Abstract
Microorganisms display an optimal temperature and hydrostatic pressure for growth. To establish the molecular basis of piezo- and psychroadaptation, we elucidated global genetic defects that give rise to susceptibility to high pressure and low temperature in Saccharomyces cerevisiae. Here we present 80 genes including 71 genes responsible for high-pressure growth and 56 responsible for low-temperature growth with a significant overlap of 47 genes. Numerous previously known cold-sensitive mutants exhibit marked high-pressure sensitivity. We identified critically important cellular functions: (i) amino acid biosynthesis, (ii) microautophagy and sorting of amino acid permease established by the exit from rapamycin-induced growth arrest/Gap1 sorting in the endosome (EGO/GSE) complex, (iii) mitochondrial functions, (iv) membrane trafficking, (v) actin organization mediated by Drs2-Cdc50, and (vi) transcription regulated by the Ccr4-Not complex. The loss of EGO/GSE complex resulted in a marked defect in amino acid uptake following high-pressure and low-temperature incubation, suggesting its role in surface delivery of amino acid permeases. Microautophagy and mitochondrial functions converge on glutamine homeostasis in the target of rapamycin (TOR) signaling pathway. The localization of actin requires numerous associated proteins to be properly delivered by membrane trafficking. In this study, we offer a novel route to gaining insights into cellular functions and the genetic network from growth properties of deletion mutants under high pressure and low temperature.
MICROORGANISMS display an optimal growth temperature and an optimal growth hydrostatic pressure. However, the basis of these profiles and the limiting factor on cell growth are poorly understood. It has been a conventional technique in yeast cell biology to analyze gene functions using high temperature- and low temperature (cold)-sensitive mutants, i.e., ts and cs, respectively. Nevertheless, little is known about why the mutation or deletion of genes limits the ability to grow under adverse conditions except for certain types of mutations. It is well-known that bacterial and yeast Saccharomyces cerevisiae mutants defective in ribosome assembly are cold sensitive, probably because ribosome assembly has high activation energy in the absence of certain subunits (Friedman et al. 1969; Bryant and Sypherd 1974; Singh et al. 1978; Ursic and Davies 1979). Some yeast mutants that have altered sensitivities to the antibiotics trichodermin and cycloheximide are also cold sensitive (Moritz et al. 1991; Dresios et al. 2000, 2001, 2003). A substitution of aspartic acid for leucine in actin at position 266 confers cold sensitivity at temperatures between 9° and 15° due to polymerization defects of actin at low temperatures (Chen et al. 1993). Deletion mutants for DRS2 and CDC50 that encode proteins localize to the late Golgi and endosomes are defective in the endocytic pathway and in organization of the actin cytoskeleton at 15° or 18°, resulting in cold-sensitive growth (Moir et al. 1982; Chen et al. 1999; Misu et al. 2003; Natarajan et al. 2004; Saito et al. 2004; Chen et al. 2006), but it is unclear which step is blocked by low temperature. Deletion of LTE1 that encodes the Cdc25 family guanine-nucleotide exchanging factor causes cells to arrest at telophase at low temperature (Shirayama et al. 1994a,b). The loss of Ccr4 and Pop2 that constitutes the Ccr4-Not transcriptional regulator (Tucker et al. 2001; Collart 2003; Denis and Chen 2003) causes marked cold sensitivity, but the reason is unknown (Hata et al. 1998). Mutants defective in tryptophan biosynthesis are cold sensitive due to decreased tryptophan uptake at low temperature (Singh and Manney 1974; Gaber et al. 1989; Chen et al. 1994). Although the cold sensitivity of these mutants is evident, their growth has not been examined at high hydrostatic pressure except in our study on tryptophan uptake.
Increasing hydrostatic pressure has an effect analogous to decreasing temperature in terms of increasing order and decreasing the fluidity of biological membranes. We demonstrated that the uptake of tryptophan is a limiting factor for yeast cell growth at high pressure as well as at low temperature (Abe and Horikoshi 2000). Wild-type strains having trp1 as a nutrient auxotrophic marker (e.g., YPH499 or W303-1A) exhibit diminished cell growth at high pressure of 25 MPa at 24° or at low temperature of 10°–15° at atmospheric pressure (∼0.1 MPa = 1 bar = 0.9869 atm = 1.0197 kg of force/cm2; to avoid confusion, MPa is used throughout) although tryptophan–prototrophic strains can efficiently grow under the same conditions. The introduction of the TRP1 gene or tryptophan permease genes TAT1 and TAT2 (Heitman et al. 1993; Schmidt et al. 1994; Beck et al. 1999) in a multicopy vector enabled cells to grow at high pressure and low temperature (Abe and Horikoshi 2000; Abe and Iida 2003). Mutations in the catalytic domain of Rsp5 ubiquitin ligase (Abe and Iida 2003), the cytoplasmic tails of Tat2 (Nagayama et al. 2004) or ubiquitin-specific protease genes DOA4, UBP6, or UBP14 (Miura and Abe 2004) result in marked stabilization of Tat2 and/or Tat1, and thereby the mutants grow at high pressure and low temperature. The effect of high pressure on tryptophan auxotrophic strains is also similar to the effect of an immunosuppressive drug, rapamycin (Beck et al. 1999), from the aspect of downregulation of Tat2 and the arrest of cell cycle in the G1 phase (Abe and Horikoshi 2000). However, high pressure and rapamycin treatment differ in terms of the regulation on Npr1 and the general amino acid permease Gap1. In contrast to a rapid dephosphorylation of Npr1 and induction of Gap1 expression upon rapamycin treatment, high pressure did not affect the phosphorylation state of Npr1, and it decreased the level of Gap1 protein, suggesting that the pressure-sensing pathway is likely to be independent of the Npr1 function (Abe and Horikoshi 2000).
To achieve a molecular understanding of cellular responses to changes in pressure and temperature, we screened the yeast deletion library (Giaever et al. 2002) to isolate mutants that were defective in growth under high pressure and low temperature. It would provide a unique clue to linking the disrupted gene and its function through thermal energy and work, by varying temperature and hydrostatic pressure, respectively. Then we would analyze the rate-limiting step on the growth of a mutant strain from the two mutually dependent energies (supplemental Figure S1 at http://www.genetics.org/supplemental/).
Our results highlight the marked similarity of the effects of high pressure and low temperature at the genomewide level. The approach and results described here offer a novel route to gaining insights into amino acid availability, nutrient sensing, membrane trafficking, or organization of macromolecules arising from presumed changes in the activation volume, and the activation energy upon the loss of genes. We focus on the role of the exit from rapamycin-induced growth arrest (EGO) complex to distinguish between responses to high pressure/low temperature and known responses to rapamycin treatment and describe that the EGO complex is involved in the regulation of amino acid uptake.
MATERIALS AND METHODS
Yeast strains, culture media, and plasmids:
The EUROSCARF yeast deletion library (cat. no. 95400.H3, Invitrogen, Carlsbad, CA) containing 4828 haploid gene deletion mutants and the parental strain BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0; wild type) were used in this study (Giaever et al. 2002). The following strains were kindly provided by C. De Virgilio of the University of Geneva Medical School, Switzerland (Dubouloz et al. 2005): Y2922 (wild type), CDV207 (ego1Δ), CDV208 (ego3Δ), and CDV209 (gtr2Δ); double-deletion mutants ego1Δnpr1Δ, ego1Δarg3Δ, ego1Δcpa2Δ, and ego1Δcbp6Δ; CDV213 (wild type, EGO1-GFP), CDV214 (wild type, GTR2-GFP), and CDV215 (wild type, EGO3-GFP). Cells were grown in YPD (1% bacto yeast extract, 2% bacto peptone, 2% d-glucose), synthetic dextrose (SD, 0.67% yeast extract nitrogen base w/o amino acids, adenine 20 mg/liter, uracil 20 mg/liter, methionine 20 mg/liter, tryptophan 40 mg/liter, histidine-HCl 20 mg/liter, leucine 90 mg/liter, lysine-HCl 30 mg/liter, 2% d-glucose) medium and synthetic complete (SC, 0.67% yeast extract nitrogen base w/o amino acids, adenine sulfate 20 mg/liter, uracil 20 mg/liter, tryptophan 40 mg/liter, histidine-HCl 20 mg/liter, leucine 90 mg/liter, lysine-HCl 30 mg/liter, arginine-HCl 20 mg/liter, methionine 20 mg/liter, tyrosine 30 mg/liter, isoleucine 30 mg/liter, phenyalanine 50 mg/liter, glutamic acid 100 mg/liter, aspartic acid 100 mg/liter, threonine 200 mg/liter, serine 400 mg/liter, 2% d-glucose) medium. YPD medium was used for the first qualitative screening. SC medium was used for the subsequent quantitative analysis. The 5× SC medium contains five-fold concentration of SD with the regular concentration of nonessential amino acids. SD medium was used for examining amino acid auxotrophy of some mutant strains.
Plasmids:
The entire coding region and its upstream and downstream noncoding region for the EGO1 allele were amplified using oligonucleotides, XbaI-EGO1-F1 (GCTCTAGAGC-AGCCTCGTTAGTGCCTTCTTCAATATCC) and XbaI-EGO1-R1 (GCTCTAGAGC-CTCTTGGTTTTTAGGATGTTTTCCCGGC). Likewise, those of the EGO3 allele were amplified using oligonucleotides, XbaI-EGO3-F1 (GCTCTAGAGC-ATGGTTGTTTACTGCACGTTGCCTTTGT) and XbaI-EGO3-R1 (GCTCTAGAGC-AAAGCTGTCATGTAGGGCCCTCTGAGCA). The resultant DNA fragments were digested with XbaI (underlined) and were inserted into YCplac33 (URA3 CEN4) to give pEGO1c and pEGO3c. pL137 (CEN4 URA3) containing GTR1 regulated by its own promoter was kindly provided by T. Sekiguchi of Kyusyu University, Japan (Nakashima et al. 1999). pCDV987 (CEN4 URA3) containing GTR2 regulated by its own promoter was kindly provided by C. De Virgilio (Dubouloz et al. 2005). YCplac33, YCplac111 (LEU2 CEN4), pRS313 (HIS3 CEN4), and pKI (LYS2 2μ) were used to confer prototrophy for uracil, leucine, histidine, and lysine together on strains of the BY4742 genetic background.
Screening of mutants defective in growth under high pressure and low temperature:
To determine screening conditions, we first compared the growth of the wild-type strain and the trp1Δ mutant in YPD medium in 96-well culture plates at pressures from 0.1 MPa to 50 MPa and temperatures from 4° to 37°. Judging from the gross yield of cell mass, a clear difference was observed in culture at 35 MPa and 24° (high-pressure condition) for 4 days and 0.1 MPa and 6° (low-temperature condition) for 4 days. Small aliquots from 4828 mutant cell cultures were transferred to fresh YPD medium in 96-well plates, followed by incubation at 24° overnight. Then, 3 μl of each preculture was transferred to fresh YPD medium in 96-well plates. After sealing the plates with sterilized plastic film, the cells were subjected to pressure of 35 MPa at 24° in hydrostatic chambers (Rigo-sha, Saitama, Japan) using a hand pump (TP200L, Teramecs, Kyoto, Japan) or to low temperature at 0.1 MPa and 6°. After 4 days, the growth-cell yields were checked visually. For more quantitative analysis, the candidate mutant cells were grown in SC medium at 0.1 MPa and 24° with vigorous shaking (150 rpm) in the exponential phase of growth (OD600 < 1.5). Then the culture was diluted with SC medium to an OD600 value of 0.15. The diluted cultures were placed in sterilized tubes and the tubes were sealed with parafilm. The culture tubes were subjected to high pressure of 25 MPa at 24° in hydrostatic chambers (PV100-360 and PV100-500, Teramecs) or to low temperature of 0.1 MPa at 15° for 20 hr. At the end of the culture period, the pressure was released and apparent optical density was measured at 600 nm (OD600ap) using a spectrophotometer. The OD600 value, which was proportional to cell density, was calculated using a conversion formula obtained by a polynomial approximation in a separate experiment using a spectrophotometer,
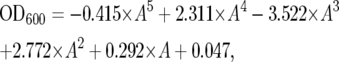 |
where A is OD600ap. For example, the OD600ap values of 0.5, 1.0, and 1.5 are comparable to the OD600 values of 0.58, 1.48, and 3.38, respectively, and are comparable to 6.96 × 106, 1.78 × 107, and 4.06 × 107 cells/ml in our experiment. The cell density of the culture was determined using hemocytometers.
Amino acid uptake assay:
The following radiolabeled amino acids were purchased from Moravek Biochemicals (Brea, CA): l-[4,5-3H] leucine (MT-672E, 2.85 TBq/mmol), l-[4,5-3H] lysine (MT-909, 1.67 TBq/mmol), and l-[2,5-3H] histidine (MT905, 1.63 TBq/mmol). Cells of the wild type, ego1Δ and gtr1Δ were incubated in SC medium at 0.1 MPa and 24°, 25 MPa and 24°, and 0.1 MPa and 15° for 5 hr. After the release from high-pressure and low-temperature incubation, the uptake of amino acid was analyzed at 0.1 MPa and 24° as described previously (Abe and Iida 2003). Data are expressed as mean values of amino acid incorporated (DPM 107 cells−1 min−1) with standard deviations obtained from three to five independent experiments.
