described an unusual temperature-sensitive and method-of-delivery-dependent RNAi defect that we observe in mut-7, rde-2, and haf-6 mutants, and we postulate that this phenotype also reflects impairments in transcriptional silencing. Upon ingestion of unc-22 food, these mutants display RNAi defects when the experiments are conducted at elevated temperatures. Normally, the unc-22 gene is an effective RNAi target in RNAi experiments using wild-type animals, and unc-22 sequences have been noted to induce cosuppression as well (Fire et al. 1991). Taken together, we postulate that unc-22 sequences have a tendency to trigger transcriptional silencing responses in these assays and that unc-22 is a particularly amenable RNAi target due to the combined activities of RISC and transcriptional silencing mechanisms. We further hypothesize that mut-7, rde-2, and haf-6 are required to maintain transcription-level silencing at elevated temperatures. On a related note, we recently reported temperature-sensitive defects in transposon silencing in rsd-6 mutants, and rsd-6 also displays temperature-sensitive RNAi defects when reared on unc-22 food (W. Han, unpublished results). Finally, we point out that chromosome-related events such as chromosome disjunction, transposon mobilization, and recombination are intrinsically sensitive to environmental conditions (Hildreth and Ulrichs 1969; Rose and Baillie 1979; Hashida et al. 2006). Thus the temperature-sensitive response to unc-22 food that we observe in mutants is likely a hallmark for genes with RNAi activities that may ultimately affect transcriptional silencing.
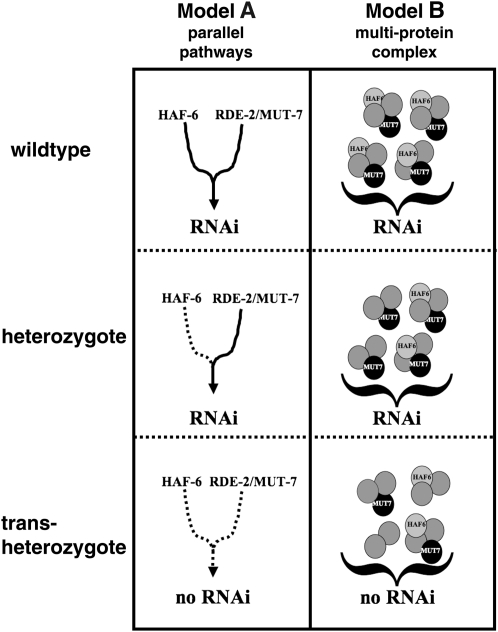
Precisely how ABCRNAi transporter and mut-7/rde-2 functions are integrated is not known; however, we can invoke at least two models on the basis of the genetic and transgene rescue data (Figure 4). In a parallel pathways model, HAF-6 and RDE-2/MUT-7 activities are predicted to reside in different pathways that converge to a common RNAi function, with a threshold level of activity from both pathways required for full RNAi effect. Alternatively, in a protein complex model, both ABCRNAi and RDE-2/MUT-7 function as a multiprotein complex that is effectual only when threshold numbers of complexes are complete. Because overexpression of one protein can rescue the RNAi defect in a mutant that is deficient for the second protein (Figure 2B), the former model is strongly favored; however, the latter model cannot be confidently eliminated. Indeed, RDE-2 and MUT-7 proteins physically associate (Tops et al. 2005).
Figure 4.—
Alternative models are predicted to help explain the genetic interactions. In a parallel pathways model, wild-type activity requires input from two pathways that converge to an RNAi function (left). In animals heterozygous for both genes, activities in multiple pathways may be reduced (left, bottom). In a protein complex model, a threshold number of complete multiprotein complexes is required for robust RNAi activity (right), while in trans-heterozygotes, a subthreshold number of complete complexes limits full activity (right, bottom). The models are not mutually exclusive; multiprotein complexes may act in pathways, and transported substrates may affect complex activity.
Thus, ABCRNAi proteins increase RNAi efficiency in the mut-7/rde-2 branch of RNAi mechanisms where they ultimately affect silencing mechanisms that act at the level of the chromosome. An important consequence of this observation resides in the potential for environmental signals to produce changes in gene expression via RNAi mechanisms that are regulated or influenced by ABCRNAi-dependent transport of specific substrates.
