Abstract
The β-carbon of the Pseudomonas aeruginosa 1244 pilin C-terminal Ser is a site of glycosylation. The present study was conducted to determine the pilin structures necessary for glycosylation. It was found that although Thr could be tolerated at the pilin C terminus, the blocking of the Ser carboxyl group with the addition of an Ala prevented glycosylation. Pilin from strain PA103 was not glycosylated by P. aeruginosa 1244, even when the C-terminal residue was converted to Ser. Substituting the disulfide loop region of strain PA103 pilin with that of strain 1244 allowed glycosylation to take place. Neither conversion of 1244 pilin disulfide loop Cys residues to Ala nor the deletion of segments of this structure prevented glycosylation. It was noted that the PA103 pilin disulfide loop environment was electronegative, whereas that of strain 1244 pilin had an overall positive charge. Insertion of a positive charge into the PA103 pilin disulfide loop of a mutant containing Ser at the C terminus allowed glycosylation to take place. Extending the “tail” region of the PA103 mutant pilin containing Ser at its terminus resulted in robust glycosylation. These results suggest that the terminal Ser is the major pilin glycosylation recognition feature and that this residue cannot be substituted at its carboxyl group. Although no other specific recognition features are present, the pilin surface must be compatible with the reaction apparatus for glycosylation to occur.
Pseudomonas aeruginosa is a Gram-negative bacterium that expresses polar filaments called type IV pili. These pili are polymers of a primarily proteinaceous subunit referred to as pilin. Although all known type IV pilins are modified by the cleavage of a leader sequence followed by the N-methylation of the exposed Phe (1), the pilin of P. aeruginosa 1244 is additionally glycosylated. This glycosylation requires the presence of PilO, an enzyme whose coding sequence is in the same operon as pilA, the pilin structural gene (2).
The first discovered examples of prokaryotic glycoproteins were archaeal S-layer proteins (3). Since this finding, numerous examples of protein glycosylation in eubacteria have been identified (4 – 8), indicating that this protein modification is distributed among all of the biological kingdoms. Proteins destined for glycosylation may be modified at a single or multiple sites along the peptide chain (7, 8) in which the glycan may be one sugar or an assortment of diverse saccharides. Oligosaccharide assembly can take place either by sequential sugar addition to the target protein or by synthesis prior to glycosylation in which the entire pre-formed glycan is transferred as a single unit (7, 8). Numerous protein/sugar linkages have been characterized involving many functional groups present on peptide chains and most of the commonly encountered monosaccharide moieties (8), in addition to exotic prokaryotic sugars such as bacillosamine, pseudaminic acid, and derivatives (7). Carbohydrates in eukaryotes and prokaryotes, alike, are usually bound to the peptide chain via the amide nitrogen of an Asn residue (N-linked) or via the functional hydroxyl group of Ser or Thr (O-linked). Some of the more uncommon glycosylation linkages include the mannose linkage to the C-2 of the indole ring of Trp observed in the mammalian cytokine interleukin 12 (8) and Tyr O-glycosylation of a surface protein of the prokaryote Acetogenium kivui (9). Besides the N-linked glycosylation system in Campylobacter jejuni (7, 10 – 13), little is known of glycan biosynthesis and transferases of prokaryotic glycosylation systems.
In P. aeruginosa 1244, a single glycan is attached to each pilin monomer in an O-linked fashion to the C-terminal residue, Ser148 (14). This residue is adjacent to the pilin disulfide loop (DSL),2 a structure previously associated with adhesion and immunogenicity (15, 16). An intriguing aspect of P. aeruginosa 1244 pilin glycosylation is that the trisaccharide used to modify pilin is structurally identical to the O-antigen repeating unit (17, 18). This suggested that the pilin glycan is a product of the O-antigen biosynthetic pathway. DiGiandomenico et al. (17) showed that strain 1244 mutants lacking functional wbpM or wbpL genes (both involved in the initial steps of O-antigen biosynthesis) synthesized no O-antigen and produced only nonglycosylated pilin. Expression of gene clusters coding for heterologous O-antigens in strain 1244 resulted in the production of pilin glycosylated with the heterologous saccharide, confirming that the source of the pilin glycan is part of the O-antigen biosynthetic pathway. PilO is not essential for O-antigen or pilin biosynthesis and is the only glycosylation-specific protein requirement in P. aeruginosa 1244 pilin glycosylation (17) suggesting that it is an oligosaccaryl-transferase.
In transferase-mediated protein glycosylation, specific interaction is necessary between this enzyme, a glycan source, and the target protein, to facilitate the enzymatic attachment of the saccharide to the designated amino acid functional group (7, 8). A consensus peptide sequence of Ser/Thr-Xaa-Asn, in which Xaa can be any residue except Pro, acts as the environment for N-linked glycosylation as observed in N-glycosylated proteins of C. jejuni, Mycobacterium tuberculosis, and Clostridium thermocellum (6, 13). A consensus sequence for O-glycosylation, other than the requisite of a Ser/Thr residue, has not been identified, indicating distinct substrate specificity for individual transferases that mediate O-linked glycosylation. Gaining an understanding of the pilin structures necessary for pilin glycosylation by P. aeruginosa 1244 would provide insight into an important biological process in which little is understood and may usher in the comprehension of O-linked glycosylation specificities of other systems.
The present study is the most comprehensive investigation of prokaryotic glycosylation substrate specificity to date. These results suggest that the C-terminal Ser of P. aeruginosa 1244 pilin is the major glycosylation recognition structure and that this residue cannot be substituted at its carboxyl group. Although no other specific recognition features are present, the pilin surface must be compatible with the reaction machinery for glycosylation to occur. Further, we present evidence that suggests that pilin folding and disulfide bond formation occurs prior to glycosylation. These findings could have future applications in vaccine engineering.
EXPERIMENTAL PROCEDURES
Culture Conditions
Bacterial strains and plasmids used in this study are listed in Table 1. Intermediate plasmids utilized for mutant construction can be found in supplemental Table S1. For general culturing, bacteria were grown aerobically at 37 °C on LB agar plates or in LB broth, shaken at 250 rpm. The cultures were grown on casamino acids yeast extract medium (2% agar, 0.75% casamino acids, 0.15% yeast extract (19)) for the preparation of cell extracts. The concentrations of antibiotics used in selective media were as follows: ampicillin at 50 μg/ml for Escherichia coli; carbenicillin at 250 μg/ml for P. aeruginosa; kanamycin at 35 μg/ml for E. coli and 100 μg/ml for P. aeruginosa; and tetracycline at 100 μg/ml for P. aeruginosa. IPTG was added to media to a final concentration of 5 mm to induce expression of genes under the control of the tac promoter.
TABLE 1.