RESULTS AND DISCUSSION
Screening of gene deletion mutants defective in growth under high pressure and low temperature:
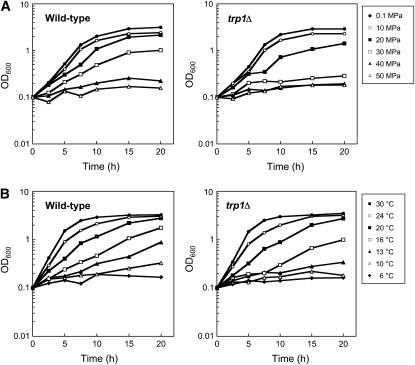
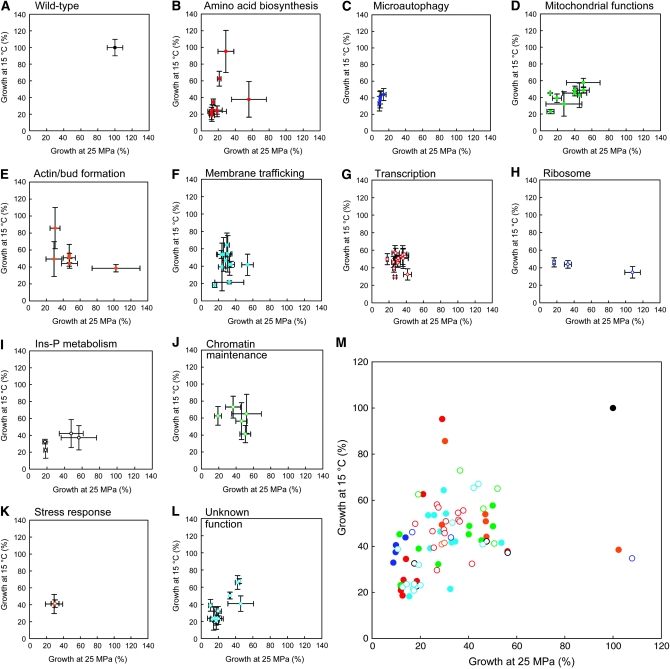
To identify genes responsible for growth under high pressure at low temperature, we screened the yeast deletion library consisting of 4828 haploid deletion mutants. All of the mutants were cultured in 96-well plates at 35 MPa and 24° or 0.1 MPa and 6° for 4 days. The first gross screening anticipated 1022 candidate strains including 779 high-pressure-sensitive mutant and 560 low-temperature-sensitive mutant strains with a significant overlap of 317 strains. To evaluate the growth of the candidates more quantitatively, it was feasible to standardize conditions by measuring the OD600 values. Figure 1 indicates the growth properties of the wild-type strain in SC medium in comparison with the trp1Δ mutant, a previously known mutant showing growth defects (Abe and Horikoshi 2000). The difference in the ability to grow between the two strains was clear at pressures between 20 and 30 MPa at 24°, and at temperatures between 13° and 16° at 0.1 MPa. Therefore, further analysis was performed under three culture conditions, 0.1 MPa and 24° (normal condition), 25 MPa and 24° (high-pressure condition), and 0.1 MPa and 15° (low-temperature condition). At 0.1 MPa and 24°, the wild-type strain grew to the OD600 value of 5.0 ± 0.14 (n = 10) in 20 hr when the culture started with an OD600 value of 0.15. We chose a cutoff value of OD600 1.0 for the normal growth condition and excluded 33 slow-growth strains from the 1022 mutants. At 25 MPa and 24°, the wild-type strain grew to an OD600 value of 2.24 ± 0.18 (n = 10) in 20 hr (45% with respect to normal growth). At 0.1 MPa and 15°, the wild-type strain grew to an OD600 value of 1.41 ± 0.14 (n = 10) in 20 hr (28.3% with respect to normal growth). The mutants exhibited various degrees of diminished growth at high pressure and low temperature. To assign high-pressure sensitivity, we set a threshold value of 22.5% with respect to the OD600 value of normal growth of individual mutants (50% with respect to the wild-type strain). To assign low-temperature sensitivity, we set a threshold value of 14.2% with respect to the OD600 value of normal growth (50% with respect to the wild-type strain). We then obtained 309 candidate strains including 152 high-pressure-sensitive mutants and 239 low-temperature-sensitive mutants. During the course of the experiments, however, we became aware that the cell density of the preculture largely affected the ability to grow at high pressure and low temperature. Once the preculture OD600 value exceeded ∼3.0, the cells resumed growth with some delay after exposure to high pressure or low temperature. Accordingly, this underestimates the ability to grow and overestimates the number of growth-deficient mutants. We refined the analysis by maintaining the preculture OD600 value at <1.5. This allowed us to identify 80 strains including 71 high-pressure-sensitive mutants and 56 low-temperature-sensitive mutants with an overlap of 47 strains (Table 1). It should be noted that most of the 80 mutants exhibited normal cell growth at high temperature of 37° except for the mot2Δ, vps45Δ, and mdj1Δ mutants (data not shown). Thus, the reasons for growth defects at high pressure and low temperature could generally differ from those at high temperature. Genes identified in the screening are involved in diverse cellular functions. In particular, genes involved in amino acid biosynthesis, microautophagy, a subset of genes involved in mitochondrial functions, and membrane trafficking are ranked in the top 20 of which deletion causes marked growth defects at high pressure and/or low temperature (Table 1, footnotes c and d). The remainders comprise genes involved in actin organization and bud formation, inositol phosphate metabolism, transcription and mRNA degradation, ribosomal functions, chromatin maintenance, stress response, and unknown open reading frames (ORFs). Next we describe their possible roles in growth at high pressure and low temperature.
Figure 1.—
Growth properties of the wild-type strain BY4742 and the trp1Δ mutant. Cells were cultured under the pressures shown at 24° (A) or at the temperatures shown at 0.1 MPa (B). The OD600ap value was measured immediately after decompression. The OD600ap values were converted into OD600 values using the formula described in materials and methods.
TABLE 1.
Genes required for growth under high-pressure and low-temperature conditions
| Name
|
OD600 at 20 hr
|
Relative growth to 0.1 MPaa
|
Relative growth to the wild typeb
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Standard | Systematic | 0.1 MPa, 24° | 25 MPa, 24° | 0.1 MPa, 15° | 25 MPa, 24° (%) | 0.1 MPa, 15° (%) | 25 MPa, 24° (% wt) | 0.1 MPa, 15° (% wt) | Description |
| Wild type | 5.0 ± 0.1 | 2.2 ± 0.2 | 1.4 ± 0.1 | 45.0 ± 4.1 | 28.3 ± 2.8 | 100.0 | 100.0 | ||
| Amino acid biosynthesis | |||||||||
| TRP1 | YDR007W | 4.7 ± 0.6 | 0.3 ± 0.1 | 0.3 ± 0.0 | 5.4 ± 1.1 | 6.4 ± 0.4 | 11.9 ± 2.6c | 22.6 ± 1.5c | Phosphoribosylanthranilate isomerase |
| TRP4 | YDR354W | 4.9 ± 0.2 | 0.3 ± 0.1 | 0.3 ± 0.1 | 5.4 ± 1.3 | 5.9 ± 2.1 | 11.9 ± 3.0c | 20.9 ± 7.5c | Anthranilate phosphoribosyl transferase |
| THR4 | YCR053W | 4.2 ± 0.6 | 0.2 ± 0.0 | 0.2 ± 0.1 | 5.7 ± 1.3 | 5.3 ± 2.0 | 12.6 ± 2.8c | 18.6 ± 7.2c | Threonine synthase |
| ARO2 | YGL148W | 5.0 ± 0.1 | 0.3 ± 0.0 | 0.4 ± 0.1 | 5.9 ± 0.5 | 7.2 ± 1.7 | 13.0 ± 1.0c | 25.4 ± 6.0c | Bifunctional chorismate synthase and flavin reductase |
| ARO1 | YDR127W | 4.7 ± 0.2 | 0.3 ± 0.0 | 0.5 ± 0.0 | 6.3 ± 0.9 | 9.8 ± 1.1 | 14.1 ± 2.0c | 34.5 ± 3.8d | Arom protein, catalyzes steps 2–6 in the biosynthesis of chorismate |
| TRP5 | YGL026C | 4.1 ± 0.2 | 0.3 ± 0.1 | 0.3 ± 0.1 | 8.3 ± 2.7 | 7.1 ± 1.4 | 18.6 ± 6.0d | 24.9 ± 4.9c | Tryptophan synthase |
| HOM3 | YER052C | 4.3 ± 0.7 | 0.4 ± 0.3 | 0.3 ± 0.1 | 8.4 ± 4.9 | 6.6 ± 1.8 | 18.7 ± 10.9d | 23.4 ± 6.5c | Aspartate kinase |
| TRP2 | YER090W | 5.2 ± 0.1 | 0.5 ± 0.0 | 0.9 ± 0.1 | 9.5 ± 1.0 | 17.7 ± 2.5 | 21.2 ± 2.2d | 62.6 ± 8.8 | Anthranilate synthase |
| LEU3 | YLR451W | 4.4 ± 0.5 | 0.6 ± 0.2 | 1.2 ± 0.2 | 13.1 ± 4.2 | 27.0 ± 7.1 | 29.1 ± 9.4 | 95.3 ± 25.2 | Zinc-finger transcription factor |
| SER1 | YOR184W | 4.6 ± 0.4 | 1.2 ± 0.5 | 0.5 ± 0.3 | 25.3 ± 9.2 | 10.7 ± 6.1 | 56.2 ± 20.4 | 37.9 ± 21.4d | 3-Phosphoserine aminotransferase |
| Microautophagy | |||||||||
| GTR2 | YGR163W | 5.1 ± 0.4 | 0.2 ± 0.1 | 0.5 ± 0.2 | 4.0 ± 0.7 | 9.3 ± 2.5 | 8.8 ± 1.6c | 32.9 ± 8.8d | Cytoplasmic GTP-binding protein |
| GTR1 | YML121W | 5.0 ± 0.2 | 0.2 ± 0.0 | 0.5 ± 0.1 | 4.4 ± 0.3 | 10.6 ± 2.8 | 9.8 ± 0.7c | 37.5 ± 9.8d | Cytoplasmic GTP-binding protein |
| EGO1 | YKR007W | 5.0 ± 0.1 | 0.2 ± 0.0 | 0.6 ± 0.1 | 4.4 ± 0.6 | 11.5 ± 2.2 | 9.9 ± 1.2c | 40.5 ± 7.9d | Component of the EGO/GSE complex |
| EGO3 | YBR077C | 5.2 ± 0.2 | 0.3 ± 0.1 | 0.6 ± 0.1 | 6.2 ± 1.3 | 12.4 ± 2.1 | 13.9 ± 2.9c | 43.8 ± 7.3 | Component of the EGO/GSE complex |
| Mitochondrial function | |||||||||
| MRPL22 | YNL177C | 4.3 ± 0.1 | 0.2 ± 0.0 | 0.5 ± 0.0 | 5.1 ± 0.1 | 12.8 ± 0.2 | 11.3 ± 0.2c | 45.2 ± 0.7 | Mitochondrial ribosomal protein of the large subunit |
| MRF1 | YGL143C | 4.3 ± 0.3 | 0.2 ± 0.1 | 0.3 ± 0.1 | 5.3 ± 1.8 | 6.5 ± 0.8 | 11.8 ± 4.1c | 23.1 ± 2.7c | Mitochondrial polypeptide chain release factor |
| CAF17 | YJR122W | 3.9 ± 0.3 | 0.3 ± 0.1 | 0.4 ± 0.1 | 8.7 ± 2.4 | 11.0 ± 1.4 | 19.3 ± 5.3d | 39.0 ± 5.1d | Mitochondrial protein that interacts with Ccr4 |
| ACO1 | YLR304C | 3.7 ± 0.2 | 0.4 ± 0.3 | 0.3 ± 0.1 | 12.3 ± 9.4 | 9.1 ± 4.2 | 27.5 ± 21.0d | 32.2 ± 14.7c | Aconitase, required for the tricarboxylic acid cycle |
| MRP51 | YPL118W | 4.1 ± 0.3 | 0.7 ± 0.3 | 0.5 ± 0.0 | 18.0 ± 6.3 | 12.8 ± 0.9 | 40.1 ± 14.0 | 45.2 ± 3.3 | Mitochondrial ribosomal protein of the large subunit |
| MRPL38 | YKL170W | 3.6 ± 0.2 | 0.6 ± 0.0 | 0.5 ± 0.1 | 18.1 ± 0.7 | 13.8 ± 1.3 | 40.3 ± 1.5 | 48.8 ± 4.6 | Mitochondrial ribosomal protein of the large subunit |
| ATP15 | YPL271W | 3.5 ± 0.0 | 0.7 ± 0.0 | 0.4 ± 0.1 | 20.4 ± 1.0 | 12.0 ± 4.2 | 45.4 ± 2.2 | 42.6 ± 14.8 | ɛ-Subunit of the F1 sector of mitochondrial F1F0 ATP synthase |
| MDJ1 | YFL016C | 2.6 ± 0.5 | 0.6 ± 0.2 | 0.4 ± 0.1 | 22.5 ± 8.8 | 16.3 ± 1.5 | 50.1 ± 19.5 | 57.7 ± 5.3 | Protein involved in folding of mitochondrially synthesized proteins |
| MSY1 | YPL097W | 4.3 ± 0.2 | 1.0 ± 0.1 | 0.6 ± 0.0 | 22.6 ± 3.0 | 13.8 ± 0.9 | 50.2 ± 6.7 | 48.7 ± 3.2 | Mitochondrial tyrosyl-tRNA synthetase |
| Actin organization/bud formation | |||||||||