The maternally deposited MUT-7/RDE-2 products are particularly long lived:
We noted that maternally deposited MUT-7/RDE-2 products were particularly long lived, as revealed in the SSNC tests, and the maternal requirement for MUT-7/RDE-2 is observed only when animals are heterozygous for ABCRNAi (Table 3). We propose that the set-aside nature of germ-line progenitors and asymmetric cell divisions during embryogenesis allows for preferential segregation of maternal MUT-7/RDE-2 gene products into germ-line progenitor cells. We further speculate that the RNAi defects observed in adult trans-heterozygotes provides evidence of a long-lived nature of maternal MUT-7/RDE-2.
The presence of functional, plasma membrane-associated ABC transporters has important applications in stem cell biology, as ABC transporters serve as phenotypic markers for stem cell populations, and expression of particular transporters can influence developmental decisions of stem cells (Bunting 2002; Lin et al. 2006). It is interesting to speculate that specific transcriptional or chromosome-related functions that could affect differentiation are afforded by ABC transporters acting through specific RNAi mechanisms in germ-line progenitors and in cultured stem cells.
A network of activity is revealed through analysis of noncomplementing pairs of ABCRNAi mutants:
A few pairs of ABCRNAi mutants were noncomplementing: some pairs of mutants failed to complement in reciprocal crosses, whereas SSNC in other sets of mutants was dependent upon the parental configuration of alleles (Table 3 and Figure 5, left section). In examples where SSNC is observed in reciprocal crosses, the results may reflect requirements for both transporters in one germ line. Alternatively, one transporter may be required in the male germ line. A SSNC that displays a parent-of-origin effect in reciprocal crosses might reflect functional redundancy for ABCRNAi genes in one gonad. Because some transporters may function as homo- and/or heterodimers, and because ABCRNAi function may not be restricted to germ-line tissue, it is not possible to draw firm conclusions regarding functional redundancy, tissue-specific requirements, or dimerization partners using the existing data. Nonetheless, we did note a pattern emerging from these data (Figure 5, right section).
Figure 5.—
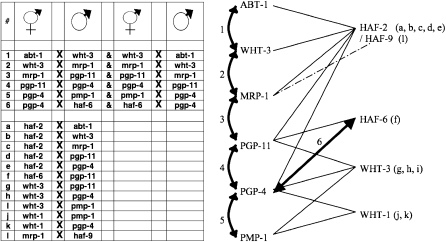
Noncomplementation patterns reveal a network of ABCRNAi transporter activity. Pairs of ABCRNAi transporters that failed to complement in reciprocal crosses are numbered; nonreciprocal, noncomplementing pairs are lettered. The diagram indicates the interconnected nature of noncomplementing pairs of ABCRNAi alleles. All 10 ABCRNAi transporter genes are represented in this diagram. haf-9 has only one connection, but haf-9 mutants do not display RNAi defects. RNAi defects in abt-1, pgp-4, and wht-3 do not complement mut-7/rde-2 when mut-7/rde-2 are used as parental males, and these ABCRNAi transporters occupy pivotal positions in the network.
All 10 ABCRNAi transporters are interconnected in a noncomplementation network with the exception of haf-9, the only ABCRNAi that does not display RNAi defects. However, the haf-9 interaction can be added to the network in a manner that is consistent with the assumption that HAF-9 functions as a heterodimer with another ABCRNAi protein. We note that both haf-9 and haf-2 fail to complement mrp-1; thus these two HAF proteins may function as heterodimers in the germ line. abt-1, wht-3, and pgp-4 are the only mutants that display SSNC when mut-7 or rde-2 are used as the parental males; furthermore, abt-1, wht-3, and pgp-4 are positioned at critical points in the noncomplementation network. abt-1 is located at the end of the network, pgp-4 has the greatest number of interacting partners, including strong interactions with haf-6, pgp-11, and pmp-1, and wht-3 is located in two positions in the network (Figure 5, right section). While additional experiments are required to fully elucidate the meaning of the noncomplementation network, the fact that all 10 ABCRNAi genes can be represented in an interconnected manner lends validity to the results of our complementation testing. We speculate that the network may represent a trafficking route for one or more substrates that are required for efficient RNAi.