Strains and plasmids used in this study
| Strains/plasmids | Description | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| 1244 | Wild type, IATS O7 | Ref. 39 |
| 1244N3 | rpoN (TcR) | Ref. 27 |
| 1244.47 | pilAO (HgR) | This study |
| 1244.2024 | pilAOBCD (KmR) | This study |
| PA103 | Wild type, IATS O11 | Ref. 40 |
| E. coli | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac | Stratagene |
| XL10-Gold | endA1 supE44 thi-1 recA1 gyrA96 relA1 lacHte | Stratagene |
| DH5α | supE44 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Invitrogen |
| HB101 | supE44 hsdS20 recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | Ref. 41 |
|
| ||
| Plasmids | ||
| pUCP26 | 4.9 kb pUC18-derived broad host-range cloning vector, TcR | Ref. 42 |
| pUCP26pilO | pUCP26 with 1244 pilO | This study |
| pMMB66EH | 8.8-kb broad host range expression vector, ApR/CbR | Ref. 43 |
| pPAC46 | pMMB66EH with 1244 pilAO | Ref. 2 |
| pPAC202∷Tn501-13 | A cosmid subclone containing a transposon insertion in 1244 pilA | Ref. 21 |
| p1244 S148T | pPAC46 with coding sequence for Ser148 of pilA mutated to Thr | This study |
| p1244 A149 | pPAC46 with the addition of an Ala and a stop codon after Ser148 of pilA | This study |
| pSD5 | pMMB66EH with PA103 pilA | This study |
| p103 P144S | pSD5 with the Pro144 codon of pilA mutated to Ser | This study |
| p103 PKS | pSD5 with codons for Asn142, Glu143, and Pro144 of pilA mutated to Pro, Lys, and Ser, respectively | This study |
| pHYBRID | pMMB66EH with the chimeric pilA coding for the first 123 residues of PA103 pilin, a Gly at residue 124, and the last 31 residues are those of 1244 pilin; this plasmid also contains pilO | This study |
| pHYB-O | pMMB66EH with the chimeric pilA from pHYBRID; this plasmid does not contain pilO | This study |
| p1244 C127A | pPAC46 with coding sequence for Cys127 of pilA mutated to Ala | This study |
| p1244 C145A | pPAC46 with coding sequence for Cys145 of pilA mutated to Ala | This study |
| p1244Δ135–139 | pPAC46 with coding sequence for residues 135–139 of pilA (AWKPN) deleted | This study |
| p1244Δ135–144 | pPAC46 with coding sequence for residues 135–144 of pilA (AWKPNYAPAN) deleted | This study |
| p1244Δ128–144 | pPAC46 with coding sequence for residues 128–144 of 1244 pilA (KITKTPTAWKPNYAPAN) deleted | This study |
| p103 INS136 | pSD5 with coding sequence for residues 135–139 of 1244 pilA (AWKPN) inserted at amino acid positions 136–141 of PA103 pilA with Pro144 (now Pro149) mutated to Ser | This study |
| p1244 S147 | pPAC46 with mutant pilA that had coding sequence for Lys 147 mutated to Ser and Ser 148 mutated to a stop codon | This study |
| p1244 S149 | pPAC46 with 1244 pilA that had the Ser148 codon mutated to Ala and the addition of Ser 149 followed by a stop codon | This study |
| p1244 S150 | pPAC46 with mutant pilA that had coding sequence for Ser148 mutated to Ala, an Ala at position 149, a Ser at position 150 followed by a stop codon | This study |
| p103 S146 | pSD5 with coding sequence for two Ala inserted into PA103 pilA at positions 144 and 145 terminating in Ser146 | This study |
Homology Modeling
Homology modeling was performed by submission of mature pilin sequences to the online 3D-PSSM program (20). DS Viewer Pro 6.0 was used to visualize tertiary structures and to generate molecular surfaces and Gasteiger charges. Fig. 1 shows the primary (21) and predicted tertiary structure of 1244 pilin derived from homology modeling.
FIGURE 1. Primary and tertiary structure of 1244 pilin.
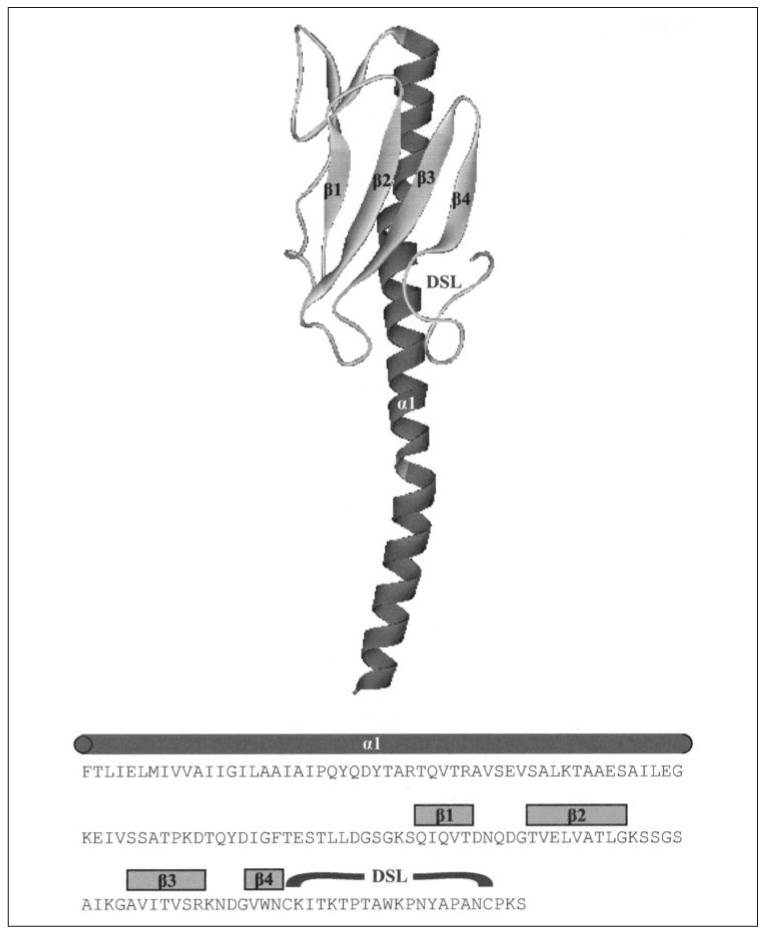
Strain 1244 pilin tertiary structure was derived from homology modeling. Predicted α-helix, β-sheets, and DSL are labeled on the model and above the corresponding amino acid sequence of mature 1244 pilin.
Generation of P. aeruginosa 1244 pilA Mutants
The deletion mutant 1244.2024 was constructed by first digesting pPAC202 (21), which contained a 7.8-kb insert harboring the pilAO operon, with PstI. This removed a 3.8-kb fragment containing the operon and left flanking insert pieces of 1.9 and 2.1 kb attached to the vector. The fragment containing the vector was ligated with a KmR cartridge, generated by digesting pUC-4K (Amersham Biosciences) with PstI, producing pPAC2024K. The cloned region of this plasmid was removed by digestion with EcoRI and ligated with pBR322 digested with the same enzyme producing pPAC3/6. This construct was introduced into P. aeruginosa 1244 by triparental mating (22) using tetracycline resistance as a selective marker. Tested clones were found to also be KmR, indicating that incorporation of the plasmid by recombination had occurred. Colony lifts probed with a strain 1244 pilin-specific monoclonal antibody produced pilin-negative clones that were TcS, CbS, and KmR, indicating the loss of vector and the functional pilA gene. The loss of pilin production was confirmed by Western blot, whereas the loss of pilA was confirmed by Southern blot using pPAC2024K insert DNA as a probe.