| LTE1 | YAL024C | 3.8 ± 0.3 | 0.5 ± 0.1 | 0.5 ± 0.2 | 13.0 ± 4.2 | 14.0 ± 5.8 | 29.0 ± 9.4 | 49.3 ± 20.4 | Putative GDP/GTP exchange factor |
| HOF1 | YMR032W | 3.9 ± 0.1 | 0.5 ± 0.1 | 0.9 ± 0.3 | 13.6 ± 2.6 | 24.3 ± 6.8 | 30.2 ± 5.8 | 85.7 ± 24.2 | Bud neck-localized, SH3 domain-containing protein |
| SLM3 | YDL033C | 4.1 ± 0.7 | 0.9 ± 0.1 | 0.6 ± 0.1 | 21.1 ± 0.6 | 15.3 ± 3.6 | 47.0 ± 1.3 | 53.9 ± 12.8 | tRNA-specific 2-thiouridylase |
| CLA4 | YNL298W | 3.9 ± 0.3 | 0.8 ± 0.1 | 0.6 ± 0.1 | 21.2 ± 3.2 | 14.4 ± 1.5 | 47.2 ± 7.1 | 50.8 ± 5.4 | Cdc42-activated signal-transducing kinase |
| CDC50 | YCR094W | 4.0 ± 0.1 | 0.8 ± 0.2 | 0.5 ± 0.1 | 21.3 ± 4.3 | 12.5 ± 1.6 | 47.4 ± 9.6 | 44.1 ± 5.6 | Endosomal protein that regulates cell polarity |
| SLM6 | YBR266C | 3.2 ± 0.9 | 1.4 ± 0.1 | 0.3 ± 0.1 | 46.0 ± 12.8 | 10.9 ± 1.2 | 102.3 ± 28.4 | 38.5 ± 4.3d | Protein with a potential role in actin cytoskeleton organization |
| Membrane trafficking | |||||||||
| VID24 | YBR105C | 4.8 ± 0.0 | 0.3 ± 0.1 | 0.2 ± 0.0 | 6.9 ± 1.2 | 5.2 ± 0.8 | 15.4 ± 2.6c | 18.3 ± 2.9c | Peripheral membrane protein located at Vid vesicles |
| VPS34 | YLR240W | 3.2 ± 0.2 | 0.3 ± 0.0 | 0.5 ± 0.0 | 10.5 ± 1.9 | 15.1 ± 1.2 | 23.3 ± 4.3d | 53.4 ± 4.4 | Phosphatidylinositol 3-kinase |
| SEC22 | YLR268W | 3.8 ± 0.1 | 0.4 ± 0.1 | 0.4 ± 0.3 | 10.8 ± 1.9 | 11.1 ± 7.8 | 24.1 ± 4.3d | 39.1 ± 27.4d | R-SNARE protein |
| PEP3 | YLR148W | 3.7 ± 0.4 | 0.4 ± 0.2 | 0.6 ± 0.2 | 11.6 ± 3.7 | 15.2 ± 5.5 | 25.7 ± 8.3d | 53.6 ± 19.4 | Vacuolar peripheral membrane protein |
| CHC1 | YGL206C | 2.5 ± 0.3 | 0.3 ± 0.0 | 0.3 ± 0.0 | 12.7 ± 1.8 | 13.1 ± 2.4 | 28.3 ± 4.0d | 46.4 ± 8.5 | Clathrin heavy chain |
| PEP5 | YMR231W | 3.5 ± 0.4 | 0.5 ± 0.1 | 0.7 ± 0.2 | 13.4 ± 1.3 | 18.2 ± 3.9 | 29.7 ± 2.8 | 64.3 ± 13.7 | Peripheral vacuolar membrane |
| VPS45 | YGL095C | 3.1 ± 0.4 | 0.4 ± 0.0 | 0.5 ± 0.2 | 13.8 ± 2.2 | 15.3 ± 5.9 | 30.7 ± 5.0 | 54.2 ± 21.0 | Protein of the Sec1/Munc-18 family |
| ERG24 | YNL280C | 4.2 ± 0.3 | 0.6 ± 0.4 | 0.3 ± 0.0 | 14.6 ± 7.6 | 6.1 ± 0.5 | 32.5 ± 16.9 | 21.5 ± 1.7c | C-14 sterol reductase, acts in ergosterol biosynthesis |
| VPS54 | YDR027C | 2.2 ± 0.5 | 0.3 ± 0.1 | 0.3 ± 0.1 | 14.8 ± 0.3 | 11.8 ± 3.4 | 33.0 ± 0.6 | 41.7 ± 12.1d | Component of the Golgi-associated retrograde protein complex |
| AKR1 | YDR264C | 3.7 ± 0.4 | 0.6 ± 0.1 | 0.4 ± 0.1 | 15.5 ± 1.8 | 11.9 ± 1.0 | 34.5 ± 4.1 | 42.0 ± 3.6d | Palmitoyl transferase |
| SAC1 | YKL212W | 4.1 ± 0.2 | 1.0 ± 0.2 | 0.5 ± 0.2 | 24.1 ± 3.0 | 11.8 ± 3.5 | 53.7 ± 6.8 | 41.6 ± 12.3d | Lipid phosphoinositide phosphatase |
| Inositol phosphate metabolism | |||||||||
| PLC1 | YPL268W | 4.5 ± 0.2 | 0.4 ± 0.1 | 0.4 ± 0.0 | 7.9 ± 1.1 | 9.2 ± 0.8 | 17.6 ± 2.4c | 32.6 ± 2.7d | Phosphoinositide-specific phospholipase C |
| ARG82 | YDR173C | 4.6 ± 0.0 | 0.4 ± 0.0 | 0.3 ± 0.1 | 8.3 ± 0.9 | 6.4 ± 2.8 | 18.4 ± 2.0d | 22.6 ± 9.8c | Inositol polyphosphate multikinase |
| PHO88 | YBR106W | 3.9 ± 0.7 | 0.8 ± 0.3 | 0.5 ± 0.2 | 21.4 ± 6.2 | 11.9 ± 4.7 | 47.5 ± 13.7 | 42.1 ± 16.6 | Probable membrane protein, involved in phosphate transport |
| KCS1 | YDR017C | 3.5 ± 0.2 | 0.9 ± 0.3 | 0.4 ± 0.1 | 25.3 ± 9.1 | 10.5 ± 4.1 | 56.3 ± 20.2 | 37.2 ± 14.3d | Inositol hexaphosphate kinase |
| Transcription/mRNA degradation | |||||||||
| SNF6 | YHL025W | 3.8 ± 0.2 | 0.3 ± 0.0 | 0.5 ± 0.1 | 8.1 ± 0.5 | 14.1 ± 1.8 | 17.9 ± 1.2d | 49.8 ± 6.3 | Subunit of the SWI/SNF chromatin remodeling complex |
| MOT2 | YER068W | 3.5 ± 1.1 | 0.4 ± 0.2 | 0.4 ± 0.1 | 11.3 ± 1.1 | 13.2 ± 1.7 | 25.1 ± 2.4d | 46.6 ± 6.1 | Component of the Ccr4-NOT transcription regulatory complex |
| POP2 | YNR052C | 3.2 ± 0.4 | 0.4 ± 0.0 | 0.4 ± 0.0 | 11.5 ± 0.8 | 11.2 ± 1.3 | 25.6 ± 1.7d | 39.4 ± 4.5d | RNase of the DEDD superfamily |
| SHE3 | YBR130C | 2.7 ± 0.3 | 0.3 ± 0.0 | 0.2 ± 0.0 | 12.1 ± 0.8 | 8.4 ± 0.3 | 26.9 ± 1.7d | 29.6 ± 1.2c | Protein that acts as an adaptor between Myo4 and the She2-mRNA complex |
| CDC73 | YLR418C | 4.2 ± 0.2 | 0.5 ± 0.0 | 0.7 ± 0.1 | 12.1 ± 0.3 | 16.4 ± 2.1 | 26.9 ± 0.6d | 58.1 ± 7.4 | Constituent of Paf1 complex with RNA polymerase II |
| RPB4 | YJL140W | 3.8 ± 0.0 | 0.5 ± 0.1 | 0.6 ± 0.1 | 12.4 ± 1.9 | 16.1 ± 1.6 | 27.6 ± 4.3d | 56.8 ± 5.5 | RNA polymerase II subunit B32 |
| HFI1 | YPL254W | 3.4 ± 0.5 | 0.5 ± 0.1 | 0.5 ± 0.1 | 13.4 ± 1.5 | 13.4 ± 1.6 | 29.8 ± 3.3 | 47.3 ± 5.5 | Adaptor protein required for structural integrity of the SAGA complex |
| PAF1 | YBR279W | 2.3 ± 0.3 | 0.3 ± 0.0 | 0.3 ± 0.1 | 13.5 ± 1.4 | 12.9 ± 1.4 | 30.1 ± 3.0 | 45.7 ± 5.0 | RNA polymerase II-associated protein |
| ELF1 | YKL160W | 5.0 ± 0.1 | 0.7 ± 0.1 | 0.7 ± 0.0 | 13.8 ± 2.2 | 14.4 ± 0.9 | 30.6 ± 4.9 | 50.8 ± 3.3 | Transcription elongation factor |
| SNF1 | YDR477W | 4.4 ± 0.2 | 0.7 ± 0.1 | 0.7 ± 0.1 | 16.1 ± 3.2 | 15.4 ± 3.0 | 35.7 ± 7.2 | 54.5 ± 10.7 | AMP-activated serine/threonine protein kinase |
| SRB5 | YGR104C | 3.7 ± 0.3 | 0.6 ± 0.2 | 0.5 ± 0.1 | 16.1 ± 2.9 | 14.6 ± 2.8 | 35.9 ± 6.5 | 51.4 ± 10.1 | Subunit of the RNA polymerase II mediator complex |
| TAF14 | YPL129W | 2.9 ± 0.3 | 0.5 ± 0.1 | 0.4 ± 0.1 | 16.3 ± 2.7 | 14.4 ± 3.2 | 36.3 ± 6.1 | 50.7 ± 11.5 | Subunit of TFIID, TFIIF, and SWI/SNF complexes |
| CCR4 | YAL021C | 4.3 ± 0.2 | 0.7 ± 0.1 | 0.7 ± 0.0 | 17.0 ± 1.3 | 15.7 ± 0.3 | 37.9 ± 2.8 | 55.6 ± 1.1 | Component of the Ccr4-Not transcriptional complex |
| SAP155 | YFR040W | 4.4 ± 0.1 | 0.8 ± 0.1 | 0.4 ± 0.1 | 18.6 ± 2.1 | 9.2 ± 1.8 | 41.5 ± 4.8 | 32.4 ± 6.3c | Protein that forms a complex with the Sit4 protein phosphatase |
| Ribosome | |||||||||
| RPL1B | YGL135W | 3.9 ± 0.1 | 0.3 ± 0.0 | 0.5 ± 0.1 | 7.5 ± 0.6 | 13.1 ± 1.5 | 16.7 ± 1.4c | 46.1 ± 5.2 | N-terminally acetylated protein component of the large ribosomal subunit |
| RPL21A | YBR191W | 3.8 ± 0.3 | 0.6 ± 0.1 | 0.5 ± 0.1 | 14.7 ± 1.8 | 12.4 ± 1.2 | 32.7 ± 4.0 | 43.8 ± 4.3 | Protein component of the large ribosomal subunit |
| RPS30B | YOR182C | 4.1 ± 0.1 | 2.0 ± 0.1 | 0.4 ± 0.1 | 48.5 ± 4.1 | 9.8 ± 1.9 | 107.9 ± 9.1 | 34.8 ± 6.6d | Protein component of the small ribosomal subunit |
| Chromatin maintenance | |||||||||
| NBP2 | YDR162C | 4.8 ± 0.2 | 0.4 ± 0.1 | 0.9 ± 0.2 | 8.6 ± 1.8 | 17.7 ± 3.1 | 19.2 ± 4.0c | 62.6 ± 10.9 | Protein involved in the HOG pathway |
| YAF9 | YNL107W | 4.4 ± 0.1 | 0.7 ± 0.2 | 0.9 ± 0.2 | 16.4 ± 3.9 | 20.6 ± 3.6 | 36.5 ± 8.7 | 72.9 ± 12.8 | Subunit of the NuA4 histone H4 acetyltransferase complex and the SWR1 complex |
| IES2 | YNL215W | 4.6 ± 0.3 | 1.0 ± 0.1 | 0.7 ± 0.3 | 20.7 ± 2.9 | 15.9 ± 6.2 | 46.2 ± 6.4 | 56.3 ± 21.7 | Associated protein with the INO80 chromatin remodeling complex |
| CGI121 | YML036W | 3.5 ± 0.2 | 0.8 ± 0.1 | 0.4 ± 0.1 | 22.8 ± 2.8 | 11.7 ± 2.9 | 50.7 ± 6.2 | 41.2 ± 10.1d | Promoting telomere uncapping and elongation and transcription |
| ARD1 | YHR013C | 4.1 ± 0.4 | 1.0 ± 0.4 | 0.8 ± 0.3 | 23.4 ± 7.7 | 18.4 ± 6.5 | 52.1 ± 17.1 | 65.0 ± 22.9 | Subunit of the N-terminal acetyltransferase NatA |
| Stress response | |||||||||
| HSP31 | YDR533C | 4.8 ± 1.1 | 0.6 ± 0.3 | 0.6 ± 0.2 | 13.0 ± 4.6 | 11.6 ± 3.2 | 28.9 ± 10.3d | 40.8 ± 11.4d | Possible chaperone and cysteine protease |
| YDJ1 | YNL064C | 1.7 ± 0.3 | 0.2 ± 0.0 | 0.2 ± 0.0 | 13.5 ± 2.1 | 11.7 ± 1.3 | 30.1 ± 4.7 | 41.5 ± 4.5d | Protein chaperone involved in regulation of the Hsp90 and Hsp70 functions |
| Unknown genes | |||||||||
| AVL9 | YLR114C | 4.6 ± 0.5 | 0.2 ± 0.0 | 0.5 ± 0.1 | 4.8 ± 0.9 | 11.0 ± 1.9 | 10.7 ± 1.9c | 38.8 ± 6.9d | ND |
| — | YDR008C | 5.0 ± 0.2 | 0.3 ± 0.1 | 0.3 ± 0.0 | 5.7 ± 1.1 | 6.3 ± 0.5 | 12.7 ± 2.5c | 22.4 ± 1.9c | Complementary to TRP1 |
| — | YKL098W | 4.6 ± 0.3 | 0.3 ± 0.2 | 0.3 ± 0.2 | 6.4 ± 3.2 | 6.7 ± 3.9 | 14.3 ± 7.1c | 23.6 ± 13.8c | ND |
| DLT1 | YMR126C | 4.7 ± 0.2 | 0.4 ± 0.1 | 0.3 ± 0.1 | 7.6 ± 1.7 | 6.1 ± 1.7 | 17.0 ± 3.8c | 21.6 ± 5.9c | ND |
| CSF1 | YLR087C | 3.3 ± 0.3 | 0.3 ± 0.0 | 0.2 ± 0.0 | 7.8 ± 1.3 | 5.9 ± 1.0 | 17.3 ± 3.0c | 20.8 ± 3.4c | Protein required for fermentation at low temperature |
| — | YHR151C | 4.6 ± 0.2 | 0.4 ± 0.1 | 0.3 ± 0.2 | 7.8 ± 2.8 | 6.7 ± 3.6 | 17.4 ± 6.2c | 23.6 ± 12.9c | ND |
| — | YPR153W | 4.2 ± 1.2 | 0.4 ± 0.1 | 0.4 ± 0.1 | 8.7 ± 1.7 | 9.0 ± 1.5 | 19.4 ± 3.9d | 31.9 ± 5.4c | ND |
| — | YBR255W | 4.8 ± 0.1 | 0.4 ± 0.1 | 0.3 ± 0.1 | 9.0 ± 2.4 | 6.5 ± 1.9 | 20.1 ± 5.4d | 23.1 ± 6.6c | ND |
| — | YGL218W | 3.0 ± 0.4 | 0.4 ± 0.1 | 0.4 ± 0.0 | 15.0 ± 0.3 | 14.2 ± 1.1 | 33.3 ± 0.7 | 50.2 ± 4.1 | ND |
| — | YDL172C | 4.6 ± 0.1 | 0.9 ± 0.0 | 0.9 ± 0.1 | 18.9 ± 1.2 | 18.5 ± 2.2 | 42.1 ± 2.7 | 65.4 ± 7.9 | ND |
| — | YDL173W | 4.7 ± 0.2 | 0.9 ± 0.0 | 0.9 ± 0.0 | 19.8 ± 0.4 | 19.0 ± 0.8 | 44.1 ± 0.9 | 67.0 ± 2.9 | ND |
| — | YDR442W | 1.7 ± 0.2 | 0.4 ± 0.1 | 0.2 ± 0.1 | 20.6 ± 6.7 | 11.6 ± 2.5 | 45.9 ± 14.8 | 40.8 ± 8.9d | ND |
ND, not determined.