Environmental conditions may influence gene expression via two conserved and interrelated networks:
Environmental conditions such as elevated temperature can influence chromosome-associated activities such as transcription, transgene silencing, recombination, meiotic chromosomal disjunction, and transposon mobilization. The additional phenotypes we observed when ABCRNAi mutants were reared at elevated temperatures (Mutator and unc-22 RNAi defects) imply an ability for ABCRNAi transporters to protect chromosome-related functions and influence transcription in suboptimal environments. On a related note, but also mRNA regulation by miRNAs has been shown to be influenced by cellular stressors (Bhattacharyya et al. 2006a,b; Peters and Meister 2007). Precisely how RNAi mechanisms respond to environmental influences is not known. Interestingly, in addition to a 3′–5′ exonuclease domain, the MUT-7 protein also has similarity to conserved proteins of unknown function that are found in several hyperthermophilic archaea and other bacteria, raising the possibility that this domain might have functional roles that are influenced by temperature. Regions similar to this conserved domain are also found in 3′–5′ exonuclease domain proteins from paramecium and humans (supplemental Figure 1 at http://www.genetics.org/supplemental/).
We speculate that a network of ABCRNAi functions may allow for multiple means of influencing mut-7/rde-2, and ultimately chromosome function and integrity, in various environmental conditions. ABC transporters and RNAi pathway components appear to be functionally connected as part of an ancient cellular mechanism that responds to the environment.
Acknowledgments
This study would not have been possible without free access to the many mutant strains generated by the C. elegans community. The authors extend many thanks to the C. elegans Gene Knockout Consortium and the National Bioresources Project in Japan for ABC gene deletion mutants; to Andrew Fire, Craig Mello, and Ron Plasterk for generating and sending mutants; to David Moore for microscopy assistance; to E. Jane Hubbard for helpful suggestions during manuscript preparation; and to the Sanger Institute and the C. elegans Genome Sequencing Project for cosmids. This work was supported by the National Institutes of Health K-INBRE Program of the National Center for Research Resources and the American Cancer Society.
References
- Bhattacharyya, S. N., R. Habermacher, U. Martine, E. I. Closs and W. Filipowicz, 2006. a Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125 1111–1124. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya, S. N., R. Habermacher, U. Martine, E. I. Closs and W. Filipowicz, 2006. b Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harbor Symp. Quant. Biol. 71 513–521. [DOI] [PubMed] [Google Scholar]
- Brooks, D. R., P. J. Appleford, L. Murray and R. E. Isaac, 2003. An essential role in molting and morphogenesis of Caenorhabditis elegans for ACN-1, a novel member of the angiotensin-converting enzyme family that lacks a metallopeptidase active site. J. Biol. Chem. 278 52340–52346. [DOI] [PubMed] [Google Scholar]
- Brunschwig, K., C. Wittmann, R. Schnabel, T. R. Burglin, H. Tobler et al., 1999. Anterior organization of the Caenorhabditis elegans embryo by the labial-like Hox gene ceh-13. Development 126 1537–1546. [DOI] [PubMed] [Google Scholar]
- Bunting, K. D., 2002. ABC transporters as phenotypic markers and functional regulators of stem cells. Stem Cells 20 11–20. [DOI] [PubMed] [Google Scholar]
- Campbell, R. B., D. A. Sinclair, M. Couling and H. W. Brock, 1995. Genetic interactions and dosage effects of Polycomb group genes of Drosophila. Mol. Gen. Genet. 246 291–300. [DOI] [PubMed] [Google Scholar]
- Dernburg, A. F., J. Zalevsky, M. P. Colaiacovo and A. M. Villeneuve, 2000. Transgene-mediated cosuppression in the C. elegans germ line. Genes Dev. 14 1578–1583. [PMC free article] [PubMed] [Google Scholar]
