Abstract
Certain bacterial protein toxins are able to insert themselves into, and at least partially across, lipid bilayer membranes in the absence of any auxiliary proteins, by using unknown mechanisms to overcome the high energy barrier presented by the hydrophobic bilayer core. We have previously shown that one such toxin, colicin Ia, translocates a large, hydrophilic part of itself completely across a lipid bilayer in conjunction with the formation of an ion-conducting channel. To address the question of whether the colicin can translocate any arbitrary amino acid sequence, we have altered the translocated segment by inserting, singly, two different foreign epitopes. Colicins containing either epitope retain significant bactericidal activity and form channels of normal conductance in planar bilayers. Furthermore, antibodies added on the side of the bilayer opposite that to which the colicin was added interact specifically with the corresponding epitopes, producing an inhibition of channel closing. Thus, the inserted epitopes are translocated along with the rest of the segment, suggesting that a surprisingly small part of colicin Ia, located elsewhere in the molecule, acts as a nonspecific protein translocator.
The movement of proteins or protein segments across membranes is a ubiquitous cellular process apparently mediated by several mechanisms, including transport through transmembrane “protein channels” (1, 2). In the case of protein export across the bacterial inner membrane, a complex of several proteins (SecA and SecYEG), as well as the proton motive force and ATP, are required (3, 4). However, such complex multicomponent systems are not essential for all instances of polypeptide translocation. A number of toxins are capable of inserting themselves into membranes in the absence of other proteins (5, 6).
The colicins are a family of plasmid-encoded protein toxins produced by some strains of Escherichia coli, which kill other E. coli strains. One group of colicins, which includes colicin Ia, forms ion-conducting channels in the inner membranes of susceptible strains (7); these colicins also form voltage-dependent ion-conducting channels in planar lipid bilayers (8–10). We recently showed that for colicin Ia, channel opening and closing is associated with the translocation, across the membrane and back, of a segment of at least 68 aa (residues 474–541) (11, 12). The translocated segment is very hydrophilic, containing 15 basic and 8 acidic residues (13) (Fig. 1), and has no conspicuous attributes that predispose it to membrane translocation. Furthermore, the translocation of this segment can be arrested at intermediate extents by attaching a large protein “anchor” at various positions within the segment or just upstream (residues 454–534); remarkably, these intermediate states still form conducting channels, albeit with substantially altered conductance and voltage-dependent gating (12). The nonuniqueness of the translocated segment, together with the channel’s tolerance for the attachment of prosthetic groups such as biotin, suggests that the colicin may be able to translocate an arbitrary amino acid sequence.
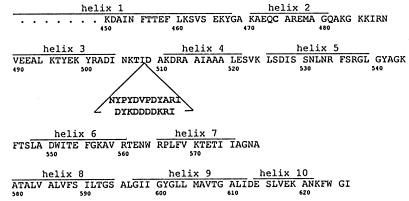
Figure 1.
Amino acid sequence (single-letter code) of the carboxyl-terminal channel-forming domain of colicin Ia (13). Helix assignments are from the x-ray crystal structure (ref. 14 and M. Wiener, personal communication). The location and sequence of the insertion in mutants HA-III (upper sequence) and FLAG-IV (lower sequence) (Table 1) are indicated. Amino acids that were mutated in the creation of the insertion (K507 → T and N509 → D) and the L474 → C mutant used are shown. The previously identified translocated segment (residues 474–541) (12) is roughly identical with helices 2–5. Helices 8 and 9 constitute the hydrophobic hairpin. Helix 6 is amphipathic.
To test this hypothesis, we prepared five mutant colicins with insertions of 9 to 12 residues in the middle of the translocated segment (Table 1 and Fig. 1). The insertions contained either the antigenic 12CA5 hemagglutinin (HA) epitope, YPYDVPDYA, from influenza virus HA protein (15), or the FLAG antigen, DYKDDDDK (16).
Table 1.
Bactericidal activity of epitope insertion mutants in colicin Ia
| Mutant | Amino acid residue preceding insert | Inserted sequence | Protein yield, mg/liter | Specific activity, killing units/mg |
|---|---|---|---|---|
| Wild type | NA | None | 30 | 1 × 107 |
| L474 → C | NA | None | 7 | 1 × 107 |
| HA-I | 511 | YPYDVPDYA | 14 | 1 × 106 |
| HA-II | 508 | CYPYDVPDYARI | 5 | 1 × 105 |
| HA-III | 508 | NYPYDVPDYARI | 8 | 1 × 106 |
| FLAG-IV | 508 | DYKDDDDKRI | 7 | 1 × 105 |
| FLAG-V | 508 | DYKDDDDKRI | 36 | 1 × 106 |
Epitope insertion mutants were created as described in Materials and Methods. The single-letter codes for the inserted sequences are indicated. The HA and FLAG epitope sequences are shown in bold. Specific activity was determined by spot-testing 10-fold dilutions on lawns of sensitive E. coli. Mutants HA-II, HA-III, FLAG-IV, and FLAG-V contained the mutations K507 → T and N509 → D. Mutants HA-II, HA-III, and FLAG-IV also contained the mutation L474 → C. Wild-type colicin Ia and the mutant L474 → C are shown for comparison.