Relative growth is represented as percentages of OD600 values at 25 MPa and 24° or 0.1 MPa and 15° compared with the OD600 values obtained at 0.1 MPa and 24° after 20 hr of culture.
Values are represented as percentages of growth relative to those obtained with the wild-type strain at 25 MPa and 24° or 0.1 MPa and 15°.
Genes ranked in the top 20 in terms of significance.
Genes ranked from 21 to 40 in terms of significance.
Amino acid biosynthesis:
The amino acid biosynthetic genes we obtained were ARO1, HOM3, THR4, ARO2, TRP4, TRP2, TRP1, TRP5, and SER1 (Table 1 and Figure 2B). On the basis of our previous findings, the sensitivity of aro1Δ, aro2Δ, trp4Δ, trp2Δ, trp1Δ, and trp5Δ mutants to high pressure and low temperature can be attributable to auxotrophy for tryptophan, and probably for phenylalanine and tyrosine. Aro1 is a pentafunctional protein that catalyzes five steps in the biosynthesis of chorismate. Chorismate is a precursor of tryptophan, tyrosine, and phenylalanine (Duncan et al. 1988). Aro2 is a bifunctional chorismate synthase and flavin reductase, which catalyzes the conversion of 5-enolpyruvylshikimate 3-phosphate to form chorismate (Jones et al. 1991). To determine amino acid auxotrophy, these mutants were cultured on SD agar plates supplemented with appropriate amino acids at 0.1 MPa and 24°. As expected, aro1Δ and aro2Δ strains required not only tryptophan but also tyrosine and phenylalanine for cell growth (data not shown). The result suggests that high pressure and low temperature impair the uptake of these aromatic amino acids. One of the unknown strains that displayed substantial high-pressure/low-temperature sensitivity was ydr008cΔ. This is a hypothetical ORF that is located on the coding strand opposite TRP1. Thus, deletion of YDR008C also results in disruption of TRP1 and hence confers high-pressure and low-temperature sensitivity. The ydr008cΔ mutant indeed displayed tryptophan auxotrophy (data not shown). For an unknown reason, deletion of TRP2 resulted in moderate sensitivity to low temperature compared with other tryptophan biosynthetic mutants.
Figure 2.—
Representation of growth defects of deletion mutants on a pressure–temperature diagram. The OD600 values are normalized to those of the wild-type strain BY4742 (A) in Table 1. Eighty mutants were classified into 11 categories: (B) amino acid biosynthesis, (C) microautophagy, (D) mitochondrial functions, (E) actin organization and bud formation, (F) membrane trafficking, (G) transcription and mRNA degradation, (H) ribosome function, (I) inositol phosphate (Ins-P) metabolism, (J) chromatin maintenance, (K) stress response, and (L) unknown function. All results are merged in M. Data are represented as mean values with standard deviations obtained from three independent experiments.
HOM3 encodes l-aspartate 4-phosphate-transferase that catalyzes the first step in the common pathway for methionine and threonine biosynthesis (Rafalski and Falco 1988). We confirmed that the hom3Δ mutant required both methionine and threonine for growth (data not shown). THR4 encodes threonine synthase that catalyzes the formation of threonine from o-phosphohomoserine (Aas and Rognes 1990; Ramos and Calderon 1994). We confirmed that the thr4Δ mutant required threonine for growth (data not shown). SER1 encodes 3-phosphoserine aminotransferase that catalyzes the formation of phosphoserine from 3-phosphohydroxypyruvate (Melcher et al. 1995). We confirmed that the ser1Δ mutant required serine for growth (data not shown). These findings suggest that, in a manner analogous to tryptophan uptake, high pressure and low temperature impair the ability to take up threonine and serine, potentially inactivating their permeases. The ser1Δ mutant exhibited more moderate sensitivity than others (Table 1 and Figure 2B). SC medium contains a higher concentration of serine (400 mg/liter) than other amino acids. Therefore, a high concentration of serine is likely to compensate for the presumed defect in serine uptake as observed in the case of tryptophan (Abe and Horikoshi 2000).
The leu3Δ mutant was defective in growth at high pressure but grew normally at low temperature reflecting a fundamental rule that pressure and temperature separately affect any reactions (Table 1 and Figure 2B). LEU3 encodes a zinc-finger transcription factor that regulates genes involved in the biosynthesis and uptake of branched-chain amino acids (Friden and Schimmel 1988; Nielsen et al. 2001). Strain BY4742 is a leucine auxotroph, and its growth depends on external leucine. Because Leu3 activates the transcription of BAP2 that encodes a leucine permease, deletion of LEU3 is likely to decrease the Bap2 protein level. Hence, it should be more difficult for leu3Δ mutant cells to take up sufficient amounts of leucine from the medium.
Microautophagy and regulation of amino acid uptake:
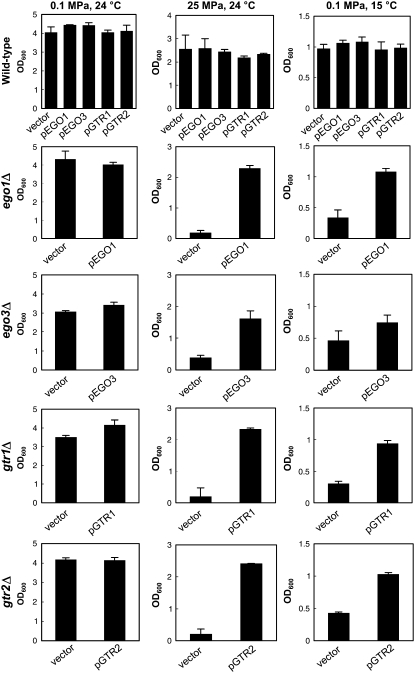
We found that genes encoding components of the EGO complex were substantially responsible for growth under high pressure and low temperature (Table 1 and Figure 2C). The EGO complex is a vacuolar membrane-associated protein complex that consists of Ego1 (also known as Meh1 and Gse2), Ego3 (also known as Slm4, Nir1, and Gse1), and Gtr2 (Dubouloz et al. 2005). It is known that cells lacking one of EGO1, EGO3, and GTR2 normally show growth arrest after the addition of rapamycin to culture medium, but no longer recover to the growth phase upon release of the rapamycin block. Deletion of EGO1, EGO3, or GTR2 resulted in marked growth defects at high pressure and low temperature. Introduction of the plasmid containing EGO1, EGO3, or GTR2 into the ego1Δ, ego3Δ, or gtr2Δ mutant, respectively, restored growth at high pressure and low temperature to the wild-type level, confirming the role of these genes (Figure 3). Gtr2 and Gtr1 are homologous GTP-binding proteins under the control of Ran/Gsp1-GTPase, which plays an essential role in nuclear macromolecular trafficking (Nakashima et al. 1999). The gtr1Δ mutant also exhibited similar growth defects (Table 1 and Figure 2C). Introduction of the plasmid containing GTR1 into the gtr1Δ mutant also restored growth at high pressure and low temperature, confirming the role of GTR1 (Figure 3). The localization of Ego1-GFP (CDV213), Ego3-GFP (CDV215), and Gtr2-GFP (CDV214) was unaffected upon shifts to high-pressure and low-temperature incubation for 10 hr (supplemental Figure S2 at http://www.genetics.org/supplemental/). Our finding suggests a novel aspect of microautophagy in terms of the regulation of growth under stressful conditions.
Figure 3.—
The EGO complex is essential for growth at high pressure and low temperature. Cells containing the specified plasmids were cultured at 0.1 MPa and 24°, 25 MPa and 24°, or 0.1 MPa and 15°. The OD600 values were measured after 20 hr of culture. pEGO1, low-copy plasmid containing EGO1 driven by its own promoter; pEGO3, low-copy plasmid containing EGO3 driven by its own promoter; pL137, low-copy plasmid containing GTR1 driven by its own promoter; pCDV987, containing GTR2 driven by its own promoter. Data are represented as mean values with standard deviations obtained from three independent experiments.
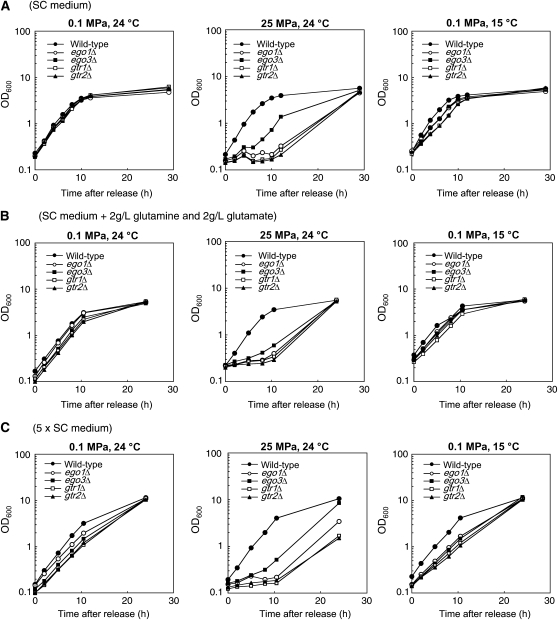
To extend our understandings of the EGO complex function, we attempted to find the difference and similarity between effects of rapamycin treatment and high pressure/low temperature with respect to growth recovery and the levels of intracellular glutamine/glutamate, key regulators in the target of rapamycin (TOR) signaling pathway. Upon the addition of rapamycin, ego1Δ, ego3Δ, and gtr2Δ mutants are known to enter into growth arrest, but fail to resume growth following rapamycin release (Dubouloz et al. 2005). While cell growth was immediately resumed following release from low-temperature incubation (0.1 MPa and 15° for 10 hr) in all strains tested, ego1Δ, gtr1Δ, and gtr2Δ mutants failed to resume growth following release from high-pressure incubation (25 MPa and 24° for 10 hr) during at least the first 12 hr (Figure 4A). Until 30 hr following the pressure release, these mutants restored the ability of growth. A similar result was obtained with the cells after being cultured for 20 hr under high pressure or low temperature (data not shown). The result suggests that the EGO complex is not essential for recovery of growth but is required for quick resumption from high-pressure-induced growth arrest as well as for growth under high pressure and low temperature. Unlike rapamycin treatment, high pressure and low temperature did not induce the formation of autophagic bodies in the vacuole when the wild-type cells were cultured in SC medium containing phenylmethylsulfonylfluoride (data not shown). It is reported that rapamycin exerts transcriptional regulations in a manner dependent on the function of Tor or Ego3/Nir1 or the shared function of Tor and Ego3/Nir1 (Huang et al. 2004). While GAT1, GAP1, GDH1, DAL2, DAL3, and DUR1 and -2 are upregulated by rapamycin treatment by 2- to 57-fold (Huang et al. 2004), these genes remained unchanged upon the shifts of growth condition to high pressure or low temperature except for downregulation of GDH1 by 2.5-fold under low temperature (the microarray data is available at Gene Expression Omnibus, accession no. GSE9136). These results suggest that cells respond to high pressure and low temperature in a distinct program from that in response to rapamycin treatment.
Figure 4.—
Cell growth of the EGO complex mutants following release from high-pressure and low-temperature incubation. (A) Cells were cultured in SC medium at 0.1 MPa and 24° following release from a 10-hr incubation at 0.1 MPa and 24°, 25 MPa and 24°, or 0.1 MPa and 15°. (B) Cells were cultured in glutamine- and glutamate-enriched SC medium at 0.1 MPa and 24° following release from a 10-hr incubation at 0.1 MPa and 24°, 25 MPa and 24° or 0.1 MPa and 15°. (C) Cells were cultured in 5× SC medium at 0.1 MPa and 24° following release from a 10-hr incubation at 0.1 MPa and 24°, 25 MPa and 24°, or 0.1 MPa and 15°. Data are confirmed by duplicate experiments.
