Abstract
We have synthesized and characterized a family of structured oligo-N-substituted-glycines (peptoids) up to 36 residues in length by using an efficient solid-phase protocol to incorporate chemically diverse side chains in a sequence-specific fashion. We investigated polypeptoids containing side chains with a chiral center adjacent to the main chain nitrogen. Some of these sequences have stable secondary structure, despite the achirality of the polymer backbone and its lack of hydrogen bond donors. In both aqueous and organic solvents, peptoid oligomers as short as five residues give rise to CD spectra that strongly resemble those of peptide α-helices. Differential scanning calorimetry and CD measurements show that polypeptoid secondary structure is highly stable and that unfolding is reversible and cooperative. Thermodynamic parameters obtained for unfolding are similar to those obtained for the α-helix to coil transitions of peptides. This class of biomimetic polymers may enable the design of self-assembling macromolecules with novel structures and functions.
Advances in bio-organic chemistry now offer accessibility to valuable new materials that integrate the structural and functional characteristics of biopolymers with the stability and diversity of synthetic polymers. Biopolymers are unique in that they are composed of specific monomer sequences and are capable of self-assembly into stable native structures, permitting a remarkable variety of functions. In contrast, most synthetic polymers do not have a specific monomer sequence but can be cheaply produced from a wide array of monomers and linking chemistries to generate many useful materials. It has been suggested that the sequence-specific polymerization of nonbiological monomers could allow the creation of synthetic polymers with stable folded structures (1, 2).
The design of novel polymers with sophisticated properties such as molecular recognition, energy transduction, and catalysis will require the precise spatial positioning of multiple chemical functionalities. The synthesis of such a polymer necessitates control of monomer sequence, control of polymer length, high coupling efficiencies, the ability to incorporate a variety of side chain types, and self-assembly into a unique conformation. Additional desired characteristics include relatively low cost, a flexible polymer backbone allowing a variety of structural motifs, cooperative conformational transitions allowing effective regulation and signaling, thermostability, structural stability in both aqueous and organic solvents, and compatibility with biological systems. In this paper we present a study of peptoids, a family of chemically diverse sequence-specific heteropolymers that may ultimately satisfy these goals.
Several methods have been developed for the synthesis of biomimetic oligomers with unnatural backbones. Many of these are structural variants of polypeptides, such as polycarbamates (3), peptide nucleic acids (4), and vinylogous polypeptides (5). Recent efforts have yielded oligomers with defined folding propensities such as oligoanthranilamides (6), vinylogous sulfonamidopeptides (7), “aedemers” (8), and oligophenylacetylenes (31). Helical secondary structures have been demonstrated in oligomers of β-amino acids (9, 10). These systems have been limited to short-chain molecules lacking chemical diversity. Typically, solution structures have been detected only in nonaqueous solvents and are stabilized by hydrogen bonding.
In contrast, polymer chemists have historically obtained helical homopolymers by incorporation of bulky side chains whose steric repulsion directs the conformation of the backbone (11). The use of chiral monomers or initiators can afford helices with a preferred screw sense. Examples of these asymmetric polymers include bulky methacrylates, isocyanides, trichloroacetaldehydes, and isocyanates (ref. 11 and references therein).
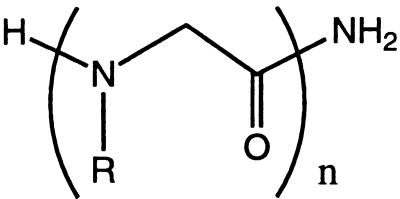
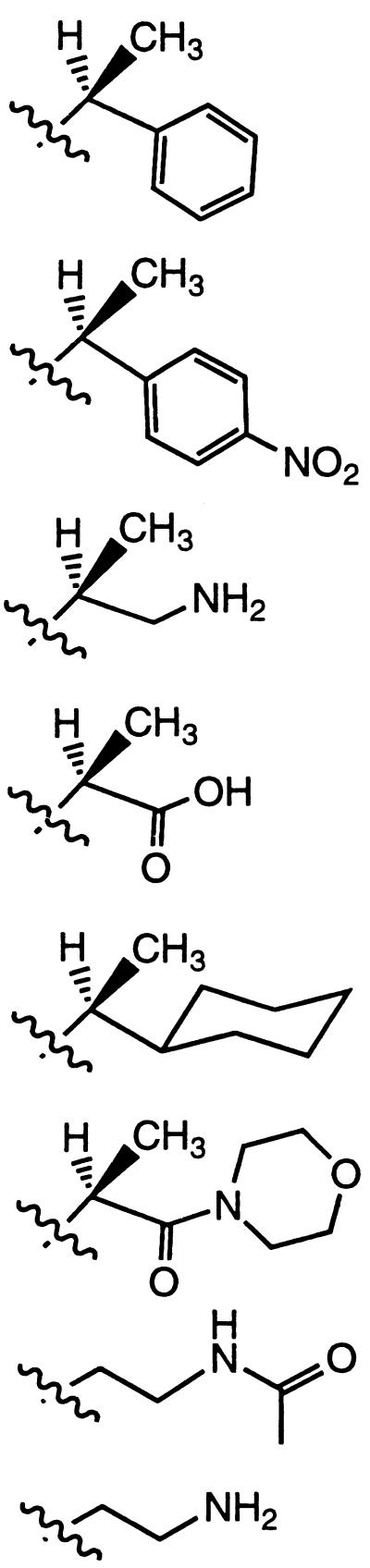
Peptoids are a family of oligomers that were developed for use in combinatorial drug discovery programs (12, 13). These non-natural oligomers are based on a polyglycine backbone, on which the side chains are appended to the amide nitrogen (Table 1). Because the side chain moiety is introduced by the reaction of primary amines, this method can yield an extremely diverse series of functionalized oligomers from readily obtainable starting materials (14, 15). Furthermore, coupling efficiencies of >98% now enable the automated synthesis of long-chain (>25 monomers) sequence-specific polymers in good yield.