MATERIALS AND METHODS
Design of Insertion Mutants.
We chose to insert the HA or FLAG epitope into the loop between helices 3 and 4 of the colicin Ia channel-forming domain, in the middle of the previously identified translocated segment. (The whole colicin was used for all experiments described in this work.) Based on a preliminary crystal structure (ref. 17 and M. Wiener, personal communication), we first made mutant HA-I (Table 1), with a 9-aa insertion between residues 511 and 512. The refined crystal structure subsequently indicated that this insertion was actually out of the loop, just within helix 4 (ref. 14 and M. Wiener, personal communication). Based on the refined structure, we inserted a 12-aa sequence for the HA epitope, or a 10-aa sequence for the FLAG epitope, between residues 508 and 509, in the middle of the loop, to produce mutants HA-II, HA-III, FLAG-IV, and FLAG-V. Mutants HA-II and HA-III differ in the first inserted residue (after 508), which is Cys in mutant HA-II and Asn in mutant HA-III. Mutants HA-II, HA-III, and FLAG-IV have some other mutations in common (see Fig. 1 and Table 1), including a change from Leu to Cys at residue 474, a change from Lys to Thr at position 507, and a change from Asn to Asp at position 509, just beyond the inserted sequence. These last two mutations are a consequence of the creation of the MfeI cloning site and are also present in mutant FLAG-V.
Construction of Insertion Mutants.
The plasmid used for creating mutants HA-II, HA-III, FLAG-IV, and FLAG-V in the colicin Ia gene was pUC19, with the colicin Ia gene, the colicin Ia immunity gene, and their promoters inserted between the unique EcoRI and BamHI sites of the vector. The starting plasmid for those mutants also contained an L474 → C mutation in the colicin Ia gene (12). A unique MfeI site that spanned codons 507–509 was created first, by using the QuikChange site-directed mutagenesis kit (Stratagene). The introduction of the MfeI site created the mutations K507 → T and N509 → D. The resulting plasmid was cut with MfeI. Linear DNA was gel-purified and treated with calf intestine alkaline phosphatase (Promega).
To create mutant HA-II, oligonucleotides 5′- AATTTGCTACCCATACGACGTCCCAGACTACGCTCG-3′ and 5′- AATTCGAGCGTAGTCTGGGACGTCGTATGGGTAGCA-3′ were treated with T4 polynucleotide kinase (Pharmacia) and annealed for 5 min at 70°C followed by 30 min at 37°C in 140 mM Mops/140 mM NaCl at a final concentration of 2.2 pmol/μl. The annealed oligonucleotides have MfeI-compatible sticky ends, a cysteine codon, TGC, followed by the DNA sequence encoding the 12CA5 HA epitope (YPYDVPDYA) (15, 18) and two additional nucleotides (CG) to correct the reading frame of the final construct. The annealed oligonucleotides were ligated [with T4 DNA ligase (BRL Life Technologies)] to the phosphatased linearized vector DNA at a 4-fold molar excess of oligonucleotide to vector. Plasmid DNA was prepared from ampicillin-resistant transformants and screened for the introduction of an AatII site (shown underlined in the insert oligonucleotide sequence) in the HA insert. DNA from colonies with the new AatII site was then sequenced to confirm the correct frame and orientation of the insertion. The amino acid sequence of the resulting plasmid is thus … Lys506Thr507Ile508CysTyrProTyrAspValProAspTyrAlaArgIleAsp509Ala510… . Residues indicated in bold were either mutated as a result of the creation of the MfeI site and re-ligation or are part of the HA epitope with an initial cysteine added. This colicin contains two cysteine residues, at position 474 and the first inserted amino acid. To eliminate the possibility of intramolecular disulfide bond formation, the cysteine at the beginning of the insert was mutated to asparagine (the original residue at position 509 of colicin Ia), by using site-directed mutagenesis (Stratagene QuikChange), thus creating mutant HA-III.
Mutant FLAG-IV was created in the same way as HA mutants II and III, but by using complementary FLAG-containing oligonucleotides with MfeI sticky ends and additional nucleotides to correct the reading frame.
Mutant HA-I was created by site-directed oligonucleotide mutagenesis of filamentous phage mp19 containing the colicin Ia and immunity protein genes, as described (19). Potential mutants were screened for insertion of an AatII site and sequenced as described above.
Preparation of Mutant Colicin Proteins.
The mutant colicins were induced and purified from E. coli strain TG1, as described previously (19, 20). Their yields and in vivo activities (21) are indicated in Table 1 and are well within the range of many of the colicin Ia mutant proteins that we have described previously (11, 12, 22).
The 10- to 12-residue insertions in mutants II through V appeared to increase the proteolytic susceptibility of the colicin. Slightly less than half of these mutant colicins was present in a truncated form, missing about 10–12 kDa from the full-length 66-kDa protein. Western blot analysis with anti-HA antibody, for mutants II and III, or with anti-FLAG, for mutants IV and V, revealed that the truncated polypeptides were missing the C-terminal end, including the HA or FLAG epitope. Because C-terminal deletions eliminate the channel-forming and bactericidal activities of channel-forming colicins, the truncated polypeptides should not have affected our measurements (23, 24). Mutant HA-I, in contrast, was not degraded.