It is reported that npr1Δ, arg3Δ, cpa2Δ, and cbp6Δ render the ability of growth following release from rapamycin treatment in the ego mutants arising from the increase in intracellular glutamine/glutamate levels (Dubouloz et al. 2005). To elucidate the role of intracellular glutamine/glutamate, we analyzed the effect of npr1Δ, arg3Δ, cpa2Δ, and cbp6Δ mutations on growth of the ego1Δ mutant (CDV207). The double mutants used in this experiment are isogenic with Y2922. The egoΔ and gtrΔ mutations also caused growth defects in strain Y2922 under high pressure and low temperature as observed in strain BY4742 (supplemental Figure 3A at http://www.genetics.org/supplemental/). In contrast to restoration of growth observed in the double mutants following release from rapamycin treatment (Dubouloz et al. 2005), the ego1Δnpr1Δ, ego1Δarg3Δ, ego1Δcpa2Δ, and ego1Δcbp6Δ mutants were unable to grow during high-pressure and low-temperature incubation, indicating that the increase in intracellular glutamine/glutamate does not compensate for the growth defects in the ego1Δ mutant (supplemental Figure 3A). Regarding growth following release from high-pressure incubation, there was a difference in property of the ego1Δ mutant between the BY4742 and Y2922 genetic background. The ego1Δ mutant in the Y2922 background (CDV207) resumed growth without delay following release from high-pressure incubation (supplemental Figure 3B) while that in the BY4742 background exhibited a significant delay (Figure 4A). This means that strain Y2922 is not applicable to validate the role of intracellular glutamine/glutamate in terms of restoration of growth following high-pressure release. Indeed, none of npr1Δ, arg3Δ, cpa2Δ, and cbp6Δ mutations facilitated growth of the ego1Δ mutant following high-pressure release (supplemental Figure 3B). Instead, we examined supply of excess glutamine and glutamate on growth of the egoΔ and gtrΔ mutants in the BY4742 genetic background. We found that glutamine and glutamate at concentrations of 2 g/liter in SC medium did not compensate for growth defects in ego1Δ, ego3Δ, gtr1Δ, and gtr2Δ mutants during high-pressure and low-temperature incubation (data not shown) and did not restore quick recovery from high-pressure release (Figure 4B). These results suggest that the EGO complex has a role in a manner independent of intracellular glutamine and glutamate levels to regulate growth under high pressure and low temperature. We assumed that the uptake of other nutritional substrates such as glucose, vitamins, or ions might be diminished in the egoΔ and gtrΔ mutants under high pressure and low temperature, and thereby the growth may be impaired. However, it is unlikely because the egoΔ and gtrΔ mutants still exhibited the growth defects in a 5× SC medium under high pressure and low temperature (data not shown) and also showed retardation of growth resumption after release from high-pressure incubation (Figure 4C).
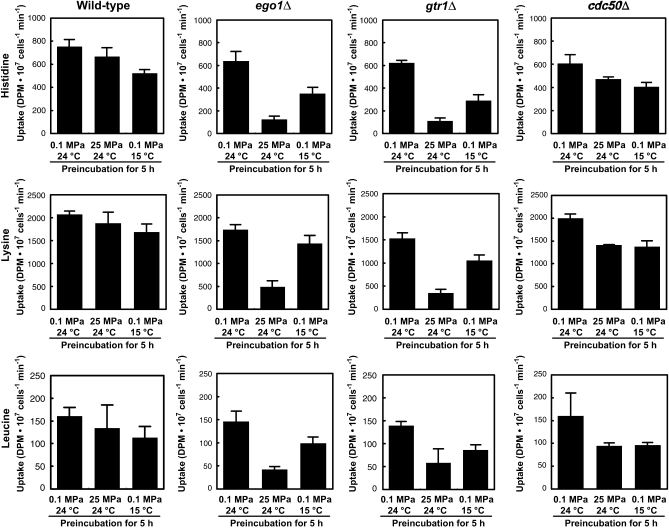
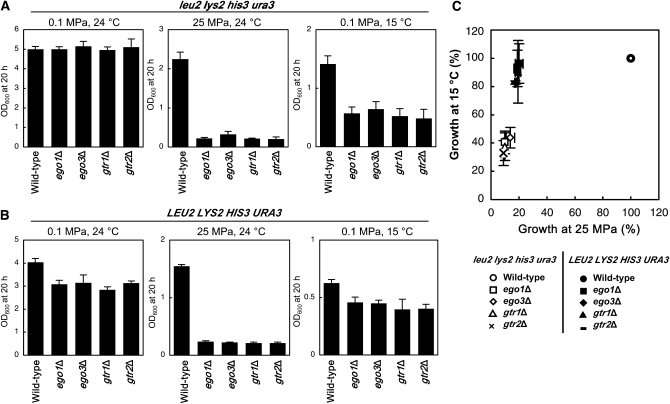
Recently, the EGO complex has been revealed to be equivalent to the GTPase-containing complex for the general amino acid permease Gap1 sorting in the endosome (GSE) complex, containing Gtr1, Gtr2, Ego3/Gse1, Ego1/Gse2, and Ltv1 (Gao and Kaiser 2006). The EGO/GSE complex plays a specific role in trafficking Gap1 out of the endosome to the plasma membrane upon a shift of growth on glutamate to urea. Deletion of individual EGO/GSE components was shown to cause a decrease in other EGO/GSE components, suggesting a hierarchical order of assembly (Gao and Kaiser 2006). This supports our finding that deletion mutants for EGO1, EGO3, GTR1, and GTR2 exhibit equivalent growth defects at high pressure and low temperature (Table 1 and Figure 2C). Gap1 is not necessary for growth at high pressure and low temperature under our nitrogen-rich culture conditions. Because strain BY4742 has amino acid auxotrophy for histidine, lysine, and leucine, there was a possibility that amino acid uptake was impaired under high pressure and low temperature when the EGO/GSE complex was lost. This could potentially be due to the lack of surface delivery of permease proteins, and thereby the mutant cells exhibited growth defects. To examine that possibility, we analyzed the uptake of histidine, lysine, and leucine at 0.1 MPa and 24° after the wild-type, ego1Δ, and gtr1Δ strains had been incubated in SC medium for 5 hr at 0.1 MPa and 24°, 25 MPa and 24°, and 0.1 MPa and 15°. For comparison, we also examined mutants of which high-pressure and low-temperature sensitivity was caused by a defect possibly unrelated to trafficking of amino acid peremeases. CDC50 encodes a protein required for actin cytoskeleton organization. The cdc50Δ mutant showed growth defects under high pressure and low temperature (Table 1 and Figure 2E, see below). Under the normal condition, there was no marked difference in the uptake of histidine between the wild-type and the mutant strains (Figure 5). While the uptake of histidine was not considerably affected by high pressure and low temperature in the wild-type strain, it was significantly diminished in the ego1Δ and gtr1Δ mutants. The effect of high pressure and low temperature in the cdc50Δ mutant was the comparable level with the wild-type strain (Figure 5). Similar results were obtained for the uptake of lysine and leucine (Figure 5). In our preliminary observation, deletions for other genes such as PLC1, CAF17, and SEC22 (see below) resulted in a slight decrease in amino acid uptake following high-pressure and low-temperature incubation, but the effect was not remarkable compared with the losses of EGO1 and GTR1 (data not shown). These findings support the hypothesis that the EGO/GSE complex is required to maintain the activity of amino acid uptake under high pressure and low temperature, probably due to the appropriate cell-surface delivery of permease proteins. Apparently, high pressure caused a more significant defect in amino acid uptake in the ego1Δ and gtr1Δ mutants than low temperature did. This would explain the differential susceptibility to high pressure and low temperature in the ego1Δ and gtr1Δ mutants in which relative growth was ∼9% (high pressure) and 37% (low temperature) compared with that of the wild-type strain (Table 1 and Figure 2C) and delay in resumption of growth following release from high-pressure incubation (Figure 4A, 25 MPa, 24°). If our hypothesis is valid, prototrophy for the nutrients should restore the ability of ego1Δ and gtr1Δ mutants to grow under high pressure and low temperature. Four plasmids containing LEU2, LYS2, HIS3, and URA3 together were introduced into individual mutants to confer nutrient prototrophy. Then the cells were cultured under high pressure and low temperature in the presence (Figure 6A) or absence (Figure 6B) of leucine, lysine, histidine, and uracil in SC medium. We found that nutrient prototrophy restored low-temperature growth to egoΔ and gtrΔ mutants, suggesting that the susceptibility to low temperature of the mutants is attributable to defects in the uptake of nutrients (Figure 6C). However, nutrient prototrophy did not restore high-pressure growth to the mutants. Accordingly, the EGO/GSE complex is likely to play a primary role in the cell-surface delivery of amino acid permeases at low temperature, but under high pressure it is likely to play multiple roles in cellular functions in addition to permease delivery. It has been shown in Gap delivery that the tyrosine-containing sequence KPRWYR within the cytosolic C-terminal domain of Gap1 is required for binding of Gap1 to Gtr2 and for the efficient sorting from the endosome to the plasma membrane (Gao and Kaiser 2006). Bap2/Bap3, Lyp1, and Hip1, which are possible amino acid permeases for leucine, lysine, and histidine, respectively, do not have KPRWYR sequence. Thus, it should be elucidated whether these permeases physically bind to any of the EGO/GSE components, or there is a protein that mediates binding of the permeases to the complex.
Figure 5.—
Amino acid uptake following release from high-pressure and low-temperature incubation. Cells of the wild type, ego1Δ, gtr1Δ, and cdc50Δ were cultured at 0.1 MPa and 24°, 25 MPa and 24°, or 0.1 MPa and 15° for 5 hr followed by the measurement of uptake of histidine, lysine, and leucine at 0.1 MPa and 24° using 3H-labeled substrates (see materials and methods). Data are represented as mean values with standard deviations obtained from three to seven independent experiments.
Figure 6.—
Cell growth of the EGO/GSE complex mutants with or without nutriment auxotrophy. The wild-type and the EGO/GSE complex mutants with (A) auxotrophy for leucine, lysine, histidine, and uracil or (B) prototrophy were cultured at 0.1 MPa and 24°, 25 MPa and 24°, or 0.1 MPa and 15°. The prototrophic strains were cultured in SC medium in the absence of leucine, lysine, histidine, and uracil for maintenance of the plasmids. The OD600 values were measured after 20 hr of culture. (C) The OD600 values are normalized to those of the wild-type strain BY4742 as shown in Figure 2. Data are represented as mean values with standard deviations obtained from three independent experiments.
Using a DNA microarray on strain BY4742 (Abe 2007a), we found that numbers of the genes encoding 24-amino-acid permeases and their homologs were downregulated upon shifts to growth under high pressure and low temperature (Table 2; the DNA microarray data is available at Gene Expression Omnibus, accession no. GSE9136). In particular, genes classified into the amino acid permease cluster I and cluster II were markedly decreased. With reduced levels of transcripts, and thereby synthesis of permease proteins could be decreased, it becomes more important to deliver the permease proteins to the cell surface to receive sufficient amounts of essential amino acids for growth. In addition, some of the genes encoding glucose/hexose transporters were also downregulated under high pressure and low temperature (Table 2). Accordingly, the level of glucose/hexose transporters in the plasma membrane might be decreased upon the loss of the EGO/GSE complex when the cells are exposed to high pressure. This might provide an account for the fact that the egoΔ and gtrΔ mutants were not rescued by conferring amino acid prototrophy (Figure 6C), but it is controversial because the 5× SC medium (containing 10% glucose) did not compensate for the growth defects of the egoΔ and gtrΔ mutants. Further biochemical analysis is necessary to validate the hypothesis that the EGO/GSE complex plays a role in the delivery of various membrane proteins, but our finding will be a good starting point to elucidate its role.
TABLE 2.
Transcription of genes encoding amino acid permeases, hexose transporters, and their homologs in response to high pressure and low temperaturea
| Ratio
|
|||
|---|---|---|---|
| Gene | HP/normal | LT/normal | |
| AAP cluster Ib | TAT1 | 0.13 | 0.22 |
| TAT2 | 0.38 | 0.46 | |
| GNP1 | 0.32 | 0.44 | |
| AGP1 | 0.53 | 0.74 | |
| BAP2 | 0.15 | 0.27 | |
| BAP3 | 0.27 | 0.67 | |
| AAP cluster IIb | HIP1 | 0.48 | 0.46 |
| GAP1 | 0.25 | 0.59 | |
| MMP1 | 0.21 | 0.35 | |
| SAM1 | 0.58 | 0.63 | |
| AAP cluster IIIb | CAN1 | 0.60 | 0.80 |
| ALP1 | 0.87 | 0.94 | |
| LYP1 | 0.78 | 0.61 | |
| AAP unclusteredb | DIP5 | 1.06 | 0.84 |
| PUT4 | 0.38 | 0.92 | |
| AGP2 | 0.76 | 1.05 | |
| AGP3 | 1.31 | 1.33 | |
| SSY1 | 1.25 | 1.22 | |
| MUP1 | 0.20 | 0.36 | |
| MUP3 | 2.69 | 1.58 | |
| TPO5 | 0.94 | 0.62 | |
| HNM1 | 0.86 | 1.03 | |
| BIO5 | 0.85 | 1.08 | |
| UGA4 | 1.04 | 1.28 | |
| Glucose/hexose transporter | HXT1 | 0.30 | 0.17 |
| HXT2 | 0.09 | 0.20 | |
| HXT3 | 0.88 | 0.74 | |
| HXT4 | 0.54 | 2.95 | |
| HXT5 | 0.20 | 0.85 | |
| HXT6 | 0.61 | 3.32 | |
| HXT7 | 0.57 | 2.96 | |
| HXT8 | 0.69 | 0.78 | |
| HXT9 | 0.99 | 1.04 | |
| HXT10 | 0.69 | 0.82 | |
| HXT11 | 1.22 | 1.27 | |
| HXT13 | 1.19 | 1.25 | |
| HXT14 | 1.01 | 0.87 | |
| HXT15 | 0.88 | 0.72 | |
| HXT16 | 0.94 | 0.94 | |
| HXT17 | 0.99 | 0.86 | |
Normal, 0.1 MPa and 24°; HP (high pressure), 25 MPa and 24°; LT (low temperature), 0.1 MPa and 15°.
Data from the Gene Expression Omnibus, accession no. GSE9136.
Classification of amino acid permeases (AAPs) according to Nelissen et al. (1997).
Mitochondrial function:
We obtained 11 deletion mutants for mitochondrial proteins in our functional screening (Table 1 and Figure 2D). In particular, deletion of MRF1, MRPL22, and CAF17 resulted in marked growth defects. The lack of MRF1 causes mitochondrial genome instability (Pel et al. 1992; Askarian-Amiri et al. 2000). In accordance with mitochondrial functions, glutamine is a key factor that may act in the TOR signaling pathway (Dubouloz et al. 2005). To test the possibility that the mitochondrial defects could result in a decrease in intracellular glutamate/glutamine levels, possibly due to the low production of 2-oxoglutarate in the tricarboxylic acid (TCA) cycle, we determined the requirement for glutamine and glutamate in the 11 mitochondrial mutants. Among them, the aco1Δ and caf17Δ mutants required either glutamate or glutamine for growth but others did not (data not shown). The result suggests that mitochondrial functions other than glutamine/glutamate synthesis also impinge on the high-pressure and low-temperature responsive pathway. ACO1 encodes aconitase that catalyzes cis-aconitate to isocitrate. In addition to glutamate auxotrophy upon its deletion (Gangloff et al. 1990), Aco1 is required for mitochondrial DNA maintenance (Chen et al. 2005). CAF17 encodes a mitochondrial protein that interacts in the two-hybrid system with Ccr4, a component of the Ccr4-Not transcriptional complex (Clark et al. 2004). Deletion of CCR4 also causes growth defects at high pressure and low temperature (see below).