The initial step for construction of the insertion mutant, strain 1244.47, involved ligation of pBR322 digested with BamHI and NheI with the 12-kb fragment produced by the digestion of pPAC202∷Tn501-13, a previously described construct (21) with BamHI and XbaI. The plasmid produced by this ligation reaction was confirmed by restriction site analysis and introduced into P. aeruginosa 1244 by triparental mating (22) using mercury resistance as a selective marker. Tested clones were found to also be CbR, indicating that incorporation of the plasmid by recombination had occurred. Approximately 2 × 105 colony-forming units of one of these clones were incubated with 2 × 106 plaque-forming units of pilus-specific bacteriophage PE69 in 200 μl of LB broth for 20 min at room temperature. Aliquots of this material were plated on LB-Hg medium where clones produced were found to be HgR and CbS, indicating the loss of vector. Disruption of the pilA gene of one of these clones, referred to as strain 1244.47, was confirmed by Southern blot using strain 1244 pilA DNA as a probe. Loss of gene function was confirmed by Western blotting of 1244.47 cell extracts using a pilin-specific monoclonal antibody as a probe.
Plasmid Construction
To analyze pilin specificity for glycosylation, cloned pilA was subjected either to site-directed mutagenesis (QuikChange mutagenesis kit) or to deletion/insertion mutagenesis (ExSite mutagenesis kit), or a hybrid pilA was constructed consisting of portions of strain PA103 and 1244 pilin coding sequence (Table 1 and Fig. 2). For a detailed description of the construction of plasmids, see the supplemental material.
FIGURE 2. Amino acid sequences of segments of wild type and mutant pilins used in this study.
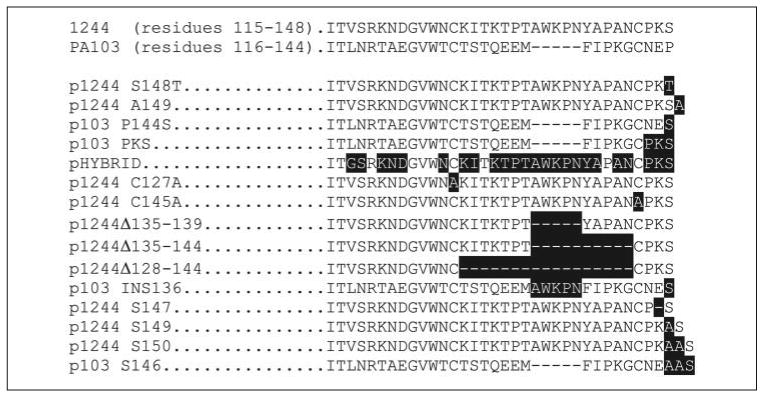
The strains and plasmids producing the pilins are shown on the left. The residues differing from the wild type strain as a result of a mutation are written in white and highlighted in black.
Western Blot Analysis
Western blotting using cell extracts was performed as previously described (21). Extracts of 1244N3/pPAC46 and 1244N3/pPAC24 (2) were used as the source of 1244 glycosylated pilin standards and 1244 nonglycosylated pilin standards, respectively. When mutant pilin was not easily detectable from cell extracts in Western blot analyses, the membrane fraction was extracted as a means to concentrate pilin for analysis. Here, the membrane fraction was harvested from 200-ml LB overnight broth cultures. The A650 of the culture was recorded followed by centrifugation at 5,000 rpm in a GSA rotor for 20 min at 4 °C. The pellet was washed twice with 200 ml of phosphate-buffered saline and was resuspended in 10 ml of lysis buffer (10 mm K2HPO4, 30 mm NaCl, 0.25% Tween 20, 10 mm β-mercaptoethanol, 10 mm EDTA, 10 mm EGTA). This material was sonicated (three 15-s bursts, on ice) and centrifuged at 6,000 rpm in an SS-34 rotor for 10 min at 4 °C to remove unbroken cells and debris. The supernatant fluid was subjected to ultracentrifugation (SW60Ti rotor) at 30,000 rpm for 1 h, after which the membrane material was located in the pellet. The membrane fraction was resuspended in 400 μl of deionized H2O. The initial A650 values taken for cell culture density were used to normalize samples for SDS-PAGE.
The cell extracts and membrane fraction samples were separated by SDS-PAGE using 16% T, Tris-glycine or 16% T, Tricine gels (23). Tricine-PAGE was used when PA103 pilin was being analyzed as this pilin resolved into sharper bands under these conditions. Separated molecules were electroblotted to nitrocellulose paper and blocked as previously described (21). Either anti-1244 pilin mouse monoclonal antibody, 5.44 (24); anti-PA103 pilin mouse monoclonal antibody, 2.97 (25); or anti-pilin polyclonal serum was used as the primary probe to detect the proteinaceous portion of pilin. To determine whether or not the pilin was glycosylated, anti-1244 pilin glycan monoclonal antibody, 11.14 (18), or anti-1244 pilin glycan polyclonal serum was used as the primary probe. To obtain the anti-1244 pilin glycan polyclonal antibodies, rabbit serum from an animal that was immunized with pure P. aeruginosa 1244 glycosylated pili was adsorbed with pure 1244 pili containing a heterologous glycan. This yielded a glycan-specific serum devoid of a pilin protein response. The monoclonal antibodies were used as a probe when epitope disruption did not result from mutagenesis of pilin genes (16, 25). Alternatively, polyclonal serum was employed. A fluorescein isothiocyanate- or alkaline phosphatase-conjugated secondary antibody (26) was used, and fluorescence was detected using a FluorImager595 (Molecular Dynamics) employing the 530 DF30 filter.
RESULTS
Structural Requirements of the Pilin Glycosylation Residue
We have previously provided evidence that the C-terminal residue, Ser148 (Fig. 2), is the glycosylation site of P. aeruginosa 1244 pilin (14). To determine whether a Thr could be tolerated at this position, the Ser148 codon of pPAC46 was changed to a Thr codon, yielding p1244 S148T (Fig. 2). When extracts of strain 1244N3, a pilAO negative mutant (27), expressing this construct were subjected to Western blot analysis (Fig. 3), it was seen that pilin containing Thr in the 148 position was capable of being glycosylated. This was determined by position of pilin migration (Fig. 3A) and by reaction with a glycan-specific antibody (Fig. 3B). These results indicate that recognition of the hydroxyl group at the glycosylation site was not hindered by the presence of the Thr methyl group.
FIGURE 3. Western blot analysis of P. aeruginosa 1244 mutant pilins.
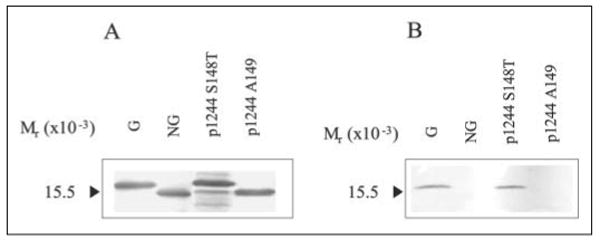
Western blot of 1244 pilin produced by P. aeruginosa 1244N3/p1244 S148T and 1244N3/p1244 A149 of cell extracts using anti-1244 pilin monoclonal antibody 5.44 (A) or anti-1244 pilin glycan monoclonal antibody 11.14 (B) as a probe. G is glycosylated 1244 pilin, and NG is nonglycosylated 1244 pilin. Pilin terminating in Thr rather than Ser was glycosylated (1244N3/p1244 S148T), whereas pilin terminating in Ala, where Ser148 was the penultimate residue, was nonglycosylated.