Mutant proteins were stored in 25 or 50 mM sodium borate/300 mM NaCl/2 mM EDTA/2 mM DTT, pH 9.0, and were stable for weeks at 4°C. Longer-term storage was at −70°C. Aliquots of mutants HA-III and FLAG-IV were biotinylated and stored as described (12).
Planar Bilayer Experiments.
Membrane formation and voltage-clamp recording were done as previously described (12, 20). Briefly, asolectin (soybean lipid) (1% in pentane) was layered on top of the aqueous solutions in two compartments separated by a Teflon partition. The partition contained a 100- to 130-μm hole, which was pretreated on each side with ≈4 μl squalene (3% in petroleum ether). After the solvents evaporated, the lipid layers were raised over the level of the hole to form the membrane (25).
In most experiments, the aqueous solutions contained 100 mM KCl/5 mM CaCl2/1 mM EDTA/20 mM of buffer (Hepes, pH 7.2, or Mes, pH 6.2). In addition, for colicin mutants with one or more cysteine residues, 5 mM DTT was generally added to ensure that the sulfhydryl groups stayed reduced. The volume of solution on each side of the membrane was ≈1 ml. The voltage is that of the cis compartment, defined as the side to which colicin was added, with respect to that of the opposite trans compartment.
For many-channel experiments, colicins were usually diluted 1:1 with 1% octyl glucoside (Calbiochem) before addition to the cis compartment; this gave more channels per μg of protein without otherwise affecting channel behavior. In single-channel experiments, octyl glucoside was omitted.
Mutant HA-I was relatively unaffected by antibody at neutral pH; it was necessary to go to pH 5.5 (using 20 mM succinate or Mes buffer) to obtain a robust effect. To increase membrane stability in these low-pH experiments, the hole in which the membrane was formed was pretreated with more squalene than usual (≈4 μl of 10% squalene on each side).
RESULTS
General Experimental Approach.
The inserted epitopes serve as potential targets for binding by appropriate mAbs. By analogy with our previous experiments using biotinylated cysteine residues as targets for streptavidin binding (11, 12, 22), we anticipated that antibody binding might affect the voltage gating of the channel formed by epitope-inserted colicin. In this way, we hoped to locate the epitope on one side or the other of a planar lipid bilayer membrane.
We inserted these epitopes in the middle of the translocated region. We chose first to use the 12CA5 HA epitope from influenza virus hemagglutinin protein, because of its relatively benign amino acid sequence. Mutants with that insertion retained in vivo activity and normal channel formation in planar bilayers (see below). Thus emboldened, we then inserted the FLAG epitope, at the same position, to ascertain whether such a highly charged sequence (seven of eight residues) would create a barrier to translocation of that region of the colicin. These mutants also retained in vivo and planar bilayer channel-forming activity (see below).
Activity of Colicin Mutants Containing Epitope Insertions.
The mutant colicins containing the epitope insertions retained significant bactericidal activity against sensitive E. coli (Table 1). Two of the insertion mutants that also bore an L474 → C mutation (HA-II and FLAG-IV) had the most severely reduced in vivo activity (by two orders of magnitude), although the L474 → C mutation itself caused no loss of activity (see Table 1). Furthermore, mutant HA-III also has the L474 → C mutation and was 10 times more active than HA-II and FLAG-IV. It is therefore not the 474C mutation, per se, that causes a loss of in vivo activity. Preincubating HA-II in 5 mM DTT to reduce potential intramolecular disulfide bonds restored activity to near that of HA-III, whereas addition of DTT had no effect on in vivo activity of either FLAG mutant (not shown). Both of the FLAG insertion mutants, as well as HA-II, had a peculiar in vivo phenotype. Normally, when colicins at low dilution (high concentration, on the order of 0.1–10 μg/ml) are spotted on a lawn of sensitive indicator cells, the spots are clear; they become increasingly turbid at greater dilutions, until finally a dilution with no detectable activity is reached. Both of the FLAG insertion mutants and HA-II, however, made turbid spots at every active dilution tested.
Bilayer Experiments.
When added to planar bilayers, all of the epitope-containing mutants formed channels with conductance and voltage-dependent gating comparable to those of wild-type colicin Ia channels. This in itself suggests that the inserted sequences are translocated across the membrane, because we have previously shown that the surrounding region is normally translocated, and that restricting its translocation prevents the formation of normal channels (12).
HA Epitope Insertions.