Actin organization, cell polarity, and membrane trafficking:
CDC50 encodes a protein required for actin cytoskeleton organization and trafficking of proteins between the Golgi complex and the endosome/vacuole. The lack of CDC50 is known to confer low-temperature sensitivity (Misu et al. 2003). This is attributable to a defect in the establishment of actin networks resulting from improper localization of regulator proteins for polarized growth such as Bni1 and Gic1 at low temperature (Saito et al. 2004). We found that the cdc50Δ mutant also exhibited growth defects at high pressure, suggesting that high pressure perturbs the localization of actin and/or associated proteins in the absence of Cdc50 (Table 1 and Figure 2E). Cdc50 is known to associate with Drs2, a P-type ATPase of the aminophospholipid translocase that functions in phospholipids asymmetry (Chen et al. 1999; Natarajan et al. 2004; Saito et al. 2004; Chen et al. 2006). The lack of DRS2 is also known to cause low-temperature sensitivity (Chen et al. 1999). We found that the drs2Δ mutant exhibited high-pressure sensitivity although it is not included in Table 1. This is because the OD600 value of the drs2Δ cell culture at 0.1 MPa and 24° reached only 0.8 in 20 hr when the culture started with an OD600 value of 0.15, and hence the drs2Δ mutant was assumed to be a slow growth strain, which was excluded from Table 1. Interestingly, deletion of YBR255W, which encodes an unknown protein but was implicated in the synthetic sick/lethal interaction with Drs2 (Schuldiner et al. 2005), caused marked growth defects at high pressure and low temperature (Table 1 and Figure 2E). Our finding is consistent with a previous report indicating that ybr255wΔ cells exhibited a slower growth rate at 15° although the growth was not examined at high pressure (Sanjuan et al. 1999). These results suggest that the Drs2-Cdc50 complex and Ybr255w redundantly have a role in establishing appropriate actin networks at high pressure and low temperature. In our unpublished observation, actin cytoskeleton visualized with rhodamine-labeled phalloidin was disorganized in the drs2Δ, cdc50Δ, and ybr255wΔ mutants with dotted distribution in the cortical region and the cytoplasm when the cells were cultured under high pressure or low temperature (our unpublished results). Therefore, the Drs2-Cdc50 complex and Ybr255w are likely to play an essential role in actin network formation at high pressure and low temperature by modulating protein trafficking appropriately. It should be elucidated whether disorganization of actin cytoskeleton is the primary cause of the growth defects in the drs2Δ, cdc50Δ, and ybr255wΔ mutants, or the consequence of diminished ability of membrane trafficking resulting from the phospholipid asymmetry defect.
MSS4 is an essential gene encoding a single phosphoinositide-4-phosphate (PI4P) 5-kinase that generates PI4,5 P2, which is required for actin cytoskeleton organization at the plasma membrane (Desrivieres et al. 1998; Homma et al. 1998). Audhya et al. (2004) identified numbers of genes encoding PI4,5P2 effectors using a genomewide lethality screen including six SLM genes. SLM4 is identical to EGO3/GSE1 (Audhya et al. 2004; Dubouloz et al. 2005; Gao and Kaiser 2006). In addition to the slm4Δ mutant, the slm3Δ mutant exhibited high-pressure and low-temperature sensitivity but the slm6Δ mutant exhibited only low-temperature sensitivity (Table 1 and Figure 2E). Slm1 and its homolog Slm2 are downstream effectors of TORC2, the Tor2 kinase-containing complex, which regulates the actin cytoskeleton organization (Audhya et al. 2004). The slm1Δslm2Δ mutant is inviable. We found that deletion mutants for SLM1, SLM2, and SLM5 grew well under high pressure and low temperature like the wild-type strain, suggesting that TORC2 may not be required for high-pressure and low-temperature growth or that there are other proteins redundant in the functions of SLM1, SLM2, and SLM5 (data not shown). The slm3Δ mutation is known to be synthetically lethal with the cdc73Δ mutation (Tong et al. 2004). CDC73 encodes a constituent of the Paf1 complex with RNA polymerase II (Shi et al. 1997; Krogan et al. 2003). Interestingly, the cdc73Δ mutant was also unable to grow under high pressure and low temperature (see below). In our first screening on 96-well plates, deletion of BOI2, ROM2, CAP2, or GIM4, which are known to display synthetic lethality with the mss4ts mutation, caused growth defects at high pressure and low temperature (data not shown).
Lte1 is a putative guanine nucleotide exchange factor required for mitotic exit at low temperature (Wickner et al. 1987; Shirayama et al. 1994a,b). Lte1 is localized to the bud cortex and bud cytoplasm from the S phase to M phase and is uniformly distributed in the G1 phase (Yoshida et al. 2003). We found that the lte1Δ mutant was defective in growth not only at low temperature but also at high pressure (Table 1 and Figure 2E). The restriction of Lte1 to the bud cortex depends on septins Cdc42, Kel1, and Cla4 but is independent of the actin cytoskeleton and microtubule formation (Seshan et al. 2002). The PAK-like protein kinase Cla4 is required for Lte1 phosphorylation and accordingly for localization (Seshan et al. 2002). Interestingly, the cla4Δ mutant was also defective in growth at high pressure and low temperature even though the effect was moderate (Table 1 and Figure 2E). HOF1 encodes a bud neck-localized Src homology 3-domain-containing protein that is required for cytokinesis and regulation of actomyosin ring dynamics and septin localization (Kamei et al. 1998; Lippincott and Li 1998). The hof1Δ mutant exhibited growth defects at high pressure (Table 1 and Figure 2E). The proteins encoded by the genes shown here have roles in the organization of macromolecules associated with a serial event in the bud neck to exit from mitosis. We speculate that these proteins mediate the cellular processes by decreasing the system volume change and energy barriers associated with the reactions.
Membrane trafficking:
Numerous genes involved in membrane trafficking appear to be responsible for growth at high pressure and low temperature (Table 1 and Figure 2F). We assume that delivery of newly synthesized proteins to appropriate locations, e.g., the bud neck, cell surface, or cell wall, is diminished at high pressure and low temperature when one of the genes listed in Table 1 is lost. ERG24 encodes the C-14 sterol reductase that catalyzes a step in ergosterol biosynthesis (Lorenz and Parks 1992). The erg24 mutants are viable but accumulate the abnormal sterol ignosterol (ergosta-8, 14 dienol) (Parks et al. 1995). Although the erg24 mutation has not been examined for growth at high pressure and low temperature, the erg6 mutation is known to confer cold sensitivity when combined with tryptophan auxotrophy (Gaber et al. 1989). In our unpublished observation, tryptophan prototrophic strains carrying the erg6Δ, erg2Δ, erg3Δ, erg4Δ, or erg5Δ mutation fail to growth under high pressure and low temperature, suggesting that the structural motif of ergosterol is required for function and/or trafficking of membrane proteins under high pressure and low temperature, and hence cell growth. Kishimoto et al. (2005) demonstrated that the cdc50 mutation is synthetically lethal with the five erg mutations. In the cdc50Δ erg3Δ mutant, sterol was predominantly detected either diffusely within the cytosol or as punctate dots with a concomitant decrease in the plasma membrane resulting from a possible defect in recycling sterols to the plasma membrane. Therefore, high pressure and low temperature could lead to the situation analogous to ergosterol deficiency in which the Cdc50-Drs2 complex is unable to function in membrane trafficking and the organization of actin cytoskeleton.
In mammalian HeLa cells, low temperature of 15° blocks protein transport at the endoplasmic reticulum-Golgi intermediate compartment (ERGIC) (Martinez-Alonso et al. 2005). At this temperature, the Golgi complex shows long tubules and newly synthesized proteins are accumulated at the ERGIC (Martinez-Alonso et al. 2005). In this sense, vesicle-mediated protein transport steps in HeLa cells are expected to have high activation energies. High pressure as well as low temperature act to increase lipid order, and hence both potentially have adverse effects on biological membranes that are adapted to atmospheric pressure and moderate temperature. Therefore, the proteins encoded by membrane trafficking genes revealed in this study are likely to contribute to decreasing the activation volume and activation energy associated with steps in vesicle budding and/or vesicle fusion in the endocytic pathway.
Transcriptional regulation and mRNA degradation:
Genes involved in transcription and mRNA degradation comprise a major class of essential genes for growth at high pressure and low temperature. The Ccr4-Not complex is a global regulator of transcription consisting of five Not proteins (Not1–Not5), Pop2 (also known as Caf1), Caf40, Caf130, and Ccr4 (Liu et al. 1998; Tucker et al. 2001; Collart 2003). Among them, Not1 is the scaffold of the complex and is essential for viability (Maillet et al. 2000). We found that deletion of CCR4, POP2, or NOT4 (also known as MOT2) resulted in substantial growth defects at high pressure and low temperature (Table 1 and Figure 2G). Low-temperature sensitivity of the ccr4Δ and pop2Δ mutants was reported previously (Hata et al. 1998). Ccr4 and Pop2 associate with a central portion of the N-terminal domain of Not1 and function as the major yeast deadenylases that play a role in mRNA degradation (Tucker et al. 2001). Meanwhile, Not4 associates with the C-terminal domain of Not1 and functions as an E3 ubiquitin ligase on ubiquitination (Albert et al. 2002). Our result suggests that the loss of one of the components destabilizes the Ccr4-Not1 complex at high pressure and low temperature, or alternatively the assembly of the complex is impaired by high pressure and low temperature.
In addition to the Ccr4-Not complex, we found that two complex forms of RNA polymerase II, one including Srb proteins (Liu et al. 1997) and the second including Paf1 and Cdc73 (Chang et al. 1999), were indispensable for growth at high pressure and low temperature (Table 1 and Figure 2G). Because double mutants paf1Δsrb5Δ, paf1Δccr4Δ, cdc73Δccr4Δ, and ccr4Δsrb5Δ are lethal, the functions of these transcriptional regulators are redundant (Chang et al. 1999). The paf1Δ and ccr4Δ mutants are sensitive to cell wall-damaging agents such as caffeine or sodium dodecyl sulfate and sensitive to high temperature of 35.5° or 38° (Chang et al. 1999). The high-temperature sensitivity of the two mutants is suppressed by the presence of 1 m sorbitol. Thus, Paf1 and Ccr4 are thought to be required for the integrity of the cell wall. In our recent study of global transcriptional profiling using a DNA microarray, the DAN/TIR cell wall mannoprotein genes were dramatically induced by high pressure and low temperature (Abe 2007a). Accordingly, the expression of these cell wall protein genes might be regulated by Paf1 and Ccr4 in response to high pressure and low temperature.
Ribosomal proteins:
The assembly of ribosomes is known to be impaired at low temperature in the absence of certain ribosomal proteins. The rpl39Δ mutation causes an increased translational error frequency at low temperature and thereby confers low temperature-sensitive growth (Dresios et al. 2000). However, no ribosomal protein mutant has been examined under high-pressure conditions. We found that the rpl1bΔ and rpl21aΔ mutants were defective in growth at both high pressure and low temperature, but the rps30bΔ mutant exhibited growth defects only at low temperature (Table 1 and Figure 2H). Oligomerization of proteins is a typical high-pressure-sensitive process because it is usually accompanied by large volume changes. Because the ribosome is an enormously large structure, its assembly is likely to be sensitive to high pressure. Rpl1b and Rpl21a could function to stabilize ribosomal structures and subunit association at high pressure. Although biochemical analysis is required to identify the role of these ribosomal proteins, our present data suggest that Rps30b has a role different from that of Rpl1b and Rpl21a in ribosomal functions in terms of differential responses to high pressure and low temperature.
Other genes and interaction networks:
Deletion mutants for PLC, ARG82, and KCS1 exhibited growth defects at high pressure and low temperature (Table 1 and Figure 2I). Plc1, Arg82, and Kcs1 catalyze steps in inositol polyphosphate metabolism and play roles in various cellular functions including negative regulation of the phosphate signal transduction PHO pathway, remodeling of the PHO5 promoter chromatin, arginine metabolism, salt stress, and cell wall integrity (Auesukaree et al. 2005). Genes involved in chromatin remodeling (IES1), telomere maintenance (ARD1), telomere uncapping and elongation (CGI121), and histone acetylation (YAF9) were obtained although the effects were moderate (Table 1 and Figure 2J). Although their contribution to growth deficiency is evident, the role of these genes is unaccountable. We recently reported that Hsp31 has a role in high-pressure growth and obtained the same result in this study (Miura et al. 2006). In addition, YDJ1 that encodes a chaperone for the regulation of HSP90 and HSP70 functions (Schumacher et al. 1996) was obtained in this study (Table 1 and Figure 2K). Ydj1/Hsp40 and Ssa1/Hsp70 are known to cooperate with Hsp104 to unfold and reactivate denatured, aggregated proteins (Glover and Lindquist 1998). Hsp104 in association with trehalose is thought to play a role in unfolding of protein aggregates caused by hydrostatic pressure although the range of pressure (∼150 MPa) is much higher than that under pure experimental conditions (Iwahashi et al. 1997, 2000). Hsp31 and Ydj1 may act to unfold some misfolded proteins caused by high pressure and low temperature.
We identified 12 unknown or uncharacterized genes in this study (Table 1 and Figure 2L). YDL172C and YDL173W are mutually overlapping in the opposite DNA strand. Thus, the deletion of one gene means that the other is also lost. We found that the ydr442wΔ mutant had tryptophan auxotrophy in our BY4742 genetic background and thereby exhibited growth defects at high pressure and low temperature. However, the ydr442wΔ mutant was tryptophan prototrophic in the BY4741 genetic background and thereby grew at high pressure and low temperature. Therefore, YDR442W is a questionable ORF. The remainder of the mutants obtained in this study are tryptophan prototrophs.