Because the strain 1244 pilin glycosylation site is at the C terminus, this amino acid, in addition to lacking a C-linked residue, contains a carboxyl group. To see whether this arrangement was necessary for glycosylation, p1244 A149, a construct in which an Ala codon was inserted after Ser148, was generated and expressed in 1244N3. Western blot analysis of cell extracts of 1244N3/p1244 A149 revealed that pilin produced was nonglycosylated as determined by apparent molecular weight (Fig. 3A) and by lack of reaction with a glycan-specific antibody (Fig. 3B). These results suggest that pilin glycosylation by strain 1244 requires that the residue being modified be at the C terminus. Such a situation would explain why none of the other Ser or Thr residues of this protein, even though likely at the protein surface, were glycosylated.
Minimal Pilin Structures Necessary for Glycosylation
To determine structures important in glycosylation of strain 1244 pilin, we replaced residues of PA103 pilin (Fig. 2), a normally nonglycosylated pilin (44), with those of 1244 pilin and assayed for glycosylation. When plasmid pSD5, which contained the pilA gene from PA103 behind a tac promoter, was expressed in strain 1244, PA103 pilin produced had the same apparent molecular weight as that of wild type (Fig. 4A) as determined by Western blot using a PA103 pilin-specific monoclonal antibody as a probe. This is not surprising because strain 1244 pilin glycosylation requires a C-terminal Ser (14), whereas PA103 pilin terminates in a Pro (Fig. 2). Site-directed mutagenesis was used to convert the C-terminal pilin residue encoded in pSD5 to Ser, thereby producing p103 P144S (Fig. 2). When this plasmid was expressed in P. aeruginosa 1244 and pilin was assayed by Western blot for modification by PilO, it was revealed that the PA103 pilin produced showed no difference in apparent molecular weight when compared with wild type PA103 pilin, indicating the absence of glycosylation (Fig. 4A). The pilin glycosylation ability of the host strain was confirmed by Western blot analysis using an anti-1244 pilin monoclonal antibody, which revealed the presence of glycosylated strain 1244 pilin (data not shown). These results indicate that other structures besides the C-terminal Ser/Thr are important for pilin glycosylation.
FIGURE 4. Western blot analysis of P. aeruginosa PA103 mutant pilins.
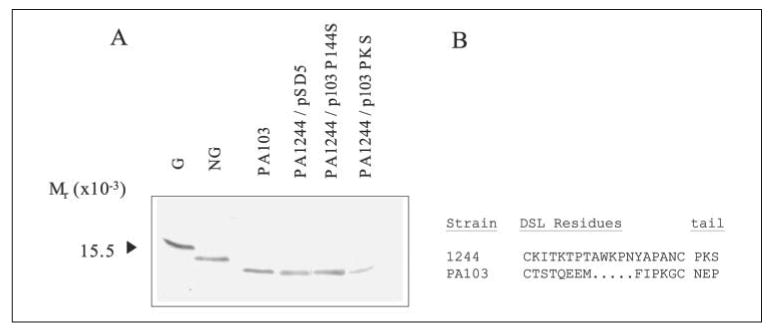
Western blot of pilin produced by P. aeruginosa 1244/pSD5, 1244/p103 P144S, and 1244/p103 PKS using anti-PA103 pilin monoclonal antibody 2.97 as a probe (A). G and NG are glycosylated and nonglycosylated pilin standards, respectively, on a separate blot that was probed with anti-1244 pilin monoclonal antibody, 5.44. Wild type PA103 pilin (pSD5), PA103 pilin with residue Pro144 (p103 P144S) mutated to Ser, as well as PA103 pilin with residues Asn142, Glu143, and Pro144 mutated to that of 1244 pilin, Pro, Lys, and Ser, respectively (p103 PKS) were all nonglycosylated in the presence of PilO. The amino acid sequences of the DSL and tail regions of strains 1244 and PA103 pilin were aligned for comparison (B).
Both 1244 and PA103 pilins contain a “tail” region that consists of three residues beyond the C-proximal DSL Cys (Fig. 4B). p103 PKS (Fig. 2) was generated to determine whether either of the remaining residues in the tail served as a recognition site for pilin glycosylation. When this plasmid was expressed in strain 1244 and extracts of these cells were subjected to Western blot analysis, it could be seen that the only PA103 pilin produced was nonglycosylated (Fig. 4A). This result suggests that structures other than the 1244 pilin tail region are needed for glycosylation.
To determine the role of the DSL in glycosylation, the loop structure of strain PA103 pilin was substituted with the analogous region of strain 1244. The plasmid constructed to test this, pHYBRID, codes for a hybrid protein consisting of the first 123 residues of PA103 pilin, the C-terminal 31 residues of 1244 pilin (which includes the entire 1244 DSL), and PilO, all behind a tac promoter (Fig. 2). This construct was expressed in 1244.47, a pilAO negative mutant, where membrane fractions were subjected to Western blot analysis (Fig. 5). The blot, when probed with a pilin-specific antibody, revealed the presence of a band with an apparent molecular weight similar to that of glycosylated pilin (Fig. 5A). When pilO was absent, this band was replaced with one corresponding with nonglycosylated pilin (Fig. 5A). In addition, in the presence of PilO, the pilin produced reacted with an antibody specific for the pilin glycan, whereas there was no reaction to the hybrid pilin expressed in the absence of PilO (Fig. 5B), indicating that the higher apparent molecular weight of the hybrid pilin was a result of the presence of the glycan (Fig. 5). These results indicate that the presence of the 1244 pilin DSL was able to facilitate the glycosylation of PA103 pilin.
FIGURE 5. Western blot analysis of P. aeruginosa hybrid pilin.
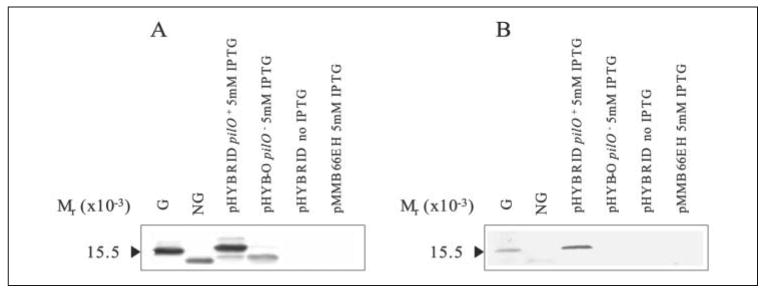
Western blot of pilin produced by P. aeruginosa 1244.47/pHYBRID and 1244.47/pHYB-O using anti-pilin polyclonal serum as a probe (A) or anti-1244 pilin glycan monoclonal antibody 11.14 (B) as a probe. G is glycosylated 1244 pilin, and NG is nonglycosylated 1244 pilin. Hybrid pilin was glycosylated in the presence of PilO (pHYBRID), whereas removal of PilO abrogated glycosylation (pHYB-O). The presence of a pilin band was found only in the presence of IPTG (A), and empty vector controls did not produce a band (pMMB66EH).
Mutational Analysis of the P. aeruginosa 1244 Pilin DSL
The finding that replacement of the DSL of PA103 pilin with that from strain 1244 supported glycosylation, whereas the PA103 pilin (even with the replacement of the tail region) did not, suggests that either the DSL of 1244 pilin contains specific structures important in the glycosylation reaction or that components of the PA103 pilin loop somehow prevent glycosylation.