We probed the location of the HA epitope (on one side of the membrane or the other) more directly by observing the effect of an HA antibody (referred to hereafter as “anti-HA”) (purified mouse monoclonal HA.11, Babco, Richmond, CA) on the gating of the mutant colicin channels. All three HA-inserted colicins gave qualitatively the same results. Channels formed by the mutant colicins opened normally when the cis compartment (the side to which colicin was added) was held at a potential of +50 mV relative to the opposite trans side, and they closed at −50 mV. The addition of anti-HA to the trans solution prevented open channels from closing. This is seen both at the macroscopic level, with many channels in the membrane (Fig. 2), and on single channels (Fig. 3). If, however, channels were exposed to trans anti-HA only in their closed state, subsequent gating was normal (not shown). Apparently, in association with channel opening, the HA epitope was translocated to the trans side, where it was bound by anti-HA; this prevented its moving back to the cis side, so the channels could not close. Several experiments demonstrated the specificity of the antibody effect: (i) control antibodies [mouse monoclonal anti-β-galactosidase (Promega) and anti-FLAG M2 (Kodak)] had no effect on HA-inserted colicins; (ii) anti-HA had no effect on wild-type channels; and (iii) the effect of trans anti-HA on the mutant colicin channels was prevented by the prior addition of excess HA peptide [YPYDVPDYA (Boehringer)] to the trans solution (not shown). Adding HA peptide after the antibody effect had already developed led to a very slow reversal of the effect, that is, to a restoration of nearly normal voltage gating (Fig. 3); presumably, the slowness of this process reflects a slow rate of dissociation of the antibody from the epitope.
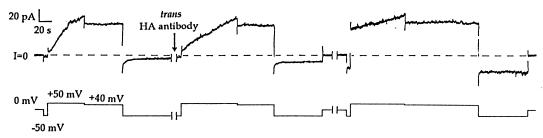
Figure 2.
The effect of trans anti-HA on HA-inserted colicin Ia. The upper trace is membrane current, and the lower trace is the voltage of the cis compartment with respect to the trans. Several minutes before the start of the record, 0.8 μg of HA-inserted colicin Ia (mutant HA-III) (plus 10 μg octyl glucoside) was added to the cis compartment, and the membrane was pulsed between positive and negative voltages to check for normal voltage gating. (For mutant HA-III, a small component of the conductance turned off slowly at −50 mV. This is seen as the small residual conductance at −50 mV, which has not yet gone to zero before the first break.) During the first break in the record (1 min), 20 μg of anti-HA was added to the trans compartment. The voltage was pulsed to +50 mV to turn on the conductance and then held at +40 mV to give the open colicin channels a longer exposure to trans antibody. Upon stepping to −50 mV, the turn-off of the conductance showed substantial inhibition. After several more voltage pulses during the second break (10 min), an even greater fraction of the conductance failed to turn off. A subsequent pulse to +50 mV and then to +40 mV resulted in a further increase in this fraction. Thus, the effect of trans anti-HA is to prevent open channels from closing, presumably because of the binding of antibody to the translocated HA epitope in the mutant colicin. The solutions on both sides of the membrane were 100 mM KCl/5 mM CaCl2/1 mM EDTA/20 mM Hepes, pH 7.2. The recording was taken from a chart record filtered at 100 Hz.
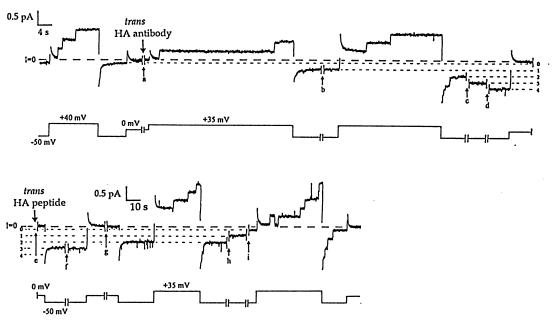
Figure 3.
The effect of trans anti-HA on HA-inserted colicin Ia at the single-channel level, and the reversal of this effect in the presence of HA peptide. For each pair of traces, the upper trace is membrane current and the lower trace is voltage. Several minutes before the start of the record, 0.5 μg of HA-inserted colicin Ia (mutant HA-II) was added to the cis compartment, and the membrane was pulsed between positive and negative voltages to confirm normal voltage gating. Near the beginning of break a (10 min), 20 μg of anti-HA was added to the trans compartment. After break a, the first pulse to +35 mV began with all channels closed, followed by the opening of two channels; when the voltage was stepped to −50 mV, instead of the rapid closing seen before antibody addition, one of the channels remained open. A second channel was locked open after break b (5 min), and a third and fourth channel were similarly locked open after channel openings during breaks c and d (2 min each). The short, dashed lines indicate the current levels at −50 mV with 0, 1, 2, 3, and 4 channels locked open. As in Fig. 2, it appears that trans anti-HA bound the translocated HA epitope in the open colicin channels and prevented the channels from closing. We attempted to reverse this effect of trans anti-HA by adding trans HA peptide to block the binding sites on the antibody. In the middle of break e (4 min.), 10 μg of HA peptide (a 68-fold molar excess) was added to the trans compartment. No additional channels were locked open over the next few hours, as the membrane was pulsed between positive and negative voltages. One of the locked-open channels closed quickly (within 2 min of peptide addition). Three channels remained open through break f (33 min), but one closed during break g (2 min) while the membrane was held at 0 mV. (Note the change in time scale after break g.) The remaining two channels closed, one at a time, during breaks h (85 min) and i (60 min). The channels subsequently showed normal voltage gating. Presumably, the locked-open colicin channels were able to close only after the antibody had dissociated from the HA epitope; the HA peptide was needed to prevent new antibody molecules from binding. Channels in the locked-open state showed greater rectification (conductance was 9 pS at +35 mV vs. 5 pS at −50 mV) than channels in the normal open state (9 pS vs. 7 pS). The solutions on both sides of the membrane were the same as in Fig. 2, with the addition of 5 mM DTT. The current transient following each voltage step was a result of the slow relaxation of the recording system at the high gain used here. The recording was taken from a chart record filtered at 10 Hz.