Table 1.
N-substituted glycine side chains
|
N-substituted glycine oligomer, or polypeptoid
| |
|---|---|
| R = Side chain* | Designator |
| Nspe = (S)-N(1-phenylethyl)glycine | |
| Nrpe = enantiomer of structure shown, (R)-N-(1-phenylethyl)glycine | |
| Nsnp = (S-N-(1-(p-nitrophenyl)ethyl)glycine | |
| Nrnp = enantiomer of structure shown, (R)-N-(1-(p-nitrophenyl)ethyl)glycine | |
| Nsam = (S)-N-(2-amino-1-methylethyl)glycine | |
| Nsce = (S)-N-(1-carboxyethyl)glycine | |
| Nsch = (S)-N-(1-cyclohexylethyl)glycine | |
| Nsme = (S)-N-(1-((morpholino)carbonyl)ethyl)glycine | |
| Nam = N-(2-acetamidoethyl)glycine | |
| Nae = N-(2-aminoethyl)glycine | |
*These structures refer to the N-substituent of a glycine monomer.
Many of the mechanisms that direct the self-assembly of biopolymer structures are lacking in peptoids. Specifically, peptoids lack amide protons, and thus no hydrogen bond network along the polymer backbone is possible. Likewise, because the main chain contains no chiral centers, peptoids have no intrinsic handedness. Previous studies of short peptoid oligomers by modeling (12, 16) and by NMR (17) have demonstrated that the tertiary amides in the peptoid backbone can result in the population of both cis and trans amide bond conformers. These findings were also observed for collagen analogs containing peptoid residues (18).
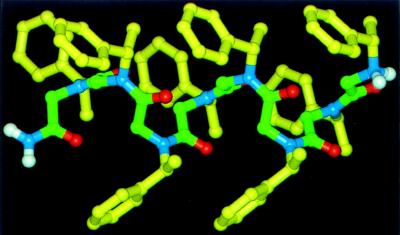
Our aim is to create biomimetic stereospecific polymers, combining strategies previously applied in the biopolymer and asymmetric polymer fields. We have investigated a family of sequence-specific peptoid polymers incorporating bulky chiral side chains. Recent modeling studies indicated that these peptoids could form a helical secondary structure (19) (Fig. 1). We report here the synthesis and characterization of this class of peptoid polymers.
Figure 1.
Molecular mechanics model of helical peptoid octamer (Nspe)8 (21).
MATERIALS AND METHODS
Peptoid Synthesis and Purification.
Solvents and reagents were purchased from commercial sources and used without further purification. Peptoid oligomers were synthesized on 50 μmol of a Rink amide resin (NovaBiochem, San Diego) at a substitution level of 0.47 mmol/g. The peptoid oligomers were synthesized by an improvement of previous methods (14, 15), which included reduced cycle times and decreased amine concentrations. Briefly, after removal of the first Fmoc group the following 90-min monomer addition cycle was performed by a robotic synthesizer and repeated until the desired length was obtained. The amino resin was bromoacetylated by adding 830 μl of 1.2 M bromoacetic acid in N,N-dimethylformamide (DMF) and 200 μl of N,N′-diisopropylcarbodiimide (DIC). The mixture was agitated for 40 min at 35°C, drained, and washed with DMF (three times with 2 ml). Next, 0.85 ml of a 1 M solution of a primary amine in DMSO was added to introduce the side chain. The mixture was agitated for 40 min at 35°C, drained, and washed with DMF (four times with 2 ml). After the last coupling the peptoid-resin was cleaved and lyophilized as described (15).
Individual peptoid oligomers were analyzed by reversed-phase HPLC on C4 columns (Vydac, 5 μm, 300 Å, 1 × 150 mm) on a Magic 2002 system (Michrom Bioresources, Auburn, CA). A linear gradient of 5–95% solvent B in solvent A over 40 min was used at a flow rate of 100 μl/min [solvent A = 0.1% trifluoroacetic acid (TFA) in water, solvent B = 0.1% TFA in acetonitrile] at a column temperature of 60°C. Preparative HPLC was performed on a Delta-Pak C4 (15 μm, 300 Å) on a Waters Prep LC3000 system (Millipore) by using the same solvent system. Peaks were eluted with a linear gradient of 20–70% solvent B in solvent A over 40 min at a flow rate of 50 ml/min. Electrospray mass spectrometry was performed on a Platform II (Micromass, Beverly, MA).
Amine Submonomers Used in Peptoid Synthesis.