- Deshpande, R., T. Inoue, J. R. Priess and R. J. Hill, 2005. lin-17/Frizzled and lin-18 regulate POP-1/TCF-1 localization and cell type specification during C. elegans vulval development. Dev. Biol. 278 118–129. [DOI] [PubMed] [Google Scholar]
- Edens, W. A., L. Sharling, G. Cheng, R. Shapira, J. M. Kinkade et al., 2001. Tyrosine cross-linking of extracellular matrix is catalyzed by Duox, a multidomain oxidase/peroxidase with homology to the phagocyte oxidase subunit gp91phox. J. Cell Biol. 154 879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann, D. M., J. N. Maloof, J. S. Simske, C. Kenyon and S. K. Kim, 1998. The beta-catenin homolog BAR-1 and LET-60 Ras coordinately regulate the Hox gene lin-39 during Caenorhabditis elegans vulval development. Development 125 3667–3680. [DOI] [PubMed] [Google Scholar]
- Feinberg, E. H., and C. P. Hunter, 2003. Transport of dsRNA into cells by the transmembrane protein SID-1. Science 301 1545–1547. [DOI] [PubMed] [Google Scholar]
- Fire, A., D. Albertson, S. W. Harrison and D. G. Moerman, 1991. Production of antisense RNA leads to effective and specific inhibition of gene expression in C. elegans muscle. Development 113 503–514. [DOI] [PubMed] [Google Scholar]
- Frand, A. R., S. Russel and G. Ruvkun, 2005. Functional genomic analysis of C. elegans molting. PLoS Biol. 3 e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, A. G., R. S. Kamath, P. Zipperlen, M. Martinez-Campos, M. Sohrmann et al., 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408 325–330. [DOI] [PubMed] [Google Scholar]
- Fukushige, T., M. G. Hawkins and J. D. McGhee, 1998. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev. Biol. 198 286–302. [PubMed] [Google Scholar]
- Fukushige, T., M. J. Hendzel, D. P. Bazett-Jones and J. D. McGhee, 1999. Direct visualization of the elt-2 gut-specific GATA factor binding to a target promoter inside the living Caenorhabditis elegans embryo. Proc. Natl. Acad. Sci. USA 96 11883–11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer, B., S. Viggiano, M. A. Hibbs, O. G. Troyanskaya and D. C. Amberg, 2007. Modeling complex genetic interactions in a simple eukaryotic genome: actin displays a rich spectrum of complex haploinsufficiencies. Genes Dev. 21 148–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashida, S. N., T. Uchiyama, C. Martin, Y. Kishima, Y. Sano et al., 2006. The temperature-dependent change in methylation of the Antirrhinum transposon Tam3 is controlled by the activity of its transposase. Plant Cell 18 104–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley, R. S., and W. D. Gilliland, 2006. Sometimes the result is not the answer: the truths and the lies that come from using the complementation test. Genetics 174 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildreth, P. E., and P. C. Ulrichs, 1969. A temperature effect on nondisjunction of the X chromosomes among eggs from aged Drosophila females. Genetica 40 191–197. [DOI] [PubMed] [Google Scholar]
- Hull, D., and L. Timmons, 2004. Methods for delivery of double-stranded RNA into Caenorhabditis elegans. Methods Mol. Biol. 265 23–58. [DOI] [PubMed] [Google Scholar]
- Jaspersen, S. L., B. J. Huneycutt, T. H. Giddings, Jr., K. A. Resing, N. G. Ahn et al., 2004. Cdc28/Cdk1 regulates spindle pole body duplication through phosphorylation of Spc42 and Mps1. Dev. Cell 7 263–274. [DOI] [PubMed] [Google Scholar]
- Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin et al., 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421 231–237. [DOI] [PubMed] [Google Scholar]
- Kelly, W. G., S. Xu, M. K. Montgomery and A. Fire, 1997. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, W. G., C. E. Schaner, A. F. Dernburg, M. H. Lee, S. K. Kim et al., 2002. X-chromosome silencing in the germline of C. elegans. Development 129 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting, R. F., and R. H. Plasterk, 2000. A genetic link between co-suppression and RNA interference in C. elegans. Nature 404 296–298. [DOI] [PubMed] [Google Scholar]