We also examined the effect of cis anti-HA on HA-inserted colicins. Anti-HA added to the cis solution prevented opening of the mutant channels (Fig. 4). This effect was slowly reversed by the addition of excess HA peptide to the cis solution. These are the results expected if the HA epitope must move away from the cis side (to the trans side) for channels to open, and if it moves back to the cis side when the channels close.
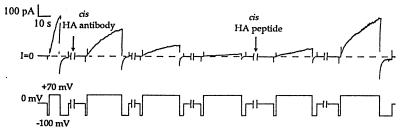
Figure 4.
The effect of cis anti-HA on HA-inserted colicin Ia, and the reversal of this effect in the presence of HA peptide. The upper trace is membrane current and the lower trace is voltage. Several minutes before the start of the record, 0.8 μg of HA-inserted colicin Ia (mutant HA-III) (plus 10 μg octyl glucoside) were added to the cis compartment, and the membrane was pulsed between positive and negative voltages to confirm normal voltage gating. (The turn-off of the conductance was fairly rapid at −100 mV.) Near the beginning of the first break in the record, 20 μg of anti-HA was added to the cis compartment. Over the next several pulses, the conductance at +70 mV declined gradually, with the rate of turn-on decreasing 60-fold. As indicated in the figure, 10 μg of HA peptide was added to the cis compartment during the fourth break. In the presence of HA peptide, the conductance slowly recovered over the next 2 hr. Thus, the binding of cis anti-HA to the HA epitope prevents channel formation by the mutant colicin. The lengths of the breaks were, in succession, 3 min, 3 min, 3 min, 10 min, and 93 min. The solutions were the same as in Fig. 3. The recording was taken from a chart record filtered at 100 Hz.
Results with mutant HA-I, with the epitope inserted after residue 511 in helix 4 (ref. 14 and M. Wiener, personal communication), were qualitatively the same as for mutants HA-II and HA-III, where the epitope, inserted after residue 508, is actually in the loop between helices 3 and 4 of the colicin’s channel-forming domain. However, with mutant HA-I, the pH had to be lowered to 5.5 to observe a robust effect of trans or cis antibody on channel opening and closing (not shown). Presumably, the HA epitope is not as exposed in mutant I as it is in mutants II and III. The lower pH may relax the structure of the colicin sufficiently, so that the epitope becomes more accessible to the antibody (26).
FLAG Epitope Insertions.
Because the colicin mutants containing HA insertions behaved normally on bilayers, we next prepared mutant colicins with the FLAG epitope, DYKDDDDK, inserted in place of the HA epitope (see Table 1). The FLAG epitope has a high density of charged residues, which might be expected to prevent translocation of this segment across the membrane. Nevertheless, when added to planar lipid bilayers, these mutants (FLAG-IV and FLAG-V) formed channels with normal single-channel conductance (not shown) and slightly altered voltage-dependent gating. Larger voltages than usual (e.g., ±90 mV) were needed to make the channels open and close quickly. Although there was little turn-on of conductance at +50 mV, if channels were first opened by pulsing to a large voltage, such as +90 mV, they stayed open when the voltage was lowered to +50 mV.
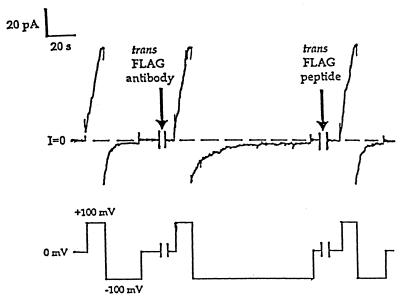
We probed the location of the FLAG epitope with an appropriate antibody (anti-FLAG M2), as we had done with HA-inserted colicins and anti-HA. The conductance induced by the FLAG-inserted colicins turned off at −100 mV with a time constant of 3–4 s. Shortly after anti-FLAG was added to the trans solution, the turn-off became slower, with a time constant of 14 s (Fig. 5). The subsequent addition of excess FLAG peptide to the trans solution rapidly restored the original fast turn-off rate. These are the results expected if anti-FLAG is in a relatively fast equilibrium with its epitope (compared with anti-HA, for instance) and dissociates from the FLAG epitope on a time scale of about 10 s. Presumably, when the antibody is bound, the epitope cannot move back across the membrane to close the channel, but when the antibody dissociates, then the channel is free to close. Thus, the slow closing rate in the presence of antibody reflects the rate of dissociation of antibody from the epitope. Consistent with this picture, the addition of anti-FLAG to the cis solution reversibly reduced the rate of channel opening at positive voltages (by about 80%) but did not completely eliminate channel activity (not shown). The specificity of the antibody effects is shown by their reversal by FLAG peptide, by the fact that control antibodies (anti-HA, anti-β-galactosidase) had no effect on FLAG-inserted colicin, and by the absence of an anti-FLAG effect on HA-inserted colicin.