N-(2-acetamidoethyl)glycine (Nam), (S)-N-(1-cyclohexylethyl)glycine (Nsch), and (R or S)-N-(1-phenylethyl)glycine (Nrpe or Nspe) were made from the amines aminoethylacetamide, (S)-1-cyclohexylethylamine, and (R or S)-1-phenylethylamine [>99% enantiomeric excess (e.e.)], respectively, which were obtained from Fluka. (R or S)-N-(1-(p-nitrophenyl)ethyl)glycine (Nrnp, Nsnp) were made from (R or S)-1-(p-nitrophenyl)ethylamine (>99% e.e.) which was obtained from Celgene (Warren, NJ). (S)-N-(1-carboxyethyl)glycine (Nsce) was made from l-O-t-butyl-alanine·hydrochloride (Bachem). N-(2-aminoethyl)glycine (Nae) was made from mono-Boc-ethylenediamine that was made by the method of Krapcho and Kuell (20). (S)-N-(2-amino-1-methylethyl)glycine (Nsam) was made from (S)-1-(Boc-amino)-2-aminopropane that was made via three steps from (S)-Z-alaninamide [briefly, the amide of (S)-Z-alaninamide was reduced to the amine with BH3, followed by protection of this amino group with di-t-butyldicarbonate and removal of the Z group by catalytic hydrogenation]. (S)-N-(1-((morpholino)carbonyl)ethylglycine (Nsmc) was made from (S)-alanylmorpholide which was made from l-Z-alanine and morpholine.
Circular Dichroism.
CD measurements were performed with a Jasco (Easton, MD) model 720 spectropolarimeter equipped with a Peltier temperature control unit. Spectra were obtained in fused quartz cells from 0.1 to 10 mm path length. Data are expressed in terms of mean residue ellipticity, [θ] (deg⋅cm2/dmol), calculated per mol of amide groups present.
Differential Scanning Calorimetry (DSC).
An MC-2 differential scanning microcalorimeter (MicroCal, Northampton, MA) was used at a scanning rate of 1°C/min under 20 psi N2. DSC was conducted in degassed 5 mM sodium citrate, 5 mM Na2HPO4, and 0.1 M NaCl (pH 4.1) at a peptoid concentration of 2.0 or 0.5 mg/ml. A minimum of six scans were conducted to ensure reproducibility. A thermal scan of buffer versus buffer was subtracted from a scan of sample versus buffer. Data analysis was conducted by using the Origins software supplied by MicroCal.
NMR Spectroscopy.
Samples for NMR experiments were prepared as ≈5 mM solutions in CD3OD. Spectra were acquired at 10°C on a Varian Unity 300 instrument. Standard one-dimensional proton spectra were collected as 8,192 data points and zero-filled to 16,384.
RESULTS
Synthesis of Sequence-Specific Peptoid Oligomers.
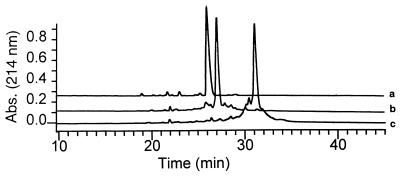
N-substituted glycine oligomers of 3–36 residues were synthesized in good yield and high purity by using bromoacetic acid and a variety of primary amines as submonomers (14, 15). We created a broad family of oligomeric peptoid products containing a specific sequence of diverse side chain moieties. These included aliphatic, aromatic, heterocyclic, cationic, and anionic groups (Table 1). Reactive side chain functionalities were protected by TFA-labile groups (Boc for amines and t-butyl esters for carboxylic acids) that were removed during cleavage of the peptoid from the resin. Crude oligomer purities ranging from 50 to 90% were determined by HPLC and largely depended on the oligomer length (Fig. 2, Table 2). Compounds were purified to >95% homogeneity by preparative reversed-phase HPLC prior to characterization. Molecular weights were confirmed by electrospray mass spectrometry and were uniformly in agreement with expected values (Table 1).
Figure 2.
Analytical HPLC traces of crude peptoid synthesis products: trace a, pentamer 1(Nspe)5; trace b, 18-mer 2 (NaeNspeNspe)6; and trace c, 36-mer 3 (NaeNspeNspe)12. Traces a and b are vertically offset for clarity. All products were purified by preparative HPLC prior to further study.
Table 2.
Peptoid oligomer structures
| Peptoid | Monomer sequence | Oligomer length | Molecular weight
|
Purity,* % |
|---|---|---|---|---|
| Calc. : Found | ||||
| 1 | (Nspe)5 | 5 | 822.6 : 822.5 | 95 |
| 2 | (NaeNspeNspe)6 | 18 | 2,550.8 : 2,550.6 | 80 |
| 3 | (NaeNspeNspe)12 | 36 | 5,084.6 : 5,084.2 | 50 |
| 4 | (NspeNspeNsce)3 | 9 | 1,370.8 : 1,370.4 | 60 |
| 5 | (NrpeNrpeNam)4 | 12 | 1,874.3 : 1,873.9 | 70 |
| 6 | (NsceNsceNspe)10 | 30 | 4,209.4 : 4,209.9 | 50 |
| 7 | Ac-Nsnp | 1 | 265.2 : 264.9 | ND |
| 8 | (Nsnp)3 | 3 | 635.4 : 635.1 | ND |
| 9 | (Nsnp)5 | 5 | 1,047.6 : 1,047.1 | 99 |
| 10 | (Nrnp)8 | 8 | 1,666.0 : 1,665.8 | 80 |
| 11 | (NspeNsce)4Nspe | 9 | 1,338.8 : 1,338.4 | 90 |
| 12 | (NsamNspeNspe)6 | 18 | 2,634.9 : 2,634.2 | 60 |
| 13 | (Nsme)12 | 12 | 2,394.4 : 2,394.5 | 95 |
| 14 | (Nsch)5 | 5 | 852.6 : 852.4 | 95 |
As determined by analytical HPLC of crude product. All compounds were purified to >95% homogeneity. ND, not determined.
