Abstract
The Escherichia coli RNA chaperone Hfq is involved in riboregulation of target mRNAs by small trans-encoded non-coding (ncRNAs). Previous structural and genetic studies revealed a RNA-binding surface on either site of the Hfq-hexamer, which suggested that one hexamer can bring together two RNAs in a pairwise fashion. The Hfq proteins of different bacteria consist of an evolutionarily conserved core, whereas there is considerable variation at the C-terminus, with the γ- and β-proteobacteria possessing the longest C-terminal extension. Using different model systems, we show that a C-terminally truncated variant of Hfq (Hfq65), comprising the conserved hexameric core of Hfq, is defective in auto- and riboregulation. Although Hfq65 retained the capacity to bind ncRNAs, and, as evidenced by fluorescence resonance energy transfer assays, to induce structural changes in the ncRNA DsrA, the truncated variant was unable to accommodate two non-complementary RNA oligonucleotides, and was defective in mRNA binding. These studies indicate that the C-terminal extension of E. coli Hfq constitutes a hitherto unrecognized RNA interaction surface with specificity for mRNAs.
INTRODUCTION
The Escherichia coli host factor I/Q (Hfq) was first described as an accessory factor of the phage Qβ replicase (1) and its importance in cellular physiology became evident when the broadly pleiotropic phenotypes of an E. coli hfq mutant were characterized (2). The observations that Hfq is involved in expression of the rpoS gene encoding the stationary sigma factor, σS (3,4), in iron metabolism (5,6), in stability control of several mRNAs (7–9) and small non-coding regulatory RNAs (ncRNAs) (10–13), in riboregulation of target mRNAs by ncRNAs (5,10,14–17) and that it acts as a virulence factor in several bacterial pathogens (18–21) has recently sparked great interest in this highly conserved bacterial protein (22).
Most data on Hfq–RNA interactions stem from studies on ncRNAs. Hfq-binding sites on ncRNAs have been demonstrated to coincide with cleavage sites of the E. coli major riboendonuclease RNase E, and Hfq was shown to protect ncRNAs from degradation (11,12). Hfq binds to OxyS, DsrA, RprA, RyhB, Spot42 and SgrS RNAs as well as to other ncRNAs identified in E. coli (10,13,14,16,17,23–27). These ncRNAs are involved in translational regulation of their cognate mRNAs. By forming RNA duplexes in the close vicinity or within the translational initiation region of target mRNAs, ncRNAs can either activate or silence translation, whereby the latter mode of action appears to be predominant (28). Hfq has been shown to stimulate in vitro annealing of Spot42 RNA with galK mRNA (16), of OxyS with fhlA mRNA (14), of RyhB with sodB mRNA (13,27) as well as that of SgrS with ptsG mRNA (17). It has been demonstrated in some cases that Hfq is dispensable once the interaction between the ncRNA and the target mRNA has taken place (13,14,29). It appears possible that the molecular mechanism by which Hfq brings about these interactions entails unfolding of ncRNAs. However, recent in vitro studies did not reveal significant changes in the secondary structure of the ncRNAs DsrA (29,30) and RyhB (27). In contrast, the Hfq RNA chaperone activity induced structural changes in the 5′untranslated regions of the RyhB target sodB (27) and of the MicA target ompA (31,32), which could facilitate the interaction of these ncRNAs with their target mRNAs. At least for ompA mRNA it was shown that the structural changes induced by Hfq prevailed upon proteolysis of the protein, another criterion, which classified Hfq as an RNA chaperone (31).
The 3D structures of the Staphylococcus aureus Hfq homologue (33), the N-terminal 72 amino acid (aa) of E. coli Hfq (34) and of Pseudomonas aeruginosa Hfq (35) revealed that it has a hexameric ring-shaped structure, and corroborated the idea that it belongs to the large family of Sm- and Sm-like proteins. These proteins are involved in RNA processing in eukaryotes and bind to various RNAs, primarily recognizing short U-rich stretches, known as SM sites (36). An A/U-rich region preceded or followed by a stem-loop structure has likewise emerged as a common RNA-binding motif for Hfq (27,30,31,37)
A prevailing question concerns the interaction of a Hfq-hexamer with RNA substrates. The X-ray structure of S. aureus Hfq complexed with a single-stranded oligoribonucleotide showed that it binds to the protein in a circular conformation around a central basic cleft of the hexameric ring (33). Mutational studies suggested that one E. coli Hfq-hexamer provides two binding surfaces. These studies implicated the known binding cavity along the inner rim made up of six potential nucleotide-binding pockets (38) as well as the proximal amino acid residues R16 and F39 (39) in the interaction with the ncRNA DsrA. In addition, in the same studies the amino acid residues Y25, I30 and K31, located on the distal site of the hexamer have been shown to be required for poly(A) binding, which culminated in the hypothesis that one Hfq-hexamer can bring together two RNAs in a pairwise fashion (38).
Escherichia coli Hfq homologues have been found in a number of Gram-negative and Gram-positive bacteria. The Hfq proteins of different organisms display an evolutionarily conserved common core consisting of amino acid residues 7–66, whereas there is considerable variation at the C-terminal end (14,16,34,40,41). The removal of 19 aa (42) had no significant effect on binding of the truncated Hfq83 protein to polyadenylated RNA.
In addition, we have previously shown that the first 65 N-terminal amino acid residues of E. coli Hfq are sufficient for hexamer formation, retain the capacity for binding of the ncRNA DsrA as well as for phage Qß replication (43).