Figure 5.
The effect of trans anti-FLAG on FLAG-inserted colicin Ia, and the reversal of this effect by FLAG peptide. The upper trace is membrane current and the lower trace is voltage. Several minutes before the start of the record, 1.5 μg of FLAG-inserted colicin Ia (mutant FLAG-IV) were added to the cis compartment, and the membrane was pulsed between ±90 or ±100 mV to establish the characteristic voltage gating of this mutant. As shown, the conductance increased rapidly at +100 mV and decreased at −100 mV. The time constant, τ, for closing at −100 mV was 3–4 s. During the first break in the record (2 min), 20 μg of anti-FLAG was added to the trans compartment. The turn-on of the conductance at +100 mV was not affected, but the turn-off rate at −100 mV was about four times slower (τ = 14 s) than before antibody addition. During the second break (5 min), 2.5 μg of FLAG peptide (a 20-fold molar excess) was added to the trans compartment. The turn-off at −100 mV was quickly restored to its original fast rate (τ = 3 s). Thus, the effect of trans anti-FLAG is to slow the closing rate of FLAG-inserted channels. The solutions were 100 mM KCl/5 mM CaCl2/1 mM EDTA/5 mM DTT/20 mM Mes, pH 6.2. The recording was taken from a chart record filtered at 30 Hz.
Biotinylated Mutants.
The above results are consistent with earlier experiments, in which we used streptavidin to probe the location of a biotin group attached to colicin Ia; we found there that a segment from residue 474 through 541 moves across the membrane when the channel opens and closes (11, 12). As a test of whether the epitope insertions perturbed this translocation, we biotinylated mutants HA-III and FLAG-IV at their unique cysteines (residue 474), as described (12). As expected, channels formed by the biotinylated mutants were prevented from closing by trans streptavidin (not shown).
DISCUSSION
The ability of colicin Ia to translocate at least 68 aa residues of itself across an otherwise unmodified lipid bilayer has been demonstrated previously by observing the effect of streptavidin on mutant channels biotinylated at various positions (12). This unexpected action could reflect either a property of the translocated region itself, or a property of some other part of the colicin molecule. That is, the translocated segment may possess some unrecognized attribute that accounts for its penchant for moving across the bilayer; or, this segment could be playing a fairly passive role as a substrate of another entity that catalyzes its translocation. If the former is true, significant alterations to its primary structure would be expected to interfere with translocation and probably with the associated properties of channel formation and voltage-dependent gating. If the latter is the case, then the translocation of this segment might be able to tolerate such changes. In this paper, we address this question by inserting strings of amino acids into the translocated segment, thereby altering its primary structure, and, at the same time, providing new target sites for water-soluble probes.
The insertion of two different foreign epitopes at the same position in the channel-forming domain of colicin Ia did not eliminate its bactericidal activity. The insertions did not alter the conductance of the mutant colicin channels in planar lipid bilayers and had, at most, minor effects on voltage-dependent channel gating. The addition of appropriate mAbs to the trans side of the planar bilayer (opposite of the side to which colicin was added) slowed or blocked channel closing; this clearly demonstrates that the inserted epitopes moved across the membrane to the trans side, where they could bind to antibody. Also, the insertions did not alter the effect of trans streptavidin on mutants biotinylated at residue 474. Evidently, the insertion of the HA or FLAG epitope does not prevent the voltage-dependent translocation of, at least, residues 474 through 509 and, presumably, the rest of the translocated segment. This confirms our suspicion that the precise sequence of the translocated segment is not critically important and suggests that the remaining part of the channel-forming domain (the last 85 residues, 542–626) is a protein translocator, capable of translocating a variety of protein sequences.
All of the insertion mutants suffered some loss of bactericidal activity relative to their “noninserted” counterparts, wild-type or L474 → C. However, their in vivo activities were well within the range of some of the cysteine mutants we have purified previously (ref. 12 and unpublished results). Both of the FLAG-insert mutants and HA-II made turbid spots on lawns of sensitive E. coli, even at very high concentrations (≈105 molecules of colicin per sensitive cell in the lawn). This behavior suggests that there was a lag in the killing of sensitive cells by these proteins. Because much of the reduction in the in vivo activity of HA-II, with two cysteine residues, relative to HA-III, which has only a single cysteine, was relieved by preincubation of HA-II with DTT, its loss of activity appears to be a result of formation of intramolecular disulfide bonds. Because there was no such effect of DTT on the killing activity of either FLAG-IV or FLAG-V, the turbid spots they form may be a reflection of the alteration in the kinetics of channel turn-on we observed on planar lipid bilayers.