Characterization of Peptoid Oligomers by CD.
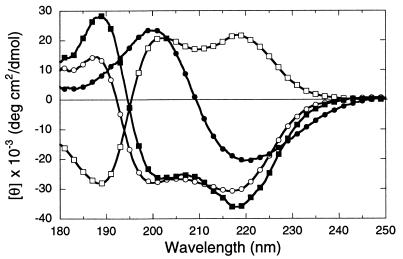
The CD spectra of peptoid oligomers of varying length, monomer composition, and sequence were obtained in aqueous solution over the 250- to 180-nm region (Fig. 3). The spectra for polypeptoids 4 and 6, which include (S)-N-(1-phenylethyl)glycine monomers, show two minima of negative ellipticity ≈218 and 202 nm, and a band of positive ellipticity ≈190 nm. The band at 218 nm corresponds to the nπ* transition of the amide chromophore (observed between 230 to 210 nm in peptides), whereas the remaining bands correspond to the high and low wavelength components of the exciton split ππ* transition (observed around 200 nm in tertiary amides) (21). Additional contributions may derive from coupled interactions between the peptide bands and the La transition of the aromatic side chains (≈210 nm for phenylalanine) (21). However, variation of aromatic content from 33% to 66% of the side chains does not substantially alter the magnitude of the ellipticity in this region (Fig. 3). Compound 5, which contains side chains of opposite chirality to 4 and 6, shows a mirror-image α-helix type CD spectrum. Peptoid homopolymers 13 and 14 comprised of chiral side chains containing cyclohexyl and morpholidyl groups yielded CD spectra lacking the double minimum profile.
Figure 3.
CD spectra of polypeptoids with varying monomer composition and length: ▪, compound 4 (NspeNspeNsce)3; □, 5 (NrpeNrpeNam)4; ○, 6 (NsceNsceNspe)10; •, 13 (Nsme)12. Peptoid concentration was 0.2 mg/ml (≈1 mM residue molarity for all species) in 10 mM sodium phosphate (pH 7.0) at 25°C.
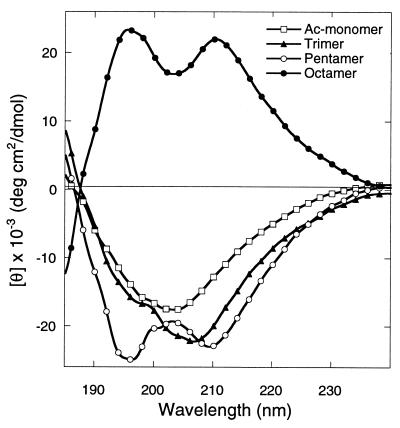
To determine the minimum chain length required to establish the characteristic helix-like CD signal, the CD spectra of compounds 7-10, corresponding to (Nsnp)n (n = 1, 3, 5) and (Nrnp)8 oligopeptoids, were obtained in acetonitrile in the region of 250 to 185 nm (Fig. 4). Compounds 7-10 all have a CD band at 204–210 nm. The pentamer and octamer display an additional band at around 195 nm, giving rise to a helix-like CD signal. Similar spectra were obtained for these compounds in methanol (22). The spectra of (Nrnp)8 (compound 10), comprised of side chains of the opposite chirality, is the mirror image of the ellipticity of (Nsnp)5 (compound 9), consistent with the data presented in Fig. 3. Comparison of spectra for 9 and 10 shows that the intensity of the signal on a per-residue basis does not increase for oligomers longer than five residues. These helix-like spectra are similar to those of peptoids 4-6 in aqueous solution, with a blue shift of the CD bands.
Figure 4.
CD spectra in acetonitrile. Ellipticities for the N-(p-nitrophenylethyl)glycine trimer 8 (Nsnp)3 (▴), pentamer 9 (Nsnp)5 (○), octamer 10 (Nrnp)8 (•), and the acetylated monomer 7 Ac-Nsnp (□) were calculated per mol of amide groups present. Helix-like CD signals are evident for the pentamer and octamer. The octamer was prepared from monomers of the opposite chirality (R vs. S). Peptoid concentration was 2 mg/ml at 25°C.
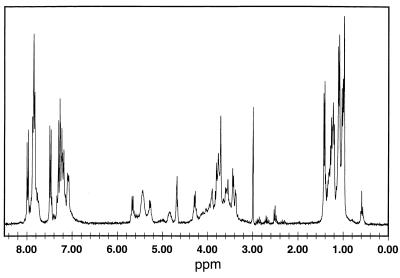
The CD spectra of compound 9 were independent of concentration in the range 50 μM to 1 mM in acetonitrile, indicating the absence of intermolecular association. Consistent with this finding, the 1H one-dimensional NMR spectra of this pentamer reveal sharp line widths (Fig. 5).