Studies on Hfq-mediated riboregulation have only been performed in E. coli and some close relatives, the Hfq protein of which contains an extended C-terminus. Here, we addressed the question whether the C-terminus of E. coli Hfq contributes to ncRNA-mediated riboregulation. Using different test systems, we show that a Hfq variant, comprising the first 65 N-terminal amino acid (Hfq65), is non-functional in hfq-auto-regulation, RyhB mediated repression of sodB mRNA as well as in DsrA-mediated stimulation of rpoS mRNA translation. Hfq65 displayed no gross defect in binding to the sRNAs RyhB and DsrA, and as evident from real-time fluorescence energy transfer (FRET) assays, Hfq65 retained the capacity to induce structural changes in DsrA. Using FRET, we further show that in contrast to full-length Hfq (Hfqwt), Hfq65 is impaired in annealing of complementary RNA oligonucleotides. In addition, while Hfqwt was able to bind two non-complementary RNA oligonucleotides on the surface, Hfq65 lacked this capacity. Moreover, Hfq65 was defective in binding to all tested mRNAs. In summary, this study showed that amino acid residues following the conserved core of Hfq are involved in mRNA binding and thereby identifies a third interaction surface with specificity for mRNAs.
MATERIALS AND METHODS
Bacterial strains and growth conditions
The E. coli strains MC4100 (44), AM111 (MC4100 hfq1::Ω) (4) and the corresponding F′ (lacIq) variants (41,44) have been described. They were grown in Luria–Bertani (LB) medium (45) or in M9 medium supplemented with 0.2% glucose, 2 mM MgSO4, 0.1 mM CaCl2 and 10 µg/ml thiamine. Where indicated, glucose was substituted by 0.2% succinate and the iron chelator 2,2′-dipyridyl (50 μM final concentration) was added to the medium. Ampicillin (100 µg/ml), kanamycin (25 µg/ml), tetracycline (30 µg/ml) or chloramphenicol (15 µg/ml) were added to the medium where appropriate to maintain plasmids.
Construction of plasmids
The plasmid pRB381 (46) derivatives pRhfq131 (37) and pRsodB-lacZ (15), which bear inducible hfq-lacZ and sodB-lacZ gene fusions, respectively, have been described. The plasmid pAHfq used for the synthesis of Hfqwt protein was constructed as follows. The hfq gene along with the lac promoter was recovered on a PvuII fragment from plasmid pUH5 (6), and ligated into the EcoRV–NruI sites of pACYC184 (New England Biolabs Ltd., UK). Plasmid pAHfq65, encoding the Hfq65 protein was constructed as follows: the fragment containing the hfq65 gene was obtained by means of PCR using the hfq forward primer (5′-GCTCTAGAAATATAATAGTTTAACTTTAAGAAGGAGATATACATATGGCTAAGGGG CAATCTTTACAAGATCCGTTCCT-3′), containing a XbaI site (italics), and the reverse primer (5′-TTTTTTGAATTCTTACTAAGACGGGACAACAGTAGAAATCG-3′), which contains two stop codons (underlined) after the triplet encoding Ser65 (bold) as well as an EcoRI site (italics). The PCR product was cleaved with XbaI and EcoRI and ligated into the corresponding sites of plasmid pUC19 (New England Biolabs Ltd., UK). From the resulting plasmid pUHfq65, the hfq65 gene was re-isolated by cleavage with PvuII and then ligated into the EcoRV and NruI sites of pACYC184, yielding plasmid pAHfq65.
The construction of the plasmids used for purification and in vivo synthesis of S. aureus Hfq (HfqSa) and Bacillus subtilis Hfq (HfqBs), respectively, is described in Supplementary Data.
The plasmids pUhfqwt, pUsod and pURyhB, which served as templates for in vitro synthesis of hfq126, sodB192 and RyhB RNAs, respectively, have been described (6,13,37).
Plasmid pUrpoS16 used for in vitro synthesis of rpoS652 mRNA was constructed using primer C16 (5′-GGGCTCTAGAGTAATACGACTCACTATAGTCGGGTGAACA-GAGTGCTAACAAAATGTTGCCG-3′), which contained a XbaI site (italics) and the T7 promoter sequence (underlined) followed by the 5′-terminal part of the 5′untranslated region of the rpoS gene (begins at the transcriptional start of rpoS mRNA when transcribed from the main P2 promoter) (47) and the reverse primer P21 (5′-AAAGAATTCCTGACAGATGCTTACTTACTCGCGGAACAG-3′), which contains an EcoRI site (italics) and is complementary to the sequence following the stop codon of the rpoS gene. The PCR product obtained with primers C16/P21 was cleaved with XbaI and EcoRI, and ligated into the corresponding sites of plasmid pUC18, resulting in plasmid pUrpos16.
Relative translational efficiency of hfq-lacZ and sodB-lacZ fusions
Cultures of AM111F′ (pRhfq131) and AM111F′ (pRsodB-lacZ) co-transformed with the compatible plasmids pACYC184 (control), pAHfq and pAHfq65, respectively, were cultivated in LB or M9 medium (Figure 2) at 37°C. At an OD600 of 0.5, the plasmid-encoded genes were induced by addition of IPTG (1 mM). After 60 min, triplicate aliquots were taken for the β-galactosidase assays and for western blot analysis to verify Hfqwt or Hfq65 production. In parallel, samples were withdrawn for isolation of total RNA to determine the respective hfq-lacZ and sodB-lacZ mRNA levels. The β-galactosidase activity was determined from triplicate samples as described (45) and the total RNA was purified by the hot phenol method (48). The averaged β-galactosidase activities obtained with the pRhfq131 and pRsodB-lacZ constructs were normalized to the corresponding hfq-lacZ and sodB-lacZ mRNA levels (=relative translational efficiencies in Figure 2A and B). The corresponding lacZ mRNA levels were determined by primer extension with AMV reverse transcriptase (Promega GmbH, Germany) using 5 μg of total RNA primed with the lacZ-specific 5′-end labelled probe (5′-GGGAAGGGCGATCGGT-3′) and normalized to the 5S rRNA levels (internal control), which were determined using primer R25 (5′-GGTGGGACCACCGCGCTACGGCCGCCAGGC-3′). The signals were visualized by a PhosphoImager (Molecular Dynamics) and quantified by ImageQuant software. Two independent sets of experiments were performed.