To examine the role of strain 1244 pilin DSL structures, we first looked at the disulfide bond to see whether its formation was essential for glycosylation. Plasmids p1244 C127A and p1244 C145A (Fig. 2), constructs that encoded mutant pilins incapable of forming the disulfide bond of the DSL, were generated and expressed in strain 1244N3. Western blot analysis of extracts of these cells showed that mutant pilin produced by either p1244 C127A or p1244 C145A was primarily glycosylated, although low levels of nonglycosylated pilin was produced from p1244 C127A (Fig. 6A). These results suggest that an intact DSL is not essential for glycosylation to occur.
FIGURE 6. Western blot analysis of P. aeruginosa 1244 mutant pilins with Cys mutation or DSL deletion.
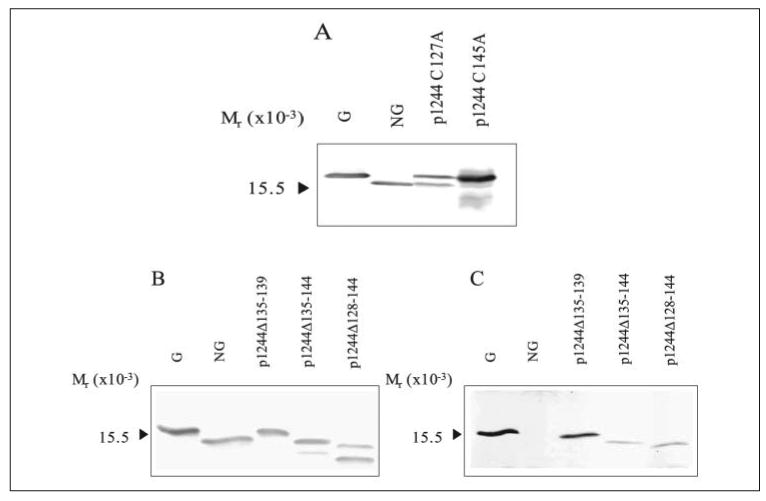
Western blot of pilin produced by P. aeruginosa 1244N3/p1244 C127A and 1244N3/p1244 C145A using anti-1244 pilin monoclonal antibody 5.44 as a probe (A). Western blot analysis of pilin produced by P. aeruginosa 1244N3/p1244Δ135–139, 1244N3/p1244Δ135–144, and 1244/p1244Δ128–144 using anti-1244 pilin monoclonal antibody 5.44 as a probe (B) or anti-1244 pilin glycan monoclonal antibody 11.14 (C) as a probe. G is glycosylated 1244 pilin, and NG is nonglycosylated 1244 pilin. All of the mutants produced some glycosylated pilin; however, p1244 C127A (A), p1244Δ135–144, and p1244Δ128–144 (B) produced some nonglycosylated pilin.
Although the disulfide bond itself was not necessary for glycosylation, there remained the possibility that specific regions of the DSL were involved in recognition of pilin by PilO. With this in mind, we deleted roughly one third, two thirds, and the entire DSL (p1244Δ135–139, p1244Δ135–144, and p1244Δ128–144 respectively; Fig. 2) from the loop region to assess the importance of specific regions of the DSL in glycosylation. These constructs were expressed in 1244N3, and Western blot analysis of cell extracts revealed that these mutants produced pilins of progressively lower apparent molecular weights (Fig. 6B), presumably because of the deletions. All three of these mutants produced pilin that reacted with the glycan-specific antibody and were therefore glycosylated (Fig. 6C). In addition, some nonglycosylated pilin was produced, especially by p1244Δ128–144, suggesting that glycosylation of the pilin lacking a DSL is less efficient than glycosylation of wild type pilin (Fig. 6B). The presence of glycosylated pilin in these mutants indicates that the specific groups or regions of the DSL are not essential in the glycosylation reaction.
Influence of DSL Charge on Pilin Glycosylation
Because deletion of the 1244 pilin DSL did not abrogate glycosylation, it is likely that the 1244 pilin DSL does not contain specific structures necessary for this reaction. Because PA103 pilin containing a 1244 pilin DSL was glycosylated, whereas PA103 pilin containing a C-terminal Ser was incapable of being glycosylated by PilO, physical properties of the PA103 pilin DSL may be responsible for preventing glycosylation in this mutant background.
Comparison of the 1244 and PA103 pilin DSLs revealed several differences, including a low degree of sequence identity (Fig. 2). Interestingly, the isoelectric point of the 1244 pilin DSL is basic (pH 9.31), whereas the pI of the PA103 pilin DSL is acidic (pH 4.53), indicating a significant difference in charge between these two DSLs. Homology modeling suggests that residues Glu133, Glu134, and Glu143 of the PA103 pilin DSL and tail region project outward from the pilin molecule, providing a negative surface, whereas the 1244 pilin is predicted to have a positive surface (Fig. 7). To determine whether DSL charge influences pilin glycosylation, DNA sequence coding for a piece of the 1244 pilin DSL previously deleted, AWKPN, was inserted into the center of the p103 P144S DSL, generating the plasmid p103 INS136 (Fig. 2). This construct was expressed in 1244.47, and membrane fractions were subjected to Western blot analysis (Fig. 8). The pilin encoded by p103 INS136 was glycosylated in the presence of pilO (Fig. 8A) as determined by the apparent molecular weight of the band that reacted with pilin-specific polyclonal rabbit serum and by the reaction with glycan-specific polyclonal rabbit antiserum (Fig. 8B). In the absence of pilO, the apparent molecular weight of pilin encoded by p103 INS136 was similar to that of nonglycosylated pilin, and the mutant pilin did not react with glycan-specific antiserum. However, Fig. 8A also shows the presence of nonglycosylated pilin being produced by the mutant expressing p103 INS136 in the presence of PilO, indicating that in this mutant, pilin glycosylation is not as efficient as in the wild type strain.
FIGURE 7. Predicted surface properties of the strain 1244 and PA103 pilin DSLs.
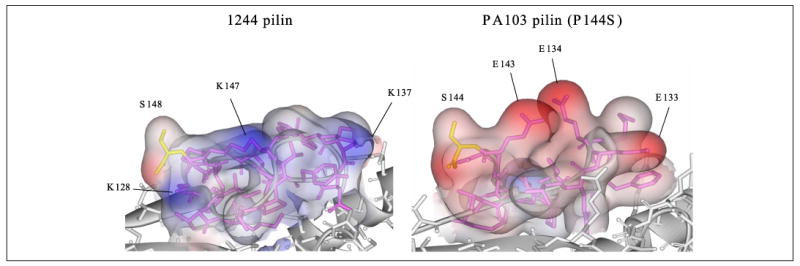
Tertiary structures of 1244 pilin and PA103 (P144S) pilin generated by homology modeling. The residues of the DSL and tail are pink, and the C-terminal residue is yellow. Molecular surface of the DSL was generated and displayed in terms of Gasteiger charges where blue is positive and red is negative.
FIGURE 8. Western blot analysis of PA103 mutant pilin with insertion.