Figure 5.
1H-NMR spectrum for a 5 mM solution of pentamer 9 in CD3OD.
Melting Studies and Determination of Thermodynamic Parameters.
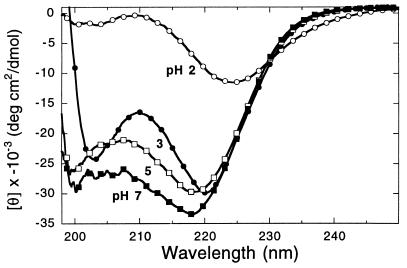
We used CD to determine whether the helical structure can be deformed as a function of pH and temperature. Some polypeptoids undergo a sharp structural transition as a function of pH. Decreasing the pH from 7 to 3 leads to only small changes in the CD spectra of the 30-mer 6 (Fig. 6). Upon further titration to pH 2, the signal around 200 nm is almost entirely abolished, and the long wavelength band is red-shifted and diminished. The spectrum at pH 2 resembles that of the Nspe monomer, indicating the absence of repeating secondary structure.
Figure 6.
The pH dependent conformational transition of 30-mer 6 as demonstrated by CD. Data were obtained at a concentration of 23 μM in 5 mM sodium citrate, 5 mM sodium phosphate buffer containing 0.1 M NaCl at 25°C. The pH value is shown near each spectrum.
CD spectra were obtained as a function of temperature for peptoid oligomers in varying solution conditions. The 9-mer 11 and 30-mer 6 show little change in ellipticity as temperature was increased from 5 to 65°C at pH 7 (Fig. 7). We utilized the pH dependent conformational transitions described above to destabilize the secondary structure sufficiently to detect melting in an experimentally accessible regime. At pH 4.1, 6 undergoes a sigmoidal thermal transition, which is largely complete within a 40°C temperature range. The original CD spectrum is regained upon return to 5°C conditions (data not shown).
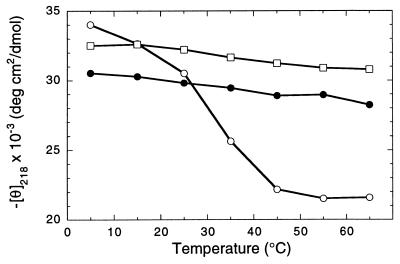
Figure 7.
Temperature dependence of θ218 between 5 and 65°C. Data were obtained at a concentration of 0.2 mg/ml. •, Peptoid 9-mer (NspeNsce)4Nspe 11 at pH 7; □, 30-mer (NsceNsceNspe)10 6 at pH 7.0; ○, 6 at pH 4.1.
To more fully investigate the nature of the melting, complete CD spectra were obtained of 6 at pH 4.1 at temperatures from 5 to 75°C (Fig. 8). The loss of signal intensity at 220 nm, and the disappearance of the 203 nm minimum over a narrow temperature range, indicate the loss of secondary structure content in a cooperative fashion. The spectra show isodichroic points at 230 and 200 nm, suggesting a two-state transition. We calculated the van’t Hoff enthalpy for the structural transition by a method similar to that described for study of a 50-residue α-helical peptide (23). The analysis assumes the peptoid is fully helical at 5°C and fully coiled at 75°C, and that ellipticity is a linear function of the amount of helical structure present. The ellipticity values at 203 and 220 nm were separately used to evaluate the fractional helical content at each temperature [fH(T)] and the van’t Hoff enthalpy of the transition, ΔHvH. The enthalpies determined by fitting to the van’t Hoff equation at each wavelength were in excellent agreement, and both equaled 0.22 kcal/mol per residue.
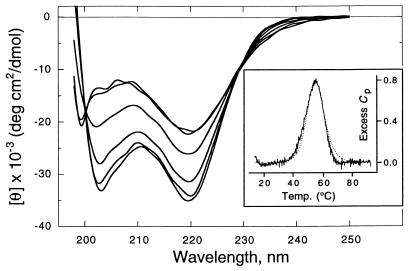
Figure 8.
Thermal unfolding of pH-destabilized 30-mer 6 in 5 mM sodium citrate, 5 mM sodium phosphate buffer, pH 4.1, containing 0.1 M NaCl. CD spectra were taken at a concentration of 23 μM. Temperature from bottom to top at 203 nm: 5, 15, 25, 35, 55, and 75°C. (Inset) DSC scan at a peptoid concentration of 460 μM. Cp in kcal/mol⋅K, scan rate was 1°C/min. ΔHcal = 1.1 kcal/mol per residue.
The thermal transition of 6 is sharp according to DSC conducted at 460 μM and pH 4.1 (Fig. 8 Inset). The excess heat capacity has a peak that is complete within a 40°C range. The peptoid transition is fully reversible and reproducible over several scans between 15 and 90°C. After subtraction of the buffer reference trace, the data were fit to a theoretical curve and the area integrated to obtain ΔHcal, 1.1 kcal/mol per residue. CD and DSC studies of 6 were both repeated at 120 μM with no variation of molar signal intensity (data not shown). Cooperative thermal unfolding is also observed in the polycationic 18-mer 12 with limited possibility of forming intramolecular hydrogen bonds through its side chains. At a concentration of 36 μM in 10 mM sodium phosphate, 10 mM sodium borate buffer (pH 8.3), this molecule undergoes complete unfolding between 25 and 65°C as monitored by CD (data not shown).