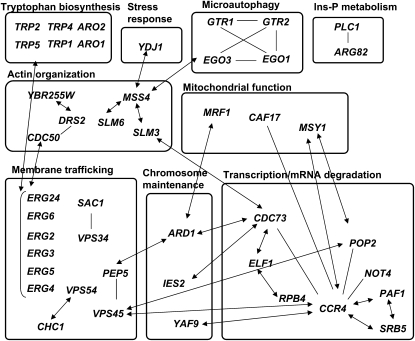
Figure 7 indicates physical and/or genetic interactions among genes responsible for high-pressure and/or low-temperature growth identified in this study according to the Saccharomyces Genome Database. Notably, 40 of the 80 genes have some interactions among themselves. This interaction map provides clues to elucidate the consequences of individual gene deletions and reveal the significance of cellular functions on growth at high pressure and low temperature.
Figure 7.—
Physical and genetic interactions among the genes identified in this study. The interactions are registered in the Saccharomyces Genome Database. Note that MSS4 is an essential gene for yeast viability and was not analyzed in this study. However, it is indicated for its importance. Arrows, genetic interactions, i.e., synthetic lethality and synthetic slow growth; solid bars, physical interactions. Ins-P, inositol phosphate.
Perspective in the study of intracellular processes as a function of pressure and temperature:
Our findings described here reveal unpredicted genes and anticipate the functional roles responsible for the growth of yeast at high pressure and low temperature, and would provide a clue to understand the survival strategies employed by deep-sea organisms. Three major classes of function we identified are (i) amino acid biosynthesis, (ii) microautophagy, and (iii) mitochondrial function. In a manner analogous to tryptophan uptake, our results suggest that the uptake of phenyalanine, tyrosine, threonine, and serine is also likely to be impaired by high pressure and low temperature, probably due to reduced activity and/or degradation of their permeases. It is necessary to analyze the stability of the permease proteins under high pressure and low temperature.
The loss of any one of the EGO/GSE constituents caused marked growth defects under high pressure and low temperature. Although the TOR signaling and high-pressure/low-temperature signaling pathways converged on the EGO complex, these were mutually different with respect to the role of intracellular glutamine/glutamate. Our finding suggests the EGO/GSE complex is required to maintain the uptake activity of leucine, lysine, and histidine under high pressure and low temperature possibly due to appropriate delivery of the permease proteins to the plasma membrane as observed in Gap1 sorting. Considering the role of microautophagy, recycling of membranes might be crucially required for growth at high pressure and low temperature. It is unlikely that recycling of cytoplasmic proteins following their uptake and degradation in the vacuole is necessary because none of the cytoplasm-to-vacuole targeting mutants showed growth defect under high pressure and low temperature. Membrane structure is susceptible to perturbations by alterations in hydrostatic pressure and temperature (see the next paragraph). The four mutants with deletions of the EGO/GSE complex grow normally at high temperature of 37°, as does the wild-type strain. In other words, the susceptibility of the mutants to pressure and temperature occurs toward increasing order and decreasing fluidity of membranes but not decreasing order and increasing fluidity caused by high temperature. Therefore, it is rationalized that the integrity of the EGO/GSE complex is crucial for the function in ordered membranes. To verify this hypothesis, a key to the answer can be provided by the measurement of biochemical activity and the endothermic nature of their reconstituted complexes in vitro as a function of pressure and temperature using differential scanning calorimetry. In addition to destabilization of preexisting macromolecular complexes, the assembly is also likely to be impaired by high pressure and low temperature. In this sense, the constituents of complexes of which the deletion causes high-pressure and low-temperature sensitivity are expected to facilitate the assembly by lowering the barriers of activation energy and activation volume that accompany any biochemical process. In view of this, pressure and temperature are effective variables to elucidate the role of individual components during the assembly of complex cellular mechanisms.
Unlike soluble monomeric proteins, membrane proteins such as permeases, channels, and pumps are likely to be susceptible to low levels of hydrostatic pressure because their functions are usually accompanied by large positive volume changes (De Smedt et al. 1979; Heremans and Wuytack 1980; Kato et al. 2002; MacDonald 2002; Abe and Iida 2003). High pressure and low temperature have an analogous effect in increasing the order of lipid bilayers (Hazel and Williams 1990; Winter 2002; Winter and Dzwolak 2004). The main phase transition temperature in artificial membranes is increased by 20°–30° at 100 MPa (Hazel and Williams 1990). This indicates that a pressure upshift of 10 MPa is equivalent to a temperature downshift of 2°–3° toward the main phase transition from liquid to gel. No clear phase transition can be observed in biological membranes, but the membranes become ordered with increasing pressure or decreasing temperature (Hazel and Williams 1990). The analogous effect of high pressure and low temperature is reflected in the growth of a tryptophan-auxotrophic strain of yeast (Abe and Horikoshi 2000). In purified pig kidney Na+/K+-ATPase, the ATP hydrolytic activity at 23.5° decreases linearly with increasing pressure toward a breakpoint at 18.5 MPa, and it decreases more rapidly over the breakpoint. A temperature upshift of 3.6° (i.e., the activity is measured at 27.1°) raises the breakpoint by 11 MPa (i.e., at 29.5 MPa) (De Smedt et al. 1979). Therefore, membrane-related cellular functions can be potentially diminished by increasing hydrostatic pressure and decreasing temperature with a pressure upshift of 10 MPa equivalent to a temperature downshift of 2°–4°. This could be the case in the loss of genes involved in microautophagy, mitochondrial function, or vesicle-mediated protein transport. In HeLa cells, protein transport is blocked at the ERGIC, and the Golgi complex exhibits long tubules containing resident glycosylation enzymes when the cells are incubated at 15° (Martinez-Alonso et al. 2005). Low temperature-induced Golgi tubulation is also observed in Vero cells but not in HepG2 or NRK cells, where instead of tubule formation, low temperature induces a shift from a typical perinuclear compact distribution to a more punctate pattern (Martinez-Alonso et al. 2005). It is interesting to examine the effect of high pressure on protein transport in various mammalian cell lines.
In our recent study, we showed that a subset of the DAN/TIR cell wall mannoprotein genes, which were well-documented to be anaerobic and cold-inducible genes (Abramova et al. 2001a,b), was dramatically upregulated by high pressure and low temperature (Abe 2007a). Abramova et al. (2001a) speculated that cells exhibited reduced membrane fluidity under hypoxia as a possible outcome of anaerobiosis and at low temperature as a result of reduced lateral diffusion and increased microviscosity. Our previous finding agrees well with those observed here in light of the effects of high pressure and low temperature on membranes.
In the present global screening, we obtained 12 functionally unknown genes responsible for growth at high pressure and low temperature including 8 highly responsible genes. Auxotrophy for tryptophan was examined in the 80 candidate mutants in the initial step, when we noted that YDR008C was encoded in the strand opposite TRP1. The loss of Ybr255w, which was anticipated to interact with Drs2, results in disorganization of the actin cytoskeleton. In this way, our approach taken in this study gives insights into assigning the functions of unknown genes to one of the categories indicated in Table 1. Experimental data on physical and/or genetic interactions in the literature are indispensable for this approach. Furthermore, our approach makes a contribution to evaluation of the results of the physical and genetic interactions obtained in co-immunoprecipitation or the yeast two-hybrid system. Genes essential for viability comprised nearly 20% of yeast genome, and they remain to be investigated in our study. The approach taken by Mnaimneh et al. (2004) will provide resources to complement the results obtained using the yeast deletion library.
High hydrostatic pressure is generally assumed to have adverse effects on biological systems, but it can serve as a useful parameter to elucidate dynamic structural changes associated with any reaction. High pressure changes the rate of reactions in a manner separable from those due to temperature. Accordingly, combined analyses using pressure and temperature offer new insights into cellular functions such as the assembly of macromolecules, membrane trafficking, or membrane protein functions. In our studies on piezophysiology, we adopt hydrostatic pressure and temperature as tools to understand complex physiological events via the volume change and activation energy associated with any reaction and equilibrium (Abe 2007b).
Acknowledgments
We thank Claudio De Virgilio and Takeshi Sekiguchi for providing plasmids and useful comments; Koki Horikoshi, Matteo Binda, Yoshiaki Kamada, Kazuma Tanaka, Masaki Mizunuma, and Akira Sakai for valuable discussions; and the Japan Society for the Promotion of Science for support (no. 18658039) to F.A.
References
- Aas, S. F., and S. E. Rognes, 1990. Nucleotide sequence of the yeast THR4 gene encoding threonine synthase. Nucleic Acids Res. 18 665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe, F., 2007. a Induction of DAN/TIR yeast cell wall mannoprotein genes in response to high hydrostatic pressure and low temperature. FEBS Lett. 581 4993–4998. [DOI] [PubMed] [Google Scholar]
- Abe, F., 2007. b Exploration of the effects of high hydrostatic pressure on microbial growth, physiology and survival: perspectives from piezophysiology. Biochem. Biosci. Biotechnol. 71 2347–2357. [DOI] [PubMed] [Google Scholar]
- Abe, F., and K. Horikoshi, 2000. Tryptophan permease gene TAT2 confers high-pressure growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 20 8093–8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe, F., and H. Iida, 2003. Pressure-induced differential regulation of the two tryptophan permeases Tat1 and Tat2 by ubiquitin ligase Rsp5 and its binding proteins, Bul1 and Bul2. Mol. Cell. Biol. 23 7566–7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramova, N., O. Sertil, S. Mehta and C. V. Lowry, 2001. a Reciprocal regulation of anaerobic and aerobic cell wall mannoprotein gene expression in Saccharomyces cerevisiae. J. Bacteriol. 183 2881–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramova, N. E., B. D. Cohen, O. Sertil, R. Kapoor, K. J. Davies et al., 2001. b Regulatory mechanisms controlling expression of the DAN/TIR mannoprotein genes during anaerobic remodeling of the cell wall in Saccharomyces cerevisiae. Genetics 157 1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert, T. K., H. Hanzawa, Y. I. Legtenberg, M. J. de Ruwe, F. A. van den Heuvel et al., 2002. Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J. 21 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askarian-Amiri, M. E., H. J. Pel, D. Guevremont, K. K. McCaughan, E. S. Poole et al., 2000. Functional characterization of yeast mitochondrial release factor 1. J. Biol. Chem. 275 17241–17248. [DOI] [PubMed] [Google Scholar]
- Audhya, A., R. Loewith, A. B. Parsons, L. Gao, M. Tabuchi et al., 2004. Genome-wide lethality screen identifies new PI4,5P2 effectors that regulate the actin cytoskeleton. EMBO J. 23 3747–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auesukaree, C., H. Tochio, M. Shirakawa, Y. Kaneko and S. Harashima, 2005. Plc1p, Arg82p, and Kcs1p, enzymes involved in inositol pyrophosphate synthesis, are essential for phosphate regulation and polyphosphate accumulation in Saccharomyces cerevisiae. J. Biol. Chem. 280 25127–25133. [DOI] [PubMed] [Google Scholar]
- Beck, T., A. Schmidt and M. N. Hall, 1999. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J. Cell Biol. 146 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant, R. E., and P. S. Sypherd, 1974. Genetic analysis of cold-sensitive ribosome maturation mutants of Escherichia coli. J. Bacteriol. 117 1082–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, M., D. French-Cornay, H. Y. Fan, H. Klein, C. L. Denis et al., 1999. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol. Cell. Biol. 19 1056–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. Y., M. F. Ingram, P. H. Rosal and T. R. Graham, 1999. Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J. Cell Biol. 147 1223–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., J. Wang, B. P. Muthusamy, K. Liu, S. Zare et al., 2006. Roles for the Drs2p-Cdc50p complex in protein transport and phosphatidylserine asymmetry of the yeast plasma membrane. Traffic 7 1503–1517. [DOI] [PubMed] [Google Scholar]
- Chen, X., R. K. Cook and P. A. Rubenstein, 1993. Yeast actin with a mutation in the “hydrophobic plug” between subdomains 3 and 4 (L266D) displays a cold-sensitive polymerization defect. J. Cell Biol. 123 1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. H., Z. Xiao and M. Fitzgerald-Hayes, 1994. SCM2, a tryptophan permease in Saccharomyces cerevisiae, is important for cell growth. Mol. Gen. Genet. 244 260–268. [DOI] [PubMed] [Google Scholar]
- Chen, X. J., X. Wang, B. A. Kaufman and R. A. Butow, 2005. Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science 307 714–717. [DOI] [PubMed] [Google Scholar]
- Clark, L. B., P. Viswanathan, G. Quigley, Y. C. Chiang, J. S. McMahon et al., 2004. Systematic mutagenesis of the leucine-rich repeat (LRR) domain of CCR4 reveals specific sites for binding to CAF1 and a separate critical role for the LRR in CCR4 deadenylase activity. J. Biol. Chem. 279 13616–13623. [DOI] [PubMed] [Google Scholar]
- Collart, M. A., 2003. Global control of gene expression in yeast by the Ccr4-Not complex. Gene 313 1–16. [DOI] [PubMed] [Google Scholar]
- de Smedt, H., R. Borghgraef, F. Ceuterick and K. Heremans, 1979. Pressure effects on lipid-protein interactions in (Na+ + K+)-ATPase. Biochim. Biophys. Acta 556 479–489. [DOI] [PubMed] [Google Scholar]
- Denis, C. L., and J. Chen, 2003. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 73 221–250. [DOI] [PubMed] [Google Scholar]