- Ketting, R. F., T. H. Haverkamp, H. G. van Luenen and R. H. Plasterk, 1999. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99 133–141. [DOI] [PubMed] [Google Scholar]
- Kim, J. K., H. W. Gabel, R. S. Kamath, M. Tewari, A. Pasquinelli et al., 2005. Functional genomic analysis of RNA interference in C. elegans. Science 308 1164–1167. [DOI] [PubMed] [Google Scholar]
- Lee, R. C., C. M. Hammell and V. Ambros, 2006. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA 12 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, R., S. Thompson and J. R. Priess, 1995. pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C. elegans embryos. Cell 83 599–609. [DOI] [PubMed] [Google Scholar]
- Lin, R., R. J. Hill and J. R. Priess, 1998. POP-1 and anterior-posterior fate decisions in C. elegans embryos. Cell 92 229–239. [DOI] [PubMed] [Google Scholar]
- Lin, T., O. Islam and K. Heese, 2006. ABC transporters, neural stem cells and neurogenesis–a different perspective. Cell Res. 16 857–871. [DOI] [PubMed] [Google Scholar]
- McKeown, C., V. Praitis and J. Austin, 1998. sma-1 encodes a betaH-spectrin homolog required for Caenorhabditis elegans morphogenesis. Development 125 2087–2098. [DOI] [PubMed] [Google Scholar]
- Moerman, D. G., G. M. Benian, R. J. Barstead, L. A. Schriefer and R. H. Waterston, 1988. Identification and intracellular localization of the unc-22 gene product of Caenorhabditis elegans. Genes Dev. 2 93–105. [DOI] [PubMed] [Google Scholar]
- Natarajan, L., B. M. Jackson, E. Szyleyko and D. M. Eisenmann, 2004. Identification of evolutionarily conserved promoter elements and amino acids required for function of the C. elegans beta-catenin homolog BAR-1. Dev. Biol. 272 536–557. [DOI] [PubMed] [Google Scholar]
- Nunes, F., M. Wolf, J. Hartmann and R. J. Paul, 2005. The ABC transporter PGP-2 from Caenorhabditis elegans is expressed in the sensory neuron pair AWA and contributes to lysosome formation and lipid storage within the intestine. Biochem. Biophys. Res. Commun. 338 862–871. [DOI] [PubMed] [Google Scholar]
- Peters, L., and G. Meister, 2007. Argonaute proteins: mediators of RNA silencing. Mol. Cell 26 611–623. [DOI] [PubMed] [Google Scholar]
- Polinko, E. S., and S. Strome, 2000. Depletion of a Cks homolog in C. elegans embryos uncovers a post-metaphase role in both meiosis and mitosis. Curr. Biol. 10 1471–1474. [DOI] [PubMed] [Google Scholar]
- Praitis, V., E. Ciccone and J. Austin, 2005. SMA-1 spectrin has essential roles in epithelial cell sheet morphogenesis in C. elegans. Dev. Biol. 283 157–170. [DOI] [PubMed] [Google Scholar]
- Robert, V. J., T. Sijen, J. van Wolfswinkel and R. H. Plasterk, 2005. Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev. 19 782–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski, T. M., G. P. Mullen, M. M. Gilbert, B. D. Williams and D. G. Moerman, 2000. The UNC-112 gene in Caenorhabditis elegans encodes a novel component of cell-matrix adhesion structures required for integrin localization in the muscle cell membrane. J. Cell Biol. 150 253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, A. M., and D. L. Baillie, 1979. The effect of temperature and parental age on recombination and nondisjunction in Caenorhabditis elegans. Genetics 92 409–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual, J. F., J. Ceron, J. Koreth, T. Hao, A. S. Nicot et al., 2004. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14 2162–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaheen, L., G. Patton and H. Fares, 2006. Suppression of the cup-5 mucolipidosis type IV-related lysosomal dysfunction by the inactivation of an ABC transporter in C. elegans. Development 133 3939–3948. [DOI] [PubMed] [Google Scholar]