There also were differences in the channel-gating properties of colicins bearing the two different epitopes. Both classes of mutant formed channels on planar lipid bilayers with conductance and voltage-dependent gating properties similar to those of wild-type colicin. However, colicins with the FLAG insert required larger voltages than wild-type or HA-insert colicins to turn their channels on and off quickly, although our results suggest that the difference is a kinetic one, rather than a change in the equilibrium voltage dependence. A possible explanation is that the highly charged FLAG insertion in these mutants elevates the energy barrier that the translocated segment must surmount to cross the membrane.
Each mAb (anti-HA and anti-FLAG M2), when added to the trans side, interfered with the closing of its corresponding epitope-inserted colicins. The apparent difference between the effects of anti-HA and anti-FLAG M2 (blocking vs. slowing of closing) is a consequence of the difference in the rates of interaction of antibody and epitope. The effects seen with anti-HA took a long time to develop (10–20 min). Once bound, the antibody dissociated still more slowly (100–200 min), even in the presence of competing peptide. (Note the lengths of the breaks in Figs. 3 and 4.) On the other hand, the effects of anti-FLAG M2 on FLAG-containing colicins developed much more rapidly and reversed very rapidly as well (<1 min). Thus, rather than “locking” channels open for hours, as does anti-HA, anti-FLAG merely slows the apparent closing rate.
Likewise, this difference in rates explains why cis anti-FLAG produced such a modest effect (reduction of channel activity) compared with cis anti-HA. Colicins bound by cis anti-HA are completely inactivated, because of its slow dissociation rate. In contrast, with cis anti-FLAG, as the few unbound channels open, the proportion of bound and unbound channels quickly reequilibrates, so that the population of unbound channels is continually replenished.
Taken together, our results suggest that the translocated segment is the substrate for a protein translocator located nearer the C-terminal end of the colicin molecule. It is not obvious how colicin Ia catalyzes the translocation of such a large, hydrophilic portion of itself across a hydrophobic membrane. A portion of the E. coli SecG protein has also been shown to undergo significant movement, back and forth across the cytoplasmic membrane, in conjunction with the ATP-driven cyclic insertion and deinsertion of SecA protein, which drives preprotein translocation (27). However, SecG protein is a small, hydrophobic protein, and its inversion is thought to consume little energy; its movement may, in fact, actually reduce the energy required for movement of SecA in the membrane (27). The gating of a variety of other channels, including the Shaker potassium channel (28), sodium channel (29), and VDAC (30), also involves movement across a membrane of part of the channel protein itself, but none of these has been shown to exhibit such extensive protein movement as has colicin Ia. Protein translocation systems must solve the energetic problem of moving a hydrophilic protein across a hydrophobic membrane. The most prominent features of the last 85 residues of colicin Ia are a hydrophobic helical hairpin and an amphipathic α-helix (Fig. 1), all of which are in an inserted state in the membrane in our model of the open channel (12, 22). These three putative transmembrane segments must somehow form a favorable pathway for an arbitrary fourth segment to snake through. This is particularly problematic, because there is no evidence to suggest that more than one colicin molecule is required to form a channel (10, 31).
Acknowledgments
We thank Michael Wiener for communicating details of the colicin Ia helix assignments prior to publication. This work was supported by National Institutes of Health Grant GM29210.
ABBREVIATION
- HA
hemagglutinin
Note Added in Proof
The association and dissociation rate constants of the FLAG- and HA-inserted mutant colicins with their respective mAbs were kindly measured for us by Jian Zhong Sun of the Howard Hughes Medical Institute of UCLA, using the biosensor technique based on surface plasmon resonance (Biacore, Pharmacia). The anti-FLAG association rate constant (2.60 × 105 M−1 s−1) was more than 200 times greater than that of anti-HA (1.16 × 103 M−1 s−1); the anti-FLAG dissociation rate constant was 1.40 × 10−3 s−1, compared with an immeasurably small dissociation rate constant for anti-HA. These measurements are consistent with our report of a rapid binding and dissociation of anti-FLAG to FLAG-inserted colicin and a very slow binding and dissociation of anti-HA to HA-inserted colicin.
Footnotes
A commentary on this article begins on page 4081.