- Desrivieres, S., F. T. Cooke, P. J. Parker and M. N. Hall, 1998. MSS4, a phosphatidylinositol-4-phosphate 5-kinase required for organization of the actin cytoskeleton in Saccharomyces cerevisiae. J. Biol. Chem. 273 15787–15793. [DOI] [PubMed] [Google Scholar]
- Dresios, J., I. L. Derkatch, S. W. Liebman and D. Synetos, 2000. Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry 39 7236–7244. [DOI] [PubMed] [Google Scholar]
- Dresios, J., P. Panopoulos, C. P. Frantziou and D. Synetos, 2001. Yeast ribosomal protein deletion mutants possess altered peptidyltransferase activity and different sensitivity to cycloheximide. Biochemistry 40 8101–8108. [DOI] [PubMed] [Google Scholar]
- Dresios, J., P. Panopoulos, K. Suzuki and D. Synetos, 2003. A dispensable yeast ribosomal protein optimizes peptidyltransferase activity and affects translocation. J. Biol. Chem. 278 3314–3322. [DOI] [PubMed] [Google Scholar]
- Dubouloz, F., O. Deloche, V. Wanke, E. Cameroni and C. De Virgilio, 2005. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol. Cell 19 15–26. [DOI] [PubMed] [Google Scholar]
- Duncan, K., R. M. Edwards and J. R. Coggins, 1988. The Saccharomyces cerevisiae ARO1 gene. An example of the co-ordinate regulation of five enzymes on a single biosynthetic pathway. FEBS Lett. 241 83–88. [DOI] [PubMed] [Google Scholar]
- Friden, P., and P. Schimmel, 1988. LEU3 of Saccharomyces cerevisiae activates multiple genes for branched-chain amino acid biosynthesis by binding to a common decanucleotide core sequence. Mol. Cell. Biol. 8 2690–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, H., P. Lu and A. Rich, 1969. Ribosomal subunits produced by cold sensitive initiation of protein synthesis. Nature 223 909–913. [DOI] [PubMed] [Google Scholar]
- Gaber, R. F., D. M. Copple, B. K. Kennedy, M. Vidal and M. Bard, 1989. The yeast gene ERG6 is required for normal membrane function but is not essential for biosynthesis of the cell-cycle-sparking sterol. Mol. Cell. Biol. 9 3447–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff, S. P., D. Marguet and G. J. Lauquin, 1990. Molecular cloning of the yeast mitochondrial aconitase gene (ACO1) and evidence of a synergistic regulation of expression by glucose plus glutamate. Mol. Cell. Biol. 10 3551–3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M., and C. A. Kaiser, 2006. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat. Cell Biol. 8 657–667. [DOI] [PubMed] [Google Scholar]
- Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles et al., 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418 387–391. [DOI] [PubMed] [Google Scholar]
- Glover, J. R., and S. Lindquist, 1998. Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell 94 73–82. [DOI] [PubMed] [Google Scholar]
- Hata, H., H. Mitsui, H. Liu, Y. Bai, C. L. Denis et al., 1998. Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics 148 571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazel, J. R., and E. E. Williams, 1990. The role of alterations in membrane lipid composition in enabling physiological adaptation of organisms to their physical environment. Prog. Lipid Res. 29 167–227. [DOI] [PubMed] [Google Scholar]
- Heitman, J., A. Koller, J. Kunz, R. Henriquez, A. Schmidt et al., 1993. The immunosuppressant FK506 inhibits amino acid import in Saccharomyces cerevisiae. Mol. Cell. Biol. 13 5010–5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heremans, K., and F. Wuytack, 1980. Pressure effect on the arrhenius discontinuity in Ca2+-ATPase from sarcoplasmic reticulum: evidence for lipid involvement. FEBS Lett. 117 161–163. [DOI] [PubMed] [Google Scholar]
- Homma, K., S. Terui, M. Minemura, H. Qadota, Y. Anraku et al., 1998. Phosphatidylinositol-4-phosphate 5-kinase localized on the plasma membrane is essential for yeast cell morphogenesis. J. Biol. Chem. 273 15779–15786. [DOI] [PubMed] [Google Scholar]
- Huang, J., H. Zhu, S. J. Haggarty, D. R. Spring, H. Hwang et al., 2004. Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips. Proc. Natl. Acad. Sci. USA 101 16594–16599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahashi, H., K. Obuchi, S. Fujii and Y. Komatsu, 1997. Effect of temperature on the role of Hsp104 and trehalose in barotolerance of Saccharomyces cerevisiae. FEBS Lett. 416 1–5. [DOI] [PubMed] [Google Scholar]
- Iwahashi, H., S. Nwaka and K. Obuchi, 2000. Evidence for contribution of neutral trehalase in barotolerance of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 66 5182–5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D. G., U. Reusser and G. H. Braus, 1991. Molecular cloning, characterization and analysis of the regulation of the ARO2 gene, encoding chorismate synthase, of Saccharomyces cerevisiae. Mol. Microbiol. 5 2143–2152. [DOI] [PubMed] [Google Scholar]
- Kamei, T., K. Tanaka, T. Hihara, M. Umikawa, H. Imamura et al., 1998. Interaction of Bnr1p with a novel Src homology 3 domain-containing Hof1p. Implication in cytokinesis in Saccharomyces cerevisiae. J. Biol. Chem. 273 28341–28345. [DOI] [PubMed] [Google Scholar]
- Kato, M., R. Hayashi, T. Tsuda and K. Taniguchi, 2002. High pressure-induced changes of biological membrane. Study on the membrane-bound Na+/K+-ATPase as a model system. Eur. J. Biochem. 269 110–118. [DOI] [PubMed] [Google Scholar]
- Kishimoto, T., T. Yamamoto and K. Tanaka, 2005. Defects in structural integrity of ergosterol and the Cdc50p-Drs2p putative phospholipid translocase cause accumulation of endocytic membranes, onto which actin patches are assembled in yeast. Mol. Biol. Cell 16 5592–5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N. J., M. C. Keogh, N. Datta, C. Sawa, O. W. Ryan et al., 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12 1565–1576. [DOI] [PubMed] [Google Scholar]
- Lippincott, J., and R. Li, 1998. Dual function of Cyk2, a Cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J. Cell Biol. 143 1947–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. Y., V. Badarinarayana, D. C. Audino, J. Rappsilber, M. Mann et al., 1998. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 17 1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. Y., J. H. Toyn, Y. C. Chiang, M. P. Draper, L. H. Johnston et al., 1997. DBF2, a cell cycle-regulated protein kinase, is physically and functionally associated with the CCR4 transcriptional regulatory complex. EMBO J. 16 5289–5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, R. T., and L. W. Parks, 1992. Cloning, sequencing, and disruption of the gene encoding sterol C-14 reductase in Saccharomyces cerevisiae. DNA Cell Biol. 11 685–692. [DOI] [PubMed] [Google Scholar]
- MacDonald, A. G., 2002. Ion channels under high pressure. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 131 587–593. [DOI] [PubMed] [Google Scholar]
- Maillet, L., C. Tu, Y. K. Hong, E. O. Shuster and M. A. Collart, 2000. The essential function of Not1 lies within the Ccr4-not complex. J. Mol. Biol. 303 131–143. [DOI] [PubMed] [Google Scholar]
- Martinez-Alonso, E., G. Egea, J. Ballesta and J. A. Martinez-Menarguez, 2005. Structure and dynamics of the Golgi complex at 15 °C: low temperature induces the formation of Golgi-derived tubules. Traffic 6 32–44. [DOI] [PubMed] [Google Scholar]
- Melcher, K., M. Rose, M. Kunzler, G. H. Braus and K. D. Entian, 1995. Molecular analysis of the yeast SER1 gene encoding 3-phosphoserine aminotransferase: regulation by general control and serine repression. Curr. Genet. 27 501–508. [DOI] [PubMed] [Google Scholar]
- Misu, K., K. Fujimura-Kamada, T. Ueda, A. Nakano, H. Katoh et al., 2003. Cdc50p, a conserved endosomal membrane protein, controls polarized growth in Saccharomyces cerevisiae. Mol. Biol. Cell 14 730–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, T., and F. Abe, 2004. Multiple ubiquitin-specific protease genes are involved in degradation of yeast tryptophan permease Tat2 at high pressure. FEMS Microbiol. Lett. 239 171–179. [DOI] [PubMed] [Google Scholar]
- Miura, T., H. Minegishi, R. Usami and F. Abe, 2006. Systematic analysis of HSP gene expression and effects on cell growth and survival at high hydrostatic pressure in Saccharomyces cerevisiae. Extremophiles 10 279–284. [DOI] [PubMed] [Google Scholar]
- Mnaimneh, S., A. P. Davierwala, J. Haynes, J. Moffat, W. T. Peng et al., 2004. Exploration of essential gene functions via titratable promoter alleles. Cell 118 31–44. [DOI] [PubMed] [Google Scholar]
- Moir, D., S. E. Stewart, B. C. Osmond and D. Botstein, 1982. Cold-sensitive cell-division-cycle mutants of yeast: isolation, properties, and pseudoreversion studies. Genetics 100 547–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz, M., B. A. Pulaski and J. L. Woolford, Jr., 1991. Assembly of 60S ribosomal subunits is perturbed in temperature-sensitive yeast mutants defective in ribosomal protein L16. Mol. Cell. Biol. 11 5681–5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagayama, A., C. Kato and F. Abe, 2004. The N- and C-terminal mutations in tryptophan permease Tat2 confer cell growth in Saccharomyces cerevisiae under high-pressure and low-temperature conditions. Extremophiles 8 143–149. [DOI] [PubMed] [Google Scholar]
- Nakashima, N., E. Noguchi and T. Nishimoto, 1999. Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics 152 853–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan, P., J. Wang, Z. Hua and T. R. Graham, 2004. Drs2p-coupled aminophospholipid translocase activity in yeast Golgi membranes and relationship to in vivo function. Proc. Natl. Acad. Sci. US A 101 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen, B., R. De Wachter and A. Goffeau, 1997. Classification of all putative permeases and other membrane plurispanners of the major facilitator superfamily encoded by the complete genome of Saccharomyces cerevisiae. FEMS Microbiol. Rev. 21 113–134. [DOI] [PubMed] [Google Scholar]
- Nielsen, P. S., B. van den Hazel, T. Didion, M. de Boer, M. Jorgensen et al., 2001. Transcriptional regulation of the Saccharomyces cerevisiae amino acid permease gene BAP2. Mol. Gen. Genet. 264 613–622. [DOI] [PubMed] [Google Scholar]
- Parks, L. W., S. J. Smith and J. H. Crowley, 1995. Biochemical and physiological effects of sterol alterations in yeast—a review. Lipids 30 227–230. [DOI] [PubMed] [Google Scholar]
- Pel, H. J., C. Maat, M. Rep and L. A. Grivell, 1992. The yeast nuclear gene MRF1 encodes a mitochondrial peptide chain release factor and cures several mitochondrial RNA splicing defects. Nucleic Acids Res. 20 6339–6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski, J. A., and S. C. Falco, 1988. Structure of the yeast HOM3 gene which encodes aspartokinase. J. Biol. Chem. 263 2146–2151. [PubMed] [Google Scholar]
- Ramos, C., and I. L. Calderon, 1994. Biochemical evidence that the Saccharomyces cerevisiae THR4 gene encodes threonine synthetase. FEBS Lett. 351 357–359. [DOI] [PubMed] [Google Scholar]
- Saito, K., K. Fujimura-Kamada, N. Furuta, U. Kato, M. Umeda et al., 2004. Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol. Biol. Cell 15 3418–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan, R., M. Leon, J. Zueco and R. Sentandreu, 1999. Basic phenotypic analysis of six novel yeast genes reveals two essential genes and one which affects the growth rate. Yeast 15 351–360. [DOI] [PubMed] [Google Scholar]
- Schmidt, A., M. N. Hall and A. Koller, 1994. Two FK506 resistance-conferring genes in Saccharomyces cerevisiae, TAT1 and TAT2, encode amino acid permeases mediating tyrosine and tryptophan uptake. Mol. Cell. Biol. 14 6597–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner, M., S. R. Collins, N. J. Thompson, V. Denic, A. Bhamidipati et al., 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123 507–519. [DOI] [PubMed] [Google Scholar]
- Schumacher, R. J., W. J. Hansen, B. C. Freeman, E. Alnemri, G. Litwack et al., 1996. Cooperative action of Hsp70, Hsp90, and DnaJ proteins in protein renaturation. Biochemistry 35 14889–14898. [DOI] [PubMed] [Google Scholar]
- Seshan, A., A. J. Bardin and A. Amon, 2002. Control of Lte1 localization by cell polarity determinants and Cdc14. Curr. Biol. 12 2098–2110. [DOI] [PubMed] [Google Scholar]
- Shi, X., M. Chang, A. J. Wolf, C. H. Chang, A. A. Frazer-Abel et al., 1997. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the srbp-containing holoenzyme. Mol. Cell. Biol. 17 1160–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama, M., Y. Matsui and A. Toh-e, 1994. a The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M phase. Mol. Cell. Biol. 14 7476–7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama, M., Y. Matsui, K. Tanaka and A. Toh-e, 1994. b Isolation of a CDC25 family gene, MSI2/LTE1, as a multicopy suppressor of ira1. Yeast 10 451–461. [DOI] [PubMed] [Google Scholar]
- Singh, A., and T. R. Manney, 1974. Genetic analysis of mutations affecting growth of Saccharomyces cerevisiae at low temperature. Genetics 77 651–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, A., T. L. Mason and R. A. Zimmermann, 1978. A cold-sensitive cytoplasmic mutation of Saccharomyces cerevisiae affecting assembly of the mitochondrial 50S ribosomal subunit. Mol. Gen. Genet. 161 143–151. [DOI] [PubMed] [Google Scholar]
- Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu et al., 2004. Global mapping of the yeast genetic interaction network. Science 303 808–813. [DOI] [PubMed] [Google Scholar]
- Tucker, M., M. A. Valencia-Sanchez, R. R. Staples, J. Chen, C. L. Denis et al., 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104 377–386. [DOI] [PubMed] [Google Scholar]
- Ursic, D., and J. Davies, 1979. A cold-sensitive mutant of Saccharomyces cerevisiae defective in ribosome processing. Mol. Gen. Genet. 175 313–323. [DOI] [PubMed] [Google Scholar]
- Wickner, R. B., T. J. Koh, J. C. Crowley, J. O'Neil and D. B. Kaback, 1987. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: Isolation of the MAK16 gene and analysis of an adjacent gene essential for growth at low temperatures. Yeast 3 51–57. [DOI] [PubMed] [Google Scholar]
- Winter, R., 2002. Synchrotron X-ray and neutron small-angle scattering of lyotropic lipid mesophases, model biomembranes and proteins in solution at high pressure. Biochim. Biophys. Acta 1595 160–184. [DOI] [PubMed] [Google Scholar]
- Winter, R., and W. Dzwolak, 2004. Temperature-pressure configurational landscape of lipid bilayers and proteins. Cell. Mol. Biol. 50 397–417. [PubMed] [Google Scholar]
- Yoshida, S., R. Ichihashi and A. Toh-e, 2003. Ras recruits mitotic exit regulator Lte1 to the bud cortex in budding yeast. J. Cell Biol. 161 889–897. [DOI] [PMC free article] [PubMed] [Google Scholar]