Figure 2.
Hfq65 is defective in auto- and riboregulation. (A) Relative translational efficiency of hfq131-lacZ mRNA in the absence of Hfq (white bar), in the presence of Hfqwt (black bar) and Hfq65 (grey bar) in strains AM111F′ (pRhfq131; pACYC184), AM111F′ (pRhfq131; pAHfq) and AMF′111(pRhfq131; pAHfq65) grown in LB medium, respectively. (B) Relative translational efficiency of sodB-lacZ mRNA in the absence of Hfq (white bar), in the presence of Hfqwt (black bar) and Hfq65 (grey bar) in strains AM111F′ (pRsodB; pACYC184), AM111F′ (pRsodB; pAHfq) and AM111F′ (pRsodB; pAHfq65) grown in M9 medium, respectively. The averaged β-galactosidase values normalized to mRNA levels obtained in the absence of Hfq was set to 1 (white bar). The values obtained in the presence of Hfqwt (black bar) and Hfq65 were normalized to the control. The experiment was performed in duplicate. The error bars represent standard deviations. Bottom: determination of the levels of Hfqwt and Hfq65 in the respective strains by quantitative immunoblotting (see Materials and Methods section). (C) Graphical representation of the σS levels in strain AM111F′ harbouring plasmid pUC18 (lane 1; control), pUHfq65 (lane 2; Hfq65) and pUH5 (lane 3; Hfqwt), respectively. The western blot analysis was carried with equal amounts of total cellular protein as described in Materials and Methods section. Only the relevant sections of the immunoblots (lower panels) showing the σS-and Hfq-specific bands are depicted. Quantification of the western blot was done with ImageQuant software. Values were normalized to the σS signal obtained in the presence of Hfqwt in strain AM111F′ (pUH5), which was set to 1. The results represent data from duplicate experiments. The error bars represent standard deviations.
Western blot analysis
The cellular levels of Hfqwt, Hfq65 and σS were determined by quantitative immunoblotting. The σS levels were determined in strain AM111F′ harbouring plasmids pUC18 (control), pUH5 (Hfqwt) and pUHfq65 (Hfq65), respectively. The strains were grown at 28°C in LB medium until they reached an OD600 of 0.3, at which time IPTG was added to a final concentration of 1 mM. At an OD600 of 1.0 equal amounts of cells were withdrawn and boiled in protein sample buffer. The Hfqwt or Hfq65 levels were determined at the times indicated in Figure 1A, concomitantly with the determination of the relative β-galactosidase values (Figure 2A and B), and the quantification of the σS levels (Figure 2C), respectively. Equal amounts of total protein were separated on 12% SDS–polyacrylamide gels and blotted to a nitrocellulose membrane. The blots were blocked with 5% dry milk in TBS buffer, and then probed with anti-Hfq (49) or anti-σS (kindly provided by Dr F. Norel, Pasteur Institute, Paris) antibodies. The antibody–antigen complexes were visualized as described previously (49). The quantification of the Hfq- or RpoS-specific bands was performed with ImageQuant software.
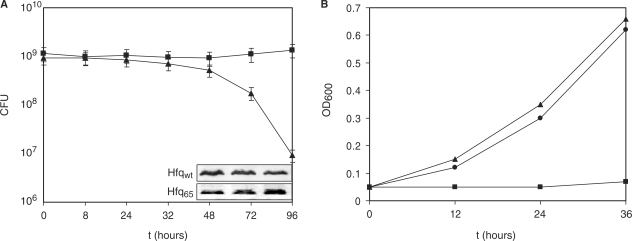
Figure 1.
Effect of Hfq65 on survival under nutrient limitation and on growth on succinate. (A) Escherichia coli strains AM111(pAHfq) (closed square) and AM111(pAHfq65) (closed triangle) were grown in LB-medium, washed with M9-minimal medium, and then resuspended in M9-minimal medium containing 0.2% glucose to 1 × 109 cells ml−1. The CFU at the indicated times was determined by plating of serial dilutions on LB plates containing kanamycin and chloramphenicol. The experiment was performed in triplicate. The error bars represent standard deviations. The inset shows the concentrations of Hfqwt and Hfq65 at different times after incubation from one representative experiment. At times 48, 72 and 96h aliquots of both strains were withdrawn, and equal amounts of total cellular protein was subjected to western blot analysis using Hfq-specific antibodies as described in Materials and Methods section. Only the Hfq-specific bands are shown. (B) Escherichia coli strains AM111(pAHfq) (closed square) and AM111(pAHfq65) (closed triangle) and AM111(pACYC184) (closed circle) were grown overnight in LB-broth, washed with M9-minimal medium and then resuspended to an OD600 of 0.05 in M9-minimal medium containing 0.2% succinate and 2,2′-dipyridyl (50 μM final concentration). Growth was followed by measuring the OD600 at the times indicated. The result of one representative experiment is shown.
RNA preparation for in vitro studies
For hfq126 mRNA synthesis, the plasmid pUhfqwt digested with AflIII was used as a template for in vitro transcription with T7 RNA polymerase (Promega). To prepare sodB192 mRNA, rpoS652 mRNA and RyhB RNA, the pUsod, pUrpoS16 and pURyhB plasmids digested with Asp718I, StuI and DraI, respectively, were used as templates. The run-off transcripts were purified on 6% polyacrylamide–8M urea gels following standard procedures. The mRNA concentration was determined by measuring the A260.