DISCUSSION
Although 25-mers have been previously synthesized in good yields in our laboratory (14), the study of longer structured molecules presented some synthetic challenges. Because we aimed to induce structure by the incorporation of hindered residues, it was necessary to strike a balance between high coupling efficiencies and strong steric influence of the side chain over the main chain. Initial efforts revealed that yields were compromised when α-branched primary alkylamines having two α substituents larger than methyl groups were incorporated into 25-mers (data not shown). However, α-methyl primary amines were efficiently incorporated (Fig. 2 and Tables 1 and 2). Importantly, this family of amines are chiral and many can be obtained in high enantiopurity from commercial sources.
The presence of these α-chiral side chains is predicted to dramatically reduce the conformational space energetically accessible to the polymer backbone. Calculations of α-chiral peptoid helices indicate the preference for a right-handed helix pitch with repeating cis-amide bonds and a limited range of φ, ψ dihedral angles (19) (Fig. 1). Green et al. (24) have investigated the effects of chiral N-pendant groups on random isocyanate polymers, which likewise have an achiral backbone. These studies demonstrated induction of helical secondary structure of specific handedness, as evidenced by characteristic CD spectra.
The present study demonstrates that peptoid oligomers containing α-chiral side chains “fold” to form stable secondary structure. The intensity of the CD signals in the UV spectral regions dominated by amide chromophore transitions (230 to 185 nm) strongly suggests a well-defined repeating main chain secondary structure (Fig. 3). The peptoid CD spectra markedly resemble those of right-handed peptide 310- or α-helices, which display a positive band around 192 nm and two minima of ellipticity around 222 and 208 nm (21). The measured ellipticity of the nπ* band of these peptoids is comparable to those of fully helical model peptides (peptoid θ218 = −2.5 to −3.5 × 104, vs. peptide θ222 = −2.8 to −3.5 × 104 deg⋅cm2/dmol) (refs. 23 and 25 and references therein). This finding strongly suggests that these peptoids exhibit helical secondary structure. Theoretical calculations indicate that helical secondary structures can display two minima between 230 and 200 nm caused by exciton splitting of the ππ* amide transition (26). It should be noted that α-helix-like “class C” CD spectra have also been associated with certain β-turn peptide structures (27). However, the magnitude of β-turn CD signals are typically of low intensity compared with the signals observed for peptide α-helices and in the present study.
The absence of a dependence of the CD signal on peptoid concentration and the sharp line-widths observed in the one-dimensional 1H NMR spectra (Fig. 5) indicate that the structure is monomeric and is not stabilized by intermolecular associations. Analysis of two-dimensional NMR spectra of α-chiral peptoid pentamers is consistent with the CD data and modeling studies (19). Based on the pattern of both intra- and inter-residue ROE cross peaks, we identified a major conformer with a helical structure and repeating cis-amide bonds (22).
We observe helix-like CD signals in a variety of peptoid sequences varying in length and in the content of charged, aromatic, and chiral residues. Helix-like CD signals were preserved in sequences comprised of 33–100% aromatic residues and 0–66% charged residues without substantial change of signal intensity. However, when the aromatic groups were removed completely, as in the homo-oligomers containing morpholido 13 and cyclohexyl 14 side chains, CD signals were observed that lacked the characteristic double minimum. Chiral secondary structure can be propagated in the presence of achiral residues; dodecamer 5, which contains 33% achiral Nam residues, retains the helix-like profile. This result should permit the synthesis of peptoid helices incorporating diverse side chains from the wide family of achiral primary amines. Peptoids composed of side chains with inverted chirality show a mirror image relationship between their CD spectra (Fig. 3). As found for random isocyanate polymers (24), this suggests that enantiomeric side chains can induce the formation of helices with an inverted screw sense.
Peptoid secondary structure is detected even in short oligomers. A study of a series of peptoid homo-oligomers of one to eight residues demonstrates a helix-like CD signal in (p-nitrophenylethyl)glycine pentamers and octamers. A maximal signal is found at the pentamer length (Fig. 4). Although peptide α-helices are seldom detected for molecules smaller than 15 residues, the finding of stable helices in short polyimides is not unprecedented. Helices have been observed in polyproline molecules as small as three to five residues on the basis of vibrational and ultraviolet CD measurements (28). A strong single minimum CD signal observed for the peptoid trimer may indicate substantial helical proclivity for these chiral monomers in the absence of the inter-residue coupling attained by a full turn of the helical structure.