References
- 1.Schatz G, Dobberstein B. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- 2.Derossi D, Calvet S, Trembleau A, Brunissen A, Chassaing G, Prochiantz A. J Biol Chem. 1996;271:18188–18193. doi: 10.1074/jbc.271.30.18188. [DOI] [PubMed] [Google Scholar]
- 3.Oliver D. Mol Microbiol. 1993;7:159–165. doi: 10.1111/j.1365-2958.1993.tb01107.x. [DOI] [PubMed] [Google Scholar]
- 4.Pugsley A P. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montecucco C, Papini E, Schiavo G. In: Sourcebook of Bacterial Protein Toxins. Alouf J E, Freer J H, editors. New York: Academic; 1991. pp. 45–56. [Google Scholar]
- 6.Lesieur C, Vécsey-Semjén, Abrami L, Fivaz M, van der Goot F G. Mol Membr Biol. 1997;14:45–64. doi: 10.3109/09687689709068435. [DOI] [PubMed] [Google Scholar]
- 7.Bourdineaud J P, Boulanger P, Lazdunski C, Letellier L. Proc Natl Acad Sci USA. 1990;87:1037–1041. doi: 10.1073/pnas.87.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schein S J, Kagan B L, Finkelstein A. Nature (London) 1978;276:159–163. doi: 10.1038/276159a0. [DOI] [PubMed] [Google Scholar]
- 9.Nogueira R A, Varanda W A. J Membr Biol. 1988;105:143–153. doi: 10.1007/BF02009167. [DOI] [PubMed] [Google Scholar]
- 10.Cramer W A, Heymann J B, Schendel S L, Deriy B N, Cohen F S, Elkins P A, Stauffacher C V. Annu Rev Biophys Biomol Struct. 1995;24:611–641. doi: 10.1146/annurev.bb.24.060195.003143. [DOI] [PubMed] [Google Scholar]
- 11.Slatin S L, Qiu X-Q, Jakes K S, Finkelstein A. Nature (London) 1994;371:158–161. doi: 10.1038/371158a0. [DOI] [PubMed] [Google Scholar]
- 12.Qiu X-Q, Jakes K S, Kienker P K, Finkelstein A, Slatin S L. J Gen Physiol. 1996;107:313–328. doi: 10.1085/jgp.107.3.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mankovich J A, Hsu C-H, Konisky J. J Bacteriol. 1986;168:228–236. doi: 10.1128/jb.168.1.228-236.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiener M, Freymann D, Ghosh P, Stroud R M. Nature (London) 1997;385:461–464. doi: 10.1038/385461a0. [DOI] [PubMed] [Google Scholar]
- 15.Wilson I A, Niman H L, Houghten R A, Cherenson A R, Connolly M L, Lerner R A. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 16.Hopp T P, Prickett K S, Price V L, Libby R T, March C J, Cerretti D P, Urdal D L, Conlon P J. Bio/Technology. 1988;6:1204–1210. [Google Scholar]
- 17.Wiener M, Freymann D, Ghosh P, Finer-Moore J, Stroud R M. Biophys J. 1996;70:A140. (abstr.). [Google Scholar]
- 18.Kolodziej P A, Young R A. Methods Enzymol. 1991;194:508–519. doi: 10.1016/0076-6879(91)94038-e. [DOI] [PubMed] [Google Scholar]
- 19.Qiu X-Q, Jakes K S, Finkelstein A, Slatin S L. J Biol Chem. 1994;269:7483–7488. [PubMed] [Google Scholar]
- 20.Jakes K S, Abrams C K, Finkelstein A, Slatin S L. J Biol Chem. 1990;265:6984–6991. [PubMed] [Google Scholar]
- 21.Jakes K S, Zinder N D, Boon T. J Biol Chem. 1974;249:438–444. [PubMed] [Google Scholar]
- 22.Kienker P K, Qiu X-Q, Slatin S L, Finkelstein A, Jakes K S. J Membr Biol. 1997;157:27–37. doi: 10.1007/s002329900213. [DOI] [PubMed] [Google Scholar]
- 23.Liu Q R, Crozel V, Levinthal F, Slatin S, Finkelstein A, Levinthal C. Proteins. 1986;1:218–229. doi: 10.1002/prot.340010304. [DOI] [PubMed] [Google Scholar]
- 24.Baty D, Knibiehler M, Verheij H, Pattus F, Shire D, Bernadac A, Lazdunski C. Proc Natl Acad Sci USA. 1987;84:1152–1156. doi: 10.1073/pnas.84.5.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montal M. Methods Enzymol. 1974;32:545–554. doi: 10.1016/0076-6879(74)32053-8. [DOI] [PubMed] [Google Scholar]
- 26.van der Goot F G, González-Mañas J M, Lakey J H, Pattus F A. Nature (London) 1991;354:408–410. doi: 10.1038/354408a0. [DOI] [PubMed] [Google Scholar]
- 27.Nishiyama K, Suzuki T, Tokuda H. Cell. 1996;85:71–81. doi: 10.1016/s0092-8674(00)81083-1. [DOI] [PubMed] [Google Scholar]
- 28.Mannuzzu L M, Moronne M M, Isacoff E Y. Science. 1996;271:213–216. doi: 10.1126/science.271.5246.213. [DOI] [PubMed] [Google Scholar]
- 29.Yang N, George A L, Jr, Horn R. Neuron. 1996;16:113–122. doi: 10.1016/s0896-6273(00)80028-8. [DOI] [PubMed] [Google Scholar]
- 30.Zizi M, Thomas L, Blachly-Dyson E, Forte M, Colombini M. J Membr Biol. 1995;144:121–129. doi: 10.1007/BF00232798. [DOI] [PubMed] [Google Scholar]
- 31.Slatin S L. Int J Biochem. 1988;20:737–744. doi: 10.1016/0020-711x(88)90058-4. [DOI] [PubMed] [Google Scholar]