Electrophoretic mobility shift assays
Hfqwt and Hfq65 proteins used in gel mobility assays were purified from AM111(pUH5) and AM111(pUH65) cells, respectively, as described (43). Gel-purified mRNAs were 5′-end labelled with [γ-32P]-ATP (Amersham Pharmacia Biotech) and again purified on 6% polyacrylamide–8M urea gels. Labelled mRNAs (5 nM) were incubated without or with increasing amounts of purified Hfqwt, Hfq65, HfqSa, or HfqBs proteins (as indicated in the legends to Figures 3C and 5 and in Supplementary Figure S3C) in a 10 μl reaction in binding buffer (10 mM Tris, pH 7.5, 60 mM NH4Cl, 5 mM β-mercaptoethanol, 2 mM MgOAc, 100 ng of yeast tRNA) for 5 min at 37°C and then for 10 min at 0°C. The samples were mixed with 40% glycerol to a final concentration of 10% and loaded on a native 5% polyacrylamide gel. Electrophoresis was performed in TAE buffer at 60 V for 12 h. Radioactive bands were visualized using a PhosphoImager.
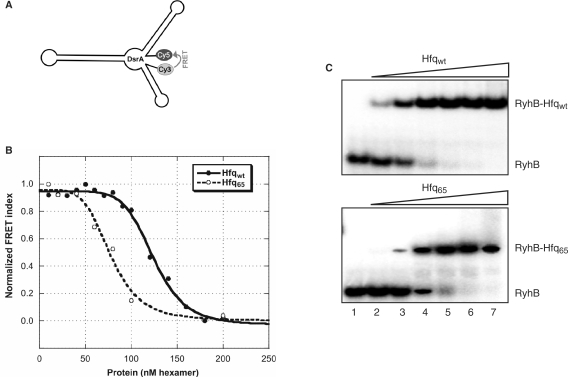
Figure 3.
Hfq65 induces structural changes in DsrA and binds to RyhB. (A) Schematic depiction of the secondary structure of DsrA as predicted by mfold (56). (B) Titration of 5 nM dual-labelled DsrA with Hfqwt or Hfq65. Binding of the protein to the RNA results in a reduction of the FRET index (calculated as FCy5/FCy3 ratio) indicating a spatial separation of the fluorophores. The data was fitted with the equilibrium binding equation y = y0 + A/(1 − (K1/2/c)nH) yielding a Kd = K1/2nH for Hfq of 1247.2 nM and 774.8 nM for Hfq65. (C) Affinity of Hfqwt and Hfq65 for RyhB as revealed by gel mobility shift assays. 5′ End-labelled RyhB (5 nM) was incubated in the absence (lane 1), in the presence of 10 nM (lane 2), 20 nM (lane 3), 40 nM (lane 4), 80 nM (lane 5), 160 nM (lane 6) and 320 nM Hfqwt-hexamer or Hfq65-hexamer, respectively.
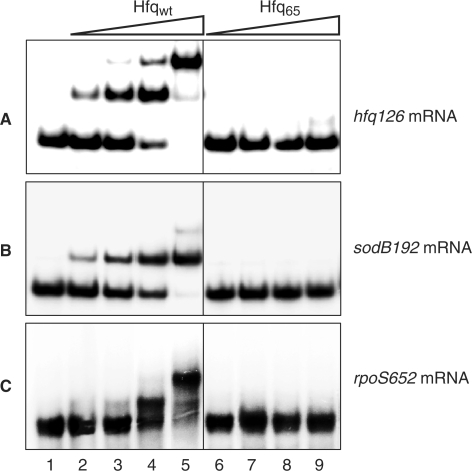
Figure 5.
Hfq65 fails to bind to hfq, sodB and rpoS mRNAs. 5′ End-labelled hfq126 mRNA (A), sodB192 mRNA (B) and rpoS652 mRNA (C) was incubated in the absence (lane 1), in the presence of 2.5 nM (lanes 2 and 6), 5 nM (lanes 3 and 7), 10 nM (lanes 4 and 8) and 20 nM (lanes 5 and 9) Hfqwt- and Hfq65-hexamer, respectively. The molar concentration of either mRNA fragment used was 5 nM.
FRET assays
This method is described in more detail in Ref. (50). Two complementary, fluorophore-tagged RNA 21-mers (Cy5-5′-AUGUGGAAAAUCUCUAGCAGU-3′ (Cy5-21R+) and Cy3-5′-ACUGCUAGAGAUUUUCCACAU-3′ (Cy3-21R−) were used in the annealing experiment shown in Figure 4A. For binding of non-complementary RNA oligonucleotides (Figure 4B), Cy5-21R+ and the RNA oligonucleotide Cy3-21R+ (Cy-3-duplex) (5′-Cy3-CUUUCAUUGGUCGGUCUCUCC-3′) were employed. The tagged RNA oligonucleotides were purchased from VBC-Biotech (Vienna, Austria). Using a Tecan GENios Pro microplate reader, the first oligoribonucleotide was injected into wells with or without Hfq protein (1 µM final Hfq–hexamer concentration), and the measurement was started with the injection of the second oligoribonucleotide. The reaction was performed in annealing buffer (50 mM Tris–HCl pH 7.5, 3 mM MgCl2 and 1 mM DTT) at 37°C. The final concentration of the RNAs was 5 nM in a volume of 40 µl. The reaction was allowed to proceed for 180 sec, and with Cy3 excited, donor and acceptor dye fluorescence emissions were measured once every second. The time-resolved ratio of the fluorescence emissions (FRET index FCy5/FCy3) was normalized to 1 at t180s and least-square fitted with Prism 4.03 (GraphPad Software Inc., San Diego, CA, USA) with the second-order reaction equation for equimolar initial reactant concentrations y = A [1 − 1/(kann t+1)]; kann=observed annealing reaction constant, A = maximum reaction amplitude. The reaction curves were fitted with y=A [1 − 1/(kdb t +1)], kdb=observed double binding reaction constant. The curves shown are representative; the observed reaction constants kann and kdb were calculated as the average of three individually fitted reactions.