CD spectra were obtained in a variety of solvents, including acetonitrile, methanol, and aqueous solution. The characteristic double minimum spectra were retained with some solvent-dependent variation of the positions of the CD bands, indicating the resilience of the helical secondary structure to environmental changes. Previous studies of synthetic “folded” polymers generally have been limited to solution studies in organic solvents (6, 9, 10). A critical difference between these “foldamers” and peptoids is the absence of hydrogen-bond formation as a driving force for peptoid secondary structure. Steric influence of the bulky chiral side chains is likely to provide a constraint that defines the conformations accessible to the peptoid backbone (19). Interactions between side chain aromatic groups, and dipole–dipole repulsion between side chains and the carbonyl of the main chain amides, may also provide for specific secondary structure. These forces may allow peptoids to retain their structure in an aqueous environment, in which intramolecular hydrogen-bond networks can be disrupted.
Peptoid secondary structure is extraordinarily resistant to thermal unfolding. The 9-mer 11 and 30-mer 6 show only a slight variation in CD signal intensity over the temperature range from 5 to 65°C in neutral aqueous solution (Fig. 7). The resistance of peptoids to proteolysis has previously been noted (29). The stability of peptoids is an obvious benefit in considering their potential applications as nanostructured materials in biological systems.
Peptoids with ionizable side chains can be destabilized in response to changes in pH conditions. Peptoid 6 shows little variation in CD signal intensity over the pH range of 7 to 3 (Fig. 6). However, further titration to pH 2 results in a greatly diminished CD signal, indicating complete unfolding. This pH-dependent conformational behavior was exploited to observe melting in an experimentally accessible regime. Specifically, at pH 4.1 compound 6 demonstrates a complete, reversible loss of helix-like CD signal over a narrow temperature range (Fig. 8). The presence of an isodichroic point near 200 nm is characteristic of a two-state helix/coil transition, as has been described by Holtzer and Holtzer (30) for polypeptides. These authors also point out that it is highly unlikely that a precise isodichroic point will be observed if side chain CD bands are making a significant contribution.
DSC demonstrates that thermal unfolding is accompanied by absorption of heat as secondary structure is melted (Fig. 8 Inset). The narrow peak in excess heat capacity is further evidence of the highly cooperative nature of peptoid unfolding. In fact, both CD and DSC studies demonstrate that 6 unfolds over a 40°C temperature range, as compared with a greater than 100°C temperature range for a 50-residue designed α-helical peptide (23).
Thermodynamic parameters obtained for the acid-destabilized melting of 30-mer 6 are similar to those determined for helix/coil transitions in peptides. DSC shows that the calorimetric enthalpy of structure deformation in peptoid helices (ΔHcal = 1.1 kcal/mol per residue) is comparable to that of α-helical peptides (ΔHcal = 1.1–1.3 kcal/mol per residue) (23), as is the van’t Hoff enthalpy derived from CD data (peptoid ΔHvH = 0.22 kcal/mol per residue, vs. peptide ΔHvH = 0.22–0.23 kcal/mol per residue) (23). Notably, the enthalpy of peptide unfolding has been ascribed to the breaking of main chain hydrogen bonds (23), which cannot be formed in peptoids. The forces that favor formation of peptoid helices are not fully understood and are currently under investigation.
The work presented here demonstrates progress toward the creation of biomimetic polymers in which a specific sequence of chemically diverse monomers are incorporated into a stable folded structure. It is notable that polypeptoids display many of the characteristics of peptide behavior such as cooperative pH- and temperature-induced unfolding in aqueous solution, despite the absence of main chain hydrogen bonds. More detailed studies of peptoid structure and dynamics are currently underway. We believe polypeptoids will be useful for investigating principles of self-assembly in macromolecules and for creating novel functional polymeric materials. We are currently evaluating the use of these materials as reagents for DNA and drug delivery. Peptoid nanostructures may prove to be of wide utility in the design and control of molecular architectures in biotic and abiotic systems.
Acknowledgments
We thank Drs. Irwin Kuntz, Gavin Dollinger, David Spellmeyer, Roland Dunbrack, Jr., and Tetsuo Uno for helpful discussions; Simon Ng for technical assistance; Dr. Bao-Lu Chen for use of the spectropolarimeter; Drs. Rusty Williams and Walter Moos for their support; and a reviewer for providing additional references on asymmetric polymerization. K.K. is supported by National Institutes of Health Training Grant GM 07175. P.A. is supported by the Medical Scientist Training Program at the University of California, San Francisco. A.E.B. was supported by a National Research Service award (Postdoctoral Fellowship 1 F32 GM 18112-01) from the National Institute of General Medical Sciences, National Institutes of Health.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: DSC, differential scanning calorimetry; TFA, trifluoroacetic acid.