- Schroeder, L. K., S. Kremer, M. J. Kramer, E. Currie, E. Kwan et al., 2007. Function of the Caenorhabditis elegans ABC transporter PGP-2 in the biogenesis of a lysosome-related fat storage organelle. Mol. Biol. Cell. 18 995–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama, M., M. C. Soto, T. Ishidate, S. Kim, K. Nakamura et al., 2006. The conserved kinases CDK-1, GSK-3, KIN-19, and MBK-2 promote OMA-1 destruction to regulate the oocyte-to-embryo transition in C. elegans. Curr. Biol. 16 47–55. [DOI] [PubMed] [Google Scholar]
- Siegfried, K. R., A. R. Kidd, 3rd, M. A. Chesney and J. Kimble, 2004. The sys-1 and sys-3 genes cooperate with Wnt signaling to establish the proximal-distal axis of the Caenorhabditis elegans gonad. Genetics 166 171–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer, F., C. Moorman, A. M. van der Linden, E. Kuijk, P. V. van den Berghe et al., 2003. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1 E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns, T., and D. Botstein, 1988. Unlinked noncomplementation: isolation of new conditional-lethal mutations in each of the tubulin genes of Saccharomyces cerevisiae. Genetics 119 249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit, A., R. Kohler, T. Marty, M. Belfiore, K. Takacs-Vellai et al., 2002. Conserved regulation of the Caenorhabditis elegans labial/Hox1 gene ceh-13. Dev. Biol. 242 96–108. [DOI] [PubMed] [Google Scholar]
- Sundaram, P., B. Echalier, W. Han, D. Hull and L. Timmons, 2006. ATP-binding cassette transporters are required for efficient RNA interference in Caenorhabditis elegans. Mol. Biol. Cell 17 3678–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara, H., M. Sarkissian, W. G. Kelly, J. Fleenor, A. Grishok et al., 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99 123–132. [DOI] [PubMed] [Google Scholar]
- Tabara, H., E. Yigit, H. Siomi and C. C. Mello, 2002. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-Box helicase to direct RNAi in C. elegans. Cell 109 861–871. [DOI] [PubMed] [Google Scholar]
- Thatcher, J. D., A. P. Fernandez, L. Beaster-Jones, C. Haun and P. G. Okkema, 2001. The Caenorhabditis elegans peb-1 gene encodes a novel DNA-binding protein involved in morphogenesis of the pharynx, vulva, and hindgut. Dev. Biol. 229 480–493. [DOI] [PubMed] [Google Scholar]
- Timmons, L., and A. Fire, 1998. Specific interference by ingested dsRNA. Nature 395 854. [DOI] [PubMed] [Google Scholar]
- Timmons, L., D. L. Court and A. Fire, 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263 103–112. [DOI] [PubMed] [Google Scholar]
- Tops, B. B., H. Tabara, T. Sijen, F. Simmer, C. C. Mello et al., 2005. RDE-2 interacts with MUT-7 to mediate RNA interference in Caenorhabditis elegans. Nucleic Acids Res. 33 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann, C., O. Bossinger, B. Goldstein, M. Fleischmann, R. Kohler et al., 1997. The expression of the C. elegans labial-like Hox gene ceh-13 during early embryogenesis relies on cell fate and on anteroposterior cell polarity. Development 124 4193–4200. [DOI] [PubMed] [Google Scholar]
- Wu, Y. C., and H. R. Horvitz, 1998. The C. elegans cell corpse engulfment gene ced-7 encodes a protein similar to ABC transporters. Cell 93 951–960. [DOI] [PubMed] [Google Scholar]
- Yigit, E., P. J. Batista, Y. Bei, K. M. Pang, C. C. Chen et al., 2006. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127 747–757. [DOI] [PubMed] [Google Scholar]
- Yook, K. J., S. R. Proulx and E. M. Jorgensen, 2001. Rules of nonallelic noncomplementation at the synapse in Caenorhabditis elegans. Genetics 158 209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z., J. A. Sheps, V. Ling, L. L. Fang and D. L. Baillie, 2004. Expression analysis of ABC transporters reveals differential functions of tandemly duplicated genes in Caenorhabditis elegans. J. Mol. Biol. 344 409–417. [DOI] [PubMed] [Google Scholar]