Figure 4.
Deletion of the Hfq C-terminus results in a decreased RNA annealing activity and abolishes the ability to bind two RNAs simultaneously. (A) Annealing of two fluorophore-labelled RNA 21-mers can be accelerated by RNA chaperones (top; 50) that can be monitored by FRET. Bottom, 5 nM of each RNA oligonucleotide Cy5-21R+ and Cy3-21R− were annealed at 37°C either in the absence or in the presence of 1 µM Hfqwt or Hfq65 hexamer. FRET was calculated as ratio of acceptor/donor emission (FCy5/FCy3), normalized to 1 at t180s, and the data was fitted with the second-order reaction equation for equimolar initial reactant concentrations y = A [1 − 1/(kann t + 1)]. Hfqwt and Hfq65 increased the observed annealing reaction constant kann 7- and 2-fold, respectively. (B) In a set-up with the non-complementary Cy5-21R+ and Cy3-21R+ (Cy3-duplex) RNA oligonucleotides, simultaneous binding of the two RNAs to a protein can be measured (top). Bottom, the time-resolved FRET index curves were fitted with y = A [1 − 1/(kdb t + 1)] to yield the dual RNA oligonucleotide binding rate constant kdb. The RNA oligonucleotides by themselves do not anneal, whereas Hfqwt can bind both RNAs simultaneously. Incubation with Hfq65 resulted in a kdb 10-fold lower than with Hfqwt, indicating that the C-terminal truncation coincides with a severely reduced capacity for simultaneous binding of the two non-complementary RNA oligonucleotides. The FRET index in this graph was not normalized to indicate the reaction amplitudes.
DsrA RNA labelled at the 5′- and 3′-ends with the fluorophores Cy3 and Cy5, respectively, was obtained from Dharmacon (USA). Five nanometre of this RNA was incubated with indicated concentrations of Hfq or Hfq65 (Figure 3B) in a buffer containing 50 mM Tris–HCl pH 7.5, 3 mM MgCl2, 1 mM DTT. After incubation at 25°C for 30 min, donor (Cy3) and acceptor (Cy5) emissions were measured, and the FRET index was calculated as FCy5/FCy3. The data were fitted in Kaleidagraph 3.51 (Synergy Software, Reading, PA, USA) with the equilibrium binding equation y=y0+A/(1 − (K1/2/c)nH); A=maximum amplitude, K1/2=dissociation constant, nH= Hill coefficient.
RESULTS
Hfq65 confers a reduced viability under nutrient limitation and does not support RyhB-mediated repression of growth on succinate
Although E. coli hfq− mutants are viable under laboratory conditions they show pronounced pleiotropic phenotypes (2). In an initial survival study, the hfq− strain AM111F′ was co-cultivated with the isogenic hfq+ strain MC4100F′ in minimal medium, and growth of both strains was followed over 4 days by scoring the total colony forming units (CFU). In contrast to MC4100F′ the CFU of AM111F′ declined rapidly from day one to day four, when the strain was close to extinction (data not shown). These data clearly indicated that Hfq is pivotal for survival of cells under nutrient limitation that can be reconciled with its requirement for riboregulation under adverse conditions (51). To test whether the C-terminus of Hfq is required for function, we therefore asked whether a truncated variant of Hfq, comprising the first N-terminal 65 aa, can sustain viability under nutrient limiting conditions. The growth and survival rates of the hfq− strain AM111, bearing the plasmid borne hfq65 allele [AM111(pAHfq65)] were compared with that of AM111 bearing the plasmid borne hfqwt gene [AM111(pAHfq)]. Both strains were grown in LB-medium, washed several times with minimal medium, and then resuspended to 1 × 109 cells/ml in M9 minimal medium supplemented with 0.2% glucose. As shown in Figure 1A, the CFU of strain AM111(pAHfq65) started to decrease ∼24 h after inoculation. While the CFU of AM111(pAHfq65) was reduced to <1% over the 4 day observation period that of AM111(pAHfq) remained constant. As the CFU was determined in the presence of the selective antibiotic, it is unlikely that the reduced viability of AM111(pAHfq65) can be attributed to plasmid loss. In addition, quantitative immunoblotting experiments after 48, 72 and 96 h did not reveal a significant difference in the level of Hfqwt and Hfq65 in the respective strains (Figure 1A).
We next sought for more direct in vivo evidence for the apparent dysfunction of Hfq65. Massé and Gottesman (5) have shown that both Hfq and the ncRNA RyhB are required for translational repression of the sdhCDAB operon, resulting in the inability to grow on succinate or fumarate as the sole carbon source. To test whether RyhB-mediated negative regulation of the sdhCDAB genes is impaired in the presence of Hfq65, the strains AM111(pAHfq65) and AM111(pAHfq) were cultured in succinate-minimal medium in the presence of the iron chelator 2,2′-dipyridyl, which is known to induce RyhB synthesis (5). As expected, the strain AM111(pAHfq) did hardly grow in the presence of succinate, whereas similar growth rates were observed for AM111(pAHfq65) and the hfq− strain AM111(pACYC184) (Figure 1B), indicating that Hfq65 is non-functional in supporting RyhB-mediated negative regulation of the sdhCDAB operon.
Hfq65 is non-functional in hfq-autoregulation, in RyhB-mediated repression of sodB mRNA and in DsrA-mediated stimulation of rpoS mRNA
Three model systems were used to further corroborate the idea that the C-terminus of Hfq is required for post-transcriptional regulation. First, we tested whether Hfq65 is affected in autogenous regulation (37). When compared to strain AM111hfq− (pACYC184; pRhfq131), co-expression of the hfq gene from plasmid pAHfq and the hfq131-lacZ reporter gene from the compatible plasmid pRhfq131 in AM111 hfq− (37) resulted in ∼50% decrease of the relative translational efficiency of the hfq131-lacZ mRNA. In contrast, Hfq65 was defective in translational autocontrol, despite comparable intracellular levels of Hfqwt and Hfq65 (Figure 2A).
Second, we employed the well-studied RyhB/sodB model system, wherein the ncRNA RyhB has been shown to repress translation initiation of sodB mRNA (encodes an iron containing superoxide-dismutase) in a Hfq-dependent manner (5,6,27,52). As shown in Figure 2B, Hfq65 was likewise defective in RyhB-mediated translational repression of a sodB-lacZ reporter gene despite comparable intracellular levels of Hfqwt and Hfq65.
Third, we asked whether Hfq65 can support DsrA-mediated stimulation of rpoS mRNA, encoding the stationary sigma factor, σS. The ncRNA DsrA has been shown to bind to the 5-untranslated region of rpoS mRNA in a Hfq-dependent manner, which leads to an exposure of the rpoS translation initiation determinants, and thereby facilitates translation (53–55). σS synthesis was assessed in the presence of either Hfq or Hfq65 at 28°C, when the level of DsrA is known to be elevated (55). In contrast to Hfqwt, Hfq65 did not stimulate σS synthesis, again suggesting that the C-terminus of Hfq is involved in regulation (Figure 2C).
Hfq65 binds ncRNAs
We have previously shown that Hfq65 binds with almost unaltered affinity to the ncRNA DsrA (43). Recent studies (30,38) suggested that DsrA binds to the inner core of the Hfq-hexamer that would explain why binding to DsrA is retained by Hfq65. It appeared possible that the molecular mechanism by which Hfq brings about ncRNA–mRNA interactions entails unfolding of ncRNAs. However, recent in vitro studies did not reveal significant changes in the secondary structure of DsrA (29,30) and RyhB (27). Nevertheless, using enzymatic probing and biophysical methods, Brescia et al. (30) suggested that Hfq might affect the tertiary structure of DsrA. Structural mapping of DsrA placed its 5′ and 3′ end at a distance of 24 consecutive nucleotides (30), which prompted us to employ a FRET assay to verify, (i) whether Hfq alters the structure of DsrA, and if so, (ii) whether this function is retained by Hfq65. Five nanomol full-length DsrA labelled with Cy3 and Cy5 at the 5′ and 3′ end (Figure 3A), respectively, was incubated at 37°C with increasing amounts of Hfq. The decrease in FRET (Figure 3B) upon binding of Hfq indicated a change in the DsrA tertiary (and possibly secondary) structure with the two 5′ and 3′-end positioned fluorophores (Figure 3A) moving apart. A similar picture was observed with Hfq65, indicating that the conserved core region of Hfq is not only capable of DsrA binding (43) but also sufficient to induce conformational changes in the ncRNA.
Hfq65 was likewise demonstrated to bind to the ncRNA RyhB. As calculated from the band-shift assays (Figure 3C), Hfq65 bound to RyhB with only a ∼2-fold lower affinity (16 nM and 35 nM for Hfqwt- and Hfq65-hexamers, respectively) when compared with Hfqwt. Thus, the C-terminus of Hfq does not contribute significantly to binding of the studied ncRNAs.
The C-terminus of Hfq provides a RNA-binding surface
We next asked whether the defects observed in Hfq65-mediated regulation (Figure 2) could be attributed to the absence of a RNA-binding surface. To test this possibility, we first used the two complementary RNA oligonucleotides, Cy5-21R+ and Cy3-21R−, which were labelled at their 5′-ends with Cy5 and Cy3, respectively. Hfq stimulated annealing of these RNA oligonucleotides with a rate constant kann of 0.034 s−1, which was ∼7-fold higher than observed for self-annealing of the two RNA oligonucleotides (Figure 4A). In contrast, the observed annealing constant for Hfq65 was ∼3-fold lower than that determined for Hfqwt, and with 0.011 the kann for Hfq65 was only approximately twice of that seen for self-annealing of the two RNA oligonucleotides (Figure 4A). Next, we used two non-complementary RNA oligonucleotides Cy5-21R+ and Cy3-21R+ (Cy3-duplex) in the FRET assay (Figure 4B). Under these conditions no significant increase in the FRET signal was observed in the presence of Hfq65 (Figure 4B), whereas an increase in FRET was observed for Hfqwt. Given the spatial considerations required for FRET under these conditions this result corroborated other data in that a Hfq-hexamer can bind two RNAs simultaneously (38,39,57). In contrast to Hfqwt, the dual rate binding constant kdb for Hfq65 was ∼10-fold lower, suggesting that Hfq65 is severely impaired in simultaneous binding of the two non-complementary oligonucleotides.
Finally, we tested whether Hfq65 is defective in binding to hfq126 mRNA, sodB192 mRNA and rpoS652 mRNA to either of which Hfqwt was shown to bind (Figure 5). As shown in Figure 5A–C, Hfq65 did not bind to either mRNA fragment, which readily explained why Hfq65 was defective in auto- as well as riboregulation.
DISCUSSION
Although an A/U-rich region adjacent to a stem-loop structure has emerged as a RNA-binding motif for Hfq (12,27,30), it remains poorly understood which sites of the Hfq-hexamer are involved in binding different RNA substrates. So far, mutational studies (38,39) revealed two independent RNA-binding surfaces, one of which is located on the proximal site of the hexamer and includes RNA interactions with the central cavity. As a poly(U) oligonucleotide competed with binding of the ncRNA DsrA (30), this binding site is apparently used by poly(U) (33) as well as by ncRNAs (30,38). Hence, this work corroborates mutational analyses (30,38), and verifies through studies on RyhB that the C-terminus is not instrumental for binding of ncRNAs. Moreover, as shown in Figure 3B, the C-terminus of Hfq is dispensable for Hfq to induce structural changes in DsrA. Taken together, these findings suggest that the evolutionary conserved common core of Hfq (34), i.e. the proximal surface of the Hfq-hexamer, specifies the ‘business surface’ for ncRNAs.
A second binding surface with a specificity for poly(A) has been located on the distal site of the hexamer with amino acid residue Y25, I30 and K31 being instrumental for binding to poly(A) stretches. In this case, poly(A) binding did not compete with DsrA binding, showing that both RNAs use indeed different binding surfaces (38). Binding of poly(A) to the distal site can also be reconciled with the unaltered binding affinity of a C-terminal Hfq deletion mutant, lacking the last 19 aa, and of Hfq65 for a polyadenylated mRNA (42) and for poly(A27) (Večerek,B., unpublished data), respectively. In addition, it has been shown that Hfq65 is proficient in phage Qβ replication and that mutations in K31 severely affect this function (43). Thus, it is possible that the distal site of Hfq is also pivotal for its function in Qβ replication. Moreover, the distal site could be involved in binding of Hfq to DNA A-tracts (58) that could explain the presence of Hfq in the nucleoid (59).
Equilibrium unfolding studies (42) indicated that the C-terminus contributes to the stability of the Hfq-hexamer. However, as Hfq65 forms stable hexamers in solution (43) and binds to ncRNAs (Figure 3) it seems less likely that the failure of Hfq65 to bind to hfq, sodB and rpoS mRNA (Figure 5) results from an altered stability. Therefore, we suggest that the C-terminus of Hfq provides a third RNA interaction surface with specificity for mRNAs, which can readily explain why negative translational autoregulation of hfq mRNA as well as Hfq-mediated riboregulation by the ncRNAs RyhB and DsrA was abolished in the presence of Hfq65 (Figure 2). Zambrano et al. (60) demonstrated that rpoS null strains died rapidly when mixed with rpoS+ strains. Therefore, the loss of viability of AM111(pHfq65) upon entry into stationary phase (Figure 1A) can at least partially be attributed to the inability of Hfq65 to mediate translational activation of rpoS by DsrA. Similarly, as Hfq65 failed to bind to sodB mRNA (Figure 5), and thus to mediate translational repression of this mRNA by RyhB (Figure 2), it is reasonable to assume that the observed growth on succinate in the presence of Hfq65 and RyhB (Figure 1B) results likewise from the failure of the truncated protein to interact with the sdhCDAB mRNA.
Both, Hfqwt and Hfq65, bound with comparable affinities to all three RNA oligonucleotides (K1/2 of ∼50–230 nM; Supplementary Figure S1), whereby the K1/2 of Hfqwt for the RNA oligonucleotides was somewhat reduced when compared with Hfq65. Assuming two independent binding sites on Hfqwt for the RNA oligonucleotides, the K1/2 for Hfqwt would represent a mean value, which could explain the reduced affinity of Hfqwt for these substrates when compared with Hfq65. In any case, these results (Supplementary Figure S1) showed that neither Hfqwt nor Hfq65 discriminate against any of the RNA oligonucleotides used, and can therefore be readily reconciled with the idea that Hfq65 fails to accommodate two RNAs. Hfq65 stimulated annealing of the two complementary RNA oligonucleotides 2-fold (Figure 4A). We speculate that this moderate acceleration of self-annealing results from a reduction of Brownian molecular motion through immobilization of one RNA oligonucleotide. Obviously, the same effect cannot stimulate FRET with non-complementary RNA oligonucleotides, as they need to be fixed in close proximity. Recent studies (38), showed that amino acid substitutions at the proximal face including alterations within the Sm2 motif, i.e. at the ‘business surface’ for ncRNAs, had a minor effect on rpoS binding, and that poly(A) did not compete with rpoS mRNA binding. Taken together with our observations that the C-terminus of Hfq is required for rpoS mRNA binding (Figure 5), and that Hfqwt stimulated FRET with non-complementary RNA oligonucleotides (Figure 4B), the available data collectively suggest that the C-terminus of Hfq forms a major and perhaps independent interaction surface for (m)RNA. A truncated version of E. coli Hfq, consisting of the first 75 aa was functional in stimulating rpoS translation (41). Therefore, the sequence between amino acid 65 and 75 of Hfq will be of prime interest to tackle the C-terminal sub-domain required for binding of mRNAs. As the binding motif for Hfq appears to be similar in ncRNAs and mRNAs (12,27,30), the molecular features underlying the preference of the C-terminus for mRNA remains to be elucidated. As yet, structural data on the C-terminus of Hfq are lacking. The PONDR (predictor of natural disordered regions) algorithm (www.pondr.com) predicts that the C-terminus of Hfq is structurally disordered (Supplementary Figure S2) that is a hallmark of RNA chaperones (61). Such flexible regions are believed to provide conformational fluctuations that facilitate intermolecular interactions. The flexibility of the C-terminus could therefore provide the molecular basis for the interaction of Hfq with many mRNA substrates or proteins. Indeed, several proteins including RNA polymerase, ribosomal protein S1 (62), RNase E (63), polyA-polymerase (PAP I) and polynucleotidephosphorylase (PNPase) (64) have been found in complex with Hfq. However, it remains to be elucidated in either case whether Hfq is in direct physical contact with these candidate proteins or whether these findings result from the spatial association of the transcriptional, translational and RNA decay machineries in bacteria.