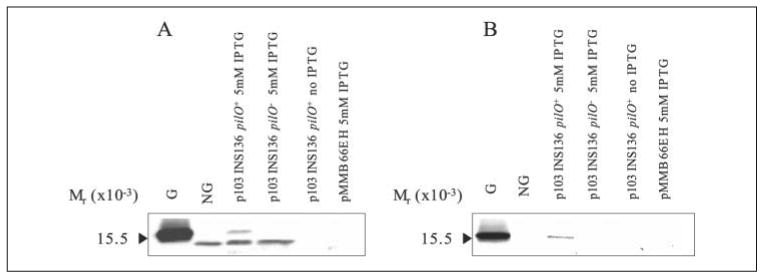
Western blot of pilin produced by P. aeruginosa 1244.47/p103 INS136 in the presence of PilO (pUCP26pilO) or without PilO (pUCP26) using anti-pilin polyclonal serum as a probe (A) or anti-1244 pilin glycan polyclonal serum (B) as a probe. G is glycosylated 1244 pilin, and NG is nonglycosylated 1244 pilin. Pilin encoded by p103 INS136 was glycosylated in the presence of PilO, whereas removal of PilO abolished glycosylation. The presence of a pilin band was consistent with the presence of IPTG (A), and empty vector controls did not produce a band (pMMB66EH).
Glycosylation of pilin produced by p103 INS136, coupled to the fact that the DSL is not essential for glycosylation of 1244 pilin, indicates the possibility that the charge provided by this sequence yielded a some-what permissive environment for pilin glycosylation. If the PA103 pilin DSL environment interferes with glycosylation, extension of the tail region could possibly position the glycosylation site far enough away from the pilus surface to allow glycan attachment. However, before testing this with PA103 pilin, it was necessary to determine whether tail length influenced glycosylation of strain 1244 pilin. The 1244 pilin tail region was extended by adding coding sequence for one or two alanines, resulting in mutant pilins terminating in CPKAS or CPKAAS, respectively (p1244 S149 and p1244 S150; Fig. 2). Constructs were expressed in strain 1244N3, and pilin glycosylation status was determined by subjecting cell extracts to Western blot analysis (Fig. 9, A and B). The glycosylated form was present in cells expressing either of these pilins as determined by the presence of a band with an apparent molecular weight similar to the glycosylated pilin standard (Fig. 9A) and reaction with the glycan-specific antibody (Fig. 9B). The presence of some nonglycosylated pilin in the mutant expressing p1244 S150 was also detected (Fig. 9A). These results provide evidence that extension of the DSL tail does not interfere with pilin glycosylation.
FIGURE 9. Western blot analysis of mutant pilins with altered tail lengths.
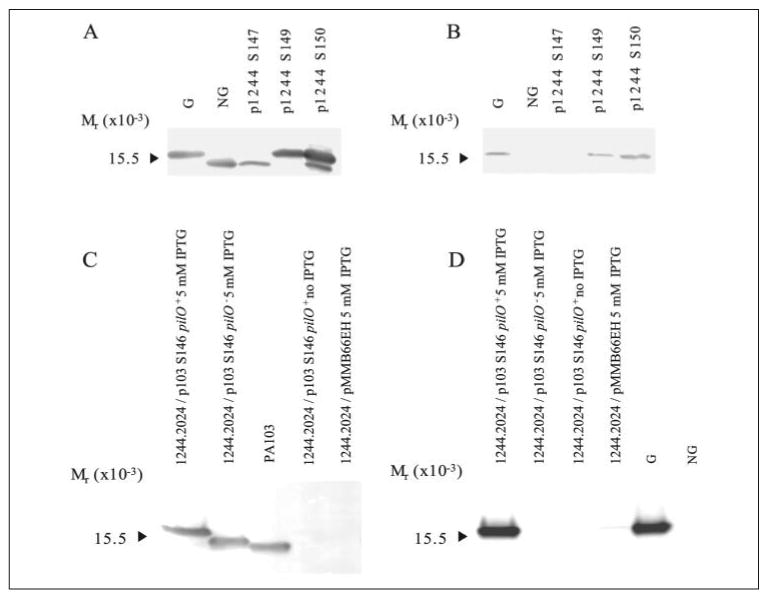
Western blot of pilin produced by P. aeruginosa 1244N3/p1244 S147, 1244N3/p1244 S149, and 1244N3/p1244 S150 using anti-1244 pilin monoclonal antibody 5.44 as a probe (A) or anti-1244 pilin glycan monoclonal antibody 11.14 (B) as a probe. G is glycosylated 1244 pilin, and NG is nonglycosylated 1244 pilin. Pilin with longer tails were glycosylated (S149 and S150), whereas pilin with a shortened tail region was not glycosylated (S147). Western blot of pilin produced by P. aeruginosa 1244.2024/p103 S146 in the presence of PilO (pUCP26pilO) or without PilO (pUCP26) using anti-PA103 pilin monoclonal antibody 2.97 as a probe (C) or anti-1244 pilin glycan polyclonal serum (D) as a probe. Pilin encoded by p103 S146 was glycosylated in the presence of PilO, whereas removal of PilO eliminated glycosylation. The presence of a pilin band was consistent with the presence of IPTG (A), and empty vector controls did not produce a band (pMMB66EH). Pilin produced by 1244.2024/p103 S146/pUCP26 was nonglycosylated; however, it has a higher molecular weight than wild type PA103 pilin because the leader sequence was not cleaved, and this mutant pilin had two additional alanines in the tail region.
Because glycosylation of these 1244 pilins occurs, the extension of the tail region of PA103 pilin terminating in Ser may position the terminal Ser far enough away from the DSL to permit glycosylation. To test this, the tail region of p103 P144S was extended by the insertion of two Ala codons between the penultimate Glu and the C-terminal Ser producing p103 S146 (Fig. 2). This plasmid was moved into strain 1244.2024 where cell extracts were subjected to Western blot analysis (Fig. 9, C and D). When pilA from p103 S146 was coexpressed with pilO, the pilin produced had a higher apparent molecular weight than when in the absence of pilO, indicating pilin glycosylation (Fig. 9C). Pilin expressed in the presence of pilO reacted with glycan-specific serum, indicating glycosylation, whereas no reaction was seen in the absence of pilO (Fig. 9D). It is also obvious that p103 S146 pilin, in the absence of pilO, had a higher apparent molecular weight when compared with wild type PA103 pilin (Fig. 9C). This is due in part to the extra Ala residues but also to the production of only prepilin in this pilD strain. Overall, these results show that extension of the C-terminal Ser puts the glycosylation site in a permissive environment.
To determine the influence of shortening tail length on glycosylation of strain 1244 pilin, we generated p1244 S147 (Fig. 2). This construct was expressed in strain 1244N3 and pilin glycosylation status was determined by subjecting cell extracts to Western blot analysis (Fig. 9, A and B). Nonglycosylated pilin was exclusively expressed by this mutant as determined by apparent molecular weight and by lack of reaction with a glycan-specific antibody (Fig. 9, A and B). These results suggest that a minimum tail length is required to allow the interaction of enzyme and substrates.