References
- 1.Dill K A. Biochemistry. 1985;24:1501–1509. doi: 10.1021/bi00327a032. [DOI] [PubMed] [Google Scholar]
- 2.Dill K A, Bromberg S, Yue K, Fiebig K M, Yee D P, Thomas P D, Chan H S. Protein Sci. 1995;4:561–602. doi: 10.1002/pro.5560040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho C Y, Moran E J, Cherry S R, Stephans J C, Fodor S P A, Adams C L, Sundaram A, Jacobs J W, Schultz P G. Science. 1993;261:1303–1305. doi: 10.1126/science.7689747. [DOI] [PubMed] [Google Scholar]
- 4.Egholm M, Buchardt O, Nielsen P E, Berg R H. J Am Chem Soc. 1992;114:1895–1897. [Google Scholar]
- 5.Hagihara M, Anthony N J, Stout T J, Clardy J, Schreiber S L. J Am Chem Soc. 1992;114:6568–6570. [Google Scholar]
- 6.Hamuro Y, Geib S J, Hamilton A D. J Am Chem Soc. 1996;118:7529–7541. [Google Scholar]
- 7.Genarri C, Salom B, Potenza D, Longari C, Fioravanzo E, Carugo O, Sardone N. Chem Eur J. 1996;2:644–655. [Google Scholar]
- 8.Lokey R S, Iverson B L. Nature (London) 1995;375:303–305. [Google Scholar]
- 9.Apella D H, Christianson L A, Karle I L, Powell D R, Gellman S H. J Am Chem Soc. 1996;118:13701–13702. [Google Scholar]
- 10.Seebach D, Overhand M, Kühnle F N M, Martinoni B, Oberer L, Hommel U, Widmer H. Helv Chim Acta. 1996;79:913–941. [Google Scholar]
- 11.Okamoto Y, Nakano T. Chem Rev. 1994;94:349–372. [Google Scholar]
- 12.Simon R J, Kania R S, Zuckermann R N, Huebner V D, Jewell D A, et al. Proc Natl Acad Sci USA. 1992;89:9367–9371. doi: 10.1073/pnas.89.20.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuckermann R N, Martin E J, Spellmeyer D C, Stauber G B, Shoemaker K R, et al. J Med Chem. 1994;37:2678–2685. doi: 10.1021/jm00043a007. [DOI] [PubMed] [Google Scholar]
- 14.Zuckermann R N, Kerr J M, Kent S B W, Moos W H. J Am Chem Soc. 1992;114:10646–10647. [Google Scholar]
- 15.Figliozzi G M, Goldsmith R, Ng S C, Banville S C, Zuckermann R N. Methods Enzymol. 1996;267:437–447. doi: 10.1016/s0076-6879(96)67027-x. [DOI] [PubMed] [Google Scholar]
- 16.Moehle K, Hofmann H-J. Biopolymers. 1996;38:781–790. doi: 10.1002/(SICI)1097-0282(199606)38:6%3C781::AID-BIP9%3E3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 17.Bradley E K, Kerr J M, Richter L S, Figliozzi G M, Goff D A, Zuckermann R N, Spellmeyer D C, Blaney J M. Mol Diversity. 1997;3:1–15. doi: 10.1023/a:1009698309407. [DOI] [PubMed] [Google Scholar]
- 18.Melacini G, Yangbo F, Goodman M. J Am Chem Soc. 1996;118:10725–10732. [Google Scholar]
- 19.Armand P, Kirshenbaum K, Falicov A, Dunbrack R L, Jr, Dill K A, Zuckermann R N, Cohen F E. Folding Design. 1997;2:369–375. doi: 10.1016/S1359-0278(97)00051-5. [DOI] [PubMed] [Google Scholar]
- 20.Krapcho A P, Kuell C S. Synth Commun. 1990;20:2559–2564. [Google Scholar]
- 21.Woody R W. In: The Peptides. Hruby V J, editor. Vol. 7. Orlando, FL: Academic; 1985. pp. 16–115. [Google Scholar]
- 22.Armand P, Kirshenbaum K, Goldsmith R A, Farr-Jones S, Barron A E, Truong K T V, Dill K A, Mierke D F, Cohen F E, Zuckermann R N, Bradley E K. Proc Natl Acad Sci USA. 1998;95:4309–4314. doi: 10.1073/pnas.95.8.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scholtz J M, Marqusee S, Baldwin R L, York E J, Stewart J M, Santoro M, Bolen D W. Proc Natl Acad Sci USA. 1991;88:2854–2858. doi: 10.1073/pnas.88.7.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green M M, Peterson N C, Sato T, Teramoto A, Cook R, Lifson S. Science. 1995;268:1860–1866. doi: 10.1126/science.268.5219.1860. [DOI] [PubMed] [Google Scholar]
- 25.Bradley E K, Thomason J F, Cohen F E, Kosen P A, Kuntz I D. J Mol Biol. 1990;215:607–622. doi: 10.1016/S0022-2836(05)80172-X. [DOI] [PubMed] [Google Scholar]
- 26.Moffitt W, Fitts D, Kirkwood J. Proc Natl Acad Sci USA. 1957;43:723–730. doi: 10.1073/pnas.43.8.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perczel A, Holl-si M. In: Circular Dichroism and the Conformational Analysis of Biomolecules. Fasman G D, editor. New York: Plenum; 1996. pp. 285–380. [Google Scholar]
- 28.Dukor R K, Keiderling T A. Biopolymers. 1991;31:1747–1761. doi: 10.1002/bip.360311409. [DOI] [PubMed] [Google Scholar]
- 29.Miller S M, Simon R J, Ng S, Zuckermann R N, Kerr J M, Moos W H. Drug Dev Res. 1995;35:20–32. [Google Scholar]
- 30.Holtzer M E, Holtzer A. Biopolymers. 1992;32:1675–1677. doi: 10.1002/bip.360321209. [DOI] [PubMed] [Google Scholar]
- 31.Nelson J C, Saven J G, Moore J S, Wolynes P G. Science. 1997;277:1793–1796. doi: 10.1126/science.277.5333.1793. [DOI] [PubMed] [Google Scholar]