As obvious from Figure 4B, Hfqwt can bind two non-complementary RNAs, whereas Hfq65 lacked this capacity. This seems somewhat at variance with band shift assays showing that Hfq only facilitates the interaction between RNAs able to base pair (14). However, the RNA oligonucleotides used here may differ in their binding requirements from the natural substrates used by Zhang et al. (14), and monitoring binding in real time in solution most likely puts less constrains on the stability of RNA–protein complexes than gel electrophoresis. Nevertheless, as Hfq65 was defective in binding two RNA oligonucleotides one binding site for these substrates appears to be provided by the C-terminus.
An extended C-terminus is only found in Hfq proteins of γ- and β-proteobacteria (42). Interestingly, Hfq-mediated riboregulation of mRNAs by ncRNAs has unequivocally been demonstrated only in E. coli (51) and Salmonella enterica (65), and was implicated in Shigella (66) and Vibrio (67) species, all of which belong to the γ-proteobacteria. In contrast, with 73 aa and 77 aa, respectively, the firmicutes B. subtilis and S. aureus possess Hfq proteins with short C-terminal extensions (40). Ectopic expression of the S. aureus or the B. subtilis hfq gene in E. coli AM111F′hfq− did not (i) inhibit growth on succinate in the presence of RyhB (Supplementary Figure S3A), was unable (ii) to support RyhB-mediated repression of sodB translation and did not (iii) result in translational auto-repression of E. coli hfq mRNA (Supplementary Figure S3B). Consistent with these observations, neither the S. aureus nor the B. subtilis Hfq protein bound to hfq126 or sodB192 mRNA (Supplementary Figure S3C). Thus, phenotypically both proteins behaved like Hfq65. Despite of the heterologous E. coli system used, these experiments could imply that there are no other inherent features in these shorter Hfq variants that would compensate for the C-terminal extension present in the Hfq proteins of γ-proteobacteria. In addition, they lend further support to the notion that an extended C-terminus is required for riboregulation. In fact, no Hfq requirement has as yet been reported for ncRNA–mRNA interactions in B. subtilis (68,69) and in S. aureus (70,71). Moreover, in contrast to a reduced resistance towards several stress conditions and an attenuated virulence phenotype reported for hfq deletion mutants of several γ-proteobacteria including Vibrio cholerae (21), P. aeruginosa (43) or Salmonella typhimurium (72), a S. aureus hfq− mutant showed no detectable defects (73). This contrasts the somewhat reduced stress tolerance and virulence reported for a hfq− mutant of Listeria monocytogenes (20) that contains likewise a Hfq protein with a short C-terminal extension. In the latter bacterium, Hfq has been shown to confer stability to at least one ncRNA (74). Taken these reports, it remains to be seen whether Hfq-mediated riboregulation, i.e. translational silencing or activation of mRNAs by ncRNAs, is confined to γ- and perhaps β-proteobacteria. On the other hand, the elucidation of the molecular function of Hfq homologues with short C-terminal tails in firmicutes remains a further challenge.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
Austrian Science Fund (FWF) within the Special Research Program 17: (F1703) to R.S. (F1720) to U.B. Funding to pay the Open Access publication charges for this article was provided by FWF.
Conflict of interest statement. None declared.
REFERENCES
- 1.Franze de Fernandez MT, Eoyang L, August JT. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature. 1968;219:588–590. doi: 10.1038/219588a0. [DOI] [PubMed] [Google Scholar]
- 2.Tsui HC, Leung HC, Winkler ME. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 3.Brown L, Elliott T. Mutations that increase expression of the rpoS gene and decrease its dependence on hfq function in Salmonella typhimurium. J. Bacteriol. 1997;179:656–662. doi: 10.1128/jb.179.3.656-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muffler A, Fischer D, Hengge-Aronis R. The RNA-binding protein HF-I, known as a host factor for phage Qbeta RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 1996;10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- 5.Massé E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl Acad. Sci. USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vecerek B, Moll I, Afonyushkin T, Kaberdin V, Bläsi U. Interaction of the RNA chaperone Hfq with mRNAs: direct and indirect roles of Hfq in iron metabolism of Escherichia coli. Mol. Microbiol. 2003;50:897–909. doi: 10.1046/j.1365-2958.2003.03727.x. [DOI] [PubMed] [Google Scholar]
- 7.Tsui HC, Feng G, Winkler ME. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J. Bacteriol. 1997;179:7476–7487. doi: 10.1128/jb.179.23.7476-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vytvytska O, Moll I, Kaberdin VR, von Gabain A, Bläsi U. Hfq (HF1) stimulates ompA mRNA decay by interfering with ribosome binding. Genes Dev. 2000;14:1109–1118. [PMC free article] [PubMed] [Google Scholar]
- 9.Folichon M, Arluison V, Pellegrini O, Huntzinger E, Regnier P, Hajnsdorf E. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 2003;31:7302–7310. doi: 10.1093/nar/gkg915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sledjeski DD, Whitman C, Zhang A. Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 2001;183:1997–2005. doi: 10.1128/JB.183.6.1997-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massé E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moll I, Afonyushkin T, Vytvytska O, Kaberdin VR, Bläsi U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA. 2003;9:1308–1