DISCUSSION
The pilin glycan of P. aeruginosa 1244 is identical to the O-antigen repeating unit of this organism and exists in a ratio of one glycan per pilin subunit (18). This structure is attached by ether linkage to the β-carbon of Ser148, the pilin C-terminal residue (14). Modification of the C terminus is unusual when compared with O-glycosylation of other prokaryotic proteins. For example, the single Neisseria gonorrhoeae pilin glycan is attached at Ser63 (28, 29), a residue in the central portion of the domain between the N-terminal α-helix and β-sheet region, referred to as the αβ-loop (30). In the C. jejuni O-linked flagellin glycosylation system, 19 separate Ser/Thr residues are modified by the addition of a pseudaminic acid moiety, none of which are located at the C terminus (12). In the present paper, we show that glycosylation of this residue is abrogated by the addition of Ala at position 149, making Ser148 the penultimate residue meaning that the C-terminal position is required. This suggests that either the exposed carboxyl group of this residue is recognized by PilO or the bulk of an additional residue prevents enzyme contact. This, in either case, would be a good mechanism for insuring glycosylation of only the terminal Ser. The value of the 1244 glycan at the pilin C terminus may be related to its proximity to the DSL, a structure that facilitates adhesion to host tissue by attachment to surface glycolipids (31). This structure, although exposed at the pilus tip, is at least partially buried between the subunits of the fiber shaft (32). We have previously shown that the glycan is located on the fiber surface where it attenuates the natural hydrophobicity of the pilus (47). It is possible that the surface location of the glycan also stabilizes the DSL and protects it against antibody recognition or proteolysis, thus maintaining pilus integrity and promoting virulence.
Neither the replacement of the PA103 pilin C-terminal residue with Ser nor exchanging the pilin tail region of this protein with that of 1244 resulted in glycosylation. However, a substitution using the complete 1244 pilin DSL region allowed glycosylation, suggesting that either the 1244 structure contained recognition elements or the PA103 pilin DSL was incompatible with glycosylation. Deletions of portions of the 1244 pilin DSL indicated that these residues were not required for glycosylation, whereas altering the charge arrangement of the PA103 pilin DSL allowed partial glycosylation to occur. These results suggest that the glycosylation apparatus is not compatible with the electronegative environment of the PA103 pilin DSL. This may be due to an influence on PilO or to repulsion from the negative charges of the undecaprenol carrier lipid or the pseudaminic acid of the O-antigen repeating unit. This interpretation is strengthened by the finding that extending the tail of the PA103 pilin (in which the terminal Pro had been replaced with Ser) resulted in a strong glycosylation response. This mutation likely positioned the glycosylation site far enough away from the negative charges of the DSL to place the terminal Ser in a permissive environment. Shortening the tail of 1244 pilin prevented glycosylation, suggesting that a minimal distance is required in this segment in the wild type strain as well, possibly because of a steric effect or to an influence on Ser flexibility. The importance of a specific environment provided by residues near the glycosylation site has been observed in several O-linked glycosylation systems. For example, in C. jejuni flagellin glycosylation, most of the modified residues were located in a hydrophobic patch of this protein, and Ser and Thr that were adjacent to charged residues were not typically glycosylated (12). In the eukaryotic mucin type O-glycosylation system (33) as well as in glycosylated cellulases of the prokaryotes, C. thermocellum and Bacteroides cellulosolvens (34), glycosylated Ser/Thr were located among regions rich in Ser/Thr/Pro, suggesting that properties of these residues provided a preferential environment for glycosylation. Altogether, these results indicate that pilin glycosylation requires only a C-terminal Ser and a compatible protein surface. In addition, an incompatible surface will allow activity if the glycosylation site is extended sufficiently. These results suggest that the tail region may function to present the glycosylation target at the minimally optimal distance from the pilin subunit. Extension of the tail region of pilin did not prevent glycosylation, indicating that molecular flexibility is well tolerated in the pilin glycosylation reaction. Further work will be required to establish whether this minimal motif can function as a glycosylation site for other proteins, or if there are pilin-specific factors, possibly related to the membrane location of both pilin and PilO.
Western blot analyses revealed that a pilD mutant, deficient in prepilin leader cleavage, is capable of pilin glycosylation (Fig. 9, C and D),3 suggesting that glycosylation occurs early in pilus biogenesis. Because 1244 pilins with mutated cysteines, as well as pilins that lacked large segments of the pilin C-proximal region, could serve as a glycosylation substrate, it was clear that an intact DSL was not required for this modification. However, shortening 1244 pilin by one residue while maintaining a C-terminal Ser prevented glycosylation. Altogether, these results suggest that, under normal conditions, pilin is folded and the DSL is formed prior to glycosylation. Previous work has shown that proteins destined for glycosylation commonly fold into their native state prior to this modification (12, 33). This is consistent with the finding that Ser/Thr residues of flagellin, predicted to be surface-exposed, were found to be glycosylated in C. jejuni, whereas buried residues were unmodified (12). Similarly, the eukaryotic mucin type O-glycosylation system utilizes only Ser/Thr residues exposed after protein folding (10).
The exploitation of this glycosylation system for vaccine engineering should be explored. Previous results from our laboratory have demonstrated that pure pili from strain 1244 induced the production of antibodies in mice against not only the proteinaceous portion of the pilin and the glycan but also against the O-antigen of this organism (14). In addition, evidence has been presented that PilO is capable of transferring an O-repeat from a number of different P. aeruginosa serotypes to 1244 pilin (17). These results suggest the potential for designing a broadly protective pilus-based vaccine specific for both the O-antigen and pilin protein. Pilin from strain PA103 is not glycosylated (47) and belongs to a structurally distinct subgrouping referred to as group II (16). We have shown that PA103 pilin can easily be engineered to act as a glycosylation substrate. Because addition of the glycosylation substrate to PA103 pilin requires only minor alteration, one would expect a minimal effect on the immunogenicity of this protein. Group I and II pilins are the most common types found among clinical isolates (35, 47). Combining these molecules with common O-antigen repeating units would target an immune response to the pilin protein and pilin glycan (inhibiting twitching motility (18)) as well as the O-antigen of lipopolysaccharide (facilitating phagocytosis). The breadth of the vaccine would be generated by increasing targets rather than by component redundancy. This means that, because this vaccine strategy involves the utilization of a small number of pilin and O-antigen types, the likelihood of antigenic suppression observed with the administration of multivalent pilus (36, 37) or O-antigen vaccines (38) would be reduced. Finally, it may be possible to engineer non-pilin proteins to act as glycosylation substrates, a strategy that would greatly widen the types of conjugate vaccines developed.
Supplementary Material
Footnotes
This work was supported by National Institutes of Health Grant AI054929 (to P. C.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and supplemental text.
The abbreviations used are: DSL, disulfide loop; IPTG, isopropyl β-d-thiogalactopyranoside; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
R. VanderWeele and P. Castric, unpublished observations.
References
- 1.Mattick JS. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 2.Castric P. Microbiology. 1995;141:1247–1254. doi: 10.1099/13500872-141-5-1247. [DOI] [PubMed] [Google Scholar]
- 3.Mescher MF, Strominger JL, Watson SW. J Bacteriol. 1974;120:945–954. doi: 10.1128/jb.120.2.945-954.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt MA, Riley L, Benz I. Trends Microbiol. 2003;11:554–561. doi: 10.1016/j.tim.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Upreti R, Kumar M, Shankar V. Proteomics. 2003;3:363–379. doi: 10.1002/pmic.200390052. [DOI] [PubMed] [Google Scholar]
- 6.Benz I, Schmidt MA. Mol Microbiol. 2002;45:267–276. doi: 10.1046/j.1365-2958.2002.03030.x. [DOI] [PubMed] [Google Scholar]
- 7.Szymanski C, Wren B. Nat Rev. 2005;3:225–237. doi: 10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- 8.Spiro R. Glycobiology. 2002;12:43R–56R. doi: 10.1093/glycob/12.4.43r. [DOI] [PubMed] [Google Scholar]
- 9.Peters J, Rudolf S, Oschkinat H, Mengele R, Sumper M, Kellermann J, Lottspeich F, Baumeister W. Biol Chem Hoppe-Seyler. 1992;373:171–176. doi: 10.1515/bchm3.1992.373.1.171. [DOI] [PubMed] [Google Scholar]
- 10.Szymanski C, Logan SM, Linton D, Wren B. Trends Microbiol. 2003;11:233–238. doi: 10.1016/s0966-842x(03)00079-9. [DOI] [PubMed] [Google Scholar]
- 11.Feldman M, Wacker M, Hernandez M, Hitchen P, Marolda C, Kowarik M, Morris HR, Dell A, Valvano M, Aebi M. Proc Natl Acad Sci U S A. 2005;102:3016–3021. doi: 10.1073/pnas.0500044102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thibault P, Logan SM, Kelly J, Brisson J, Ewing C, Trust TJ, Guerry P. J Biol Chem. 2001;276:34862–34870. doi: 10.1074/jbc.M104529200. [DOI] [PubMed] [Google Scholar]
- 13.Young N, Brisson J, Kelly J, Watson D, Tessier L, Lanthier P, Jarrell H, Cadotte N, St Michael F, Aberg E, Szymanski C. J Biol Chem. 2002;277:42530–42539. doi: 10.1074/jbc.M206114200. [DOI] [PubMed] [Google Scholar]
- 14.Comer JE, Marshall MA, Blanch VJ, Deal CD, Castric P. Infect Immun. 2002;70:2837–2845. doi: 10.1128/IAI.70.6.2837-2845.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee KK, Sheth HB, Wong WY, Sherburne R, Paranchych W, Hodges RS, Lingwood CA, Krivan H, Irvin RT. Mol Microbiol. 1994;11:705–713. doi: 10.1111/j.1365-2958.1994.tb00348.x. [DOI] [PubMed] [Google Scholar]
- 16.Castric PA, Deal CD. Infect Immun. 1994;62:371–376. doi: 10.1128/iai.62.2.371-376.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DiGiandomenico A, Matewish MJ, Bisaillon A, Stehle JR, Lam JS, Castric P. Mol Microbiol. 2002;46:519–530. doi: 10.1046/j.1365-2958.2002.03171.x. [DOI] [PubMed] [Google Scholar]
- 18.Castric P, Cassels FJ, Carlson RW. J Biol Chem. 2001;276:26479–26485. doi: 10.1074/jbc.M102685200. [DOI] [PubMed] [Google Scholar]
- 19.Silipigni-Fusco J. PhD Thesis. University of Pittsburgh; 1987. Studies on the Role of Somatic Pili as Virulence and Immunity Factors in the Pathogenicity of Pseudomonas aeruginosa. [Google Scholar]
- 20.Kelley L, MacCallum R, Sternberg M. J Mol Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- 21.Castric PA, Sidberry HF, Sadoff JC. Mol Gen Genet. 1989;216:75–80. doi: 10.1007/BF00332233. [DOI] [PubMed] [Google Scholar]
- 22.Ruvkun G, Ausubel FM. Nature. 1981;289:85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- 23.Schagger H, von Jagow G. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 24.Sidberry H, Kaufman B, Wright D, Sadoff J. J Immunol Methods. 1985;76:299–305. doi: 10.1016/0022-1759(85)90307-2. [DOI] [PubMed] [Google Scholar]
- 25.Saiman L, Sadoff J, Pyle M, Prince A. Infect Immun. 1989;57:2764–2770. doi: 10.1128/iai.57.9.2764-2770.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chia JS, Chang LY, Shun CT, Chang YY, Chen JY. Infect Immun. 2001;69:6987–6998. doi: 10.1128/IAI.69.11.6987-6998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramphal R, Koo L, Ishimoto KS, Totten PA, Lara JC, Lory S. Infect Immun. 1991;59:1307–1311. doi: 10.1128/iai.59.4.1307-1311.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parge HE, Forest KT, Hickey MJ, Christensen DE, Getzoff ED, Tainer JA. Nature. 1995;378:32–38. doi: 10.1038/378032a0. [DOI] [PubMed] [Google Scholar]
- 29.Stimson E, Virji M, Makepeace K, Dell A, Morris HR, Payne G, Saunders JR, Jennings MP, Barker S, Panico M, Blench I, Moxon ER. Mol Microbiol. 1995;17:1201–1214. doi: 10.1111/j.1365-2958.1995.mmi_17061201.x. [DOI] [PubMed] [Google Scholar]
- 30.Craig L, Pique ME, Tainer JA. Nat Rev. 2004;2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 31.Sheth HB, Lee KK, Wong WY, Srivastava G, Hindsgaul O, Hodges RS, Paranchych W, Irvin RT. Mol Microbiol. 1994;11 doi: 10.1111/j.1365-2958.1994.tb00349.x. [DOI] [PubMed] [Google Scholar]
- 32.Lee KK, Doig P, Irvin RT, Paranchych W, Hodges RS. Mol Microbiol. 1989;3:1493–1499. doi: 10.1111/j.1365-2958.1989.tb00135.x. [DOI] [PubMed] [Google Scholar]
- 33.Van den Steen P, Rudd P, Dwek R, Opdenakker G. Clin Rev Biochem Mol Biol. 1998;33:151–208. doi: 10.1080/10409239891204198. [DOI] [PubMed] [Google Scholar]
- 34.Gerwig G, Kamerling J, Vliegenthart J, Morag E, Lamed R, Bayer E. J Biol Chem. 1993;268:26956–26960. [PubMed] [Google Scholar]
- 35.Kus J, Tullis E, Cvitkovitch D, Burrows LL. Microbiology. 2004;150:1315–1326. doi: 10.1099/mic.0.26822-0. [DOI] [PubMed] [Google Scholar]
- 36.Hunt J, Jackson D, Brown L, Wood P, Stewart D. Vaccine. 1994;12:457–464. doi: 10.1016/0264-410x(94)90125-2. [DOI] [PubMed] [Google Scholar]
- 37.O'Meara T, Egerton J, Raadsma H. Immunol Cell Biol. 1993;71:473–488. doi: 10.1038/icb.1993.53. [DOI] [PubMed] [Google Scholar]
- 38.Hatano K, Boisot S, DesJardins D, Wright D, Brisker J, Pier GB. Infect Immun. 1994;62:3608–3616. doi: 10.1128/iai.62.9.3608-3616.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramphal R, Sadoff JC, Pyle M, Silipigni JD. Infect Immun. 1984;44:38–40. doi: 10.1128/iai.44.1.38-40.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu PV. J Infect Dis. 1973;128:506–513. doi: 10.1093/infdis/128.4.506. [DOI] [PubMed] [Google Scholar]
- 41.Bolivar F, Backman K. Methods Enzymol. 1979;68:245–267. doi: 10.1016/0076-6879(79)68018-7. [DOI] [PubMed] [Google Scholar]
- 42.West S, Schweizer H, Dall C, Sample A, Runyen-Janecky LJ. Gene (Amst) 1994;128:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 43.Furste JP, Pansegrau W, Blocker FR, Scholz P, Bagdasarian M, Lanka E. Gene (Amst) 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 44.Smedley JG, III, Jewell E, Roguskie J, Horzempa J, Syboldt A, Stolz DB, Castric P. Infect Immun. 2005;73:7922–7931. doi: 10.1128/IAI.73.12.7922-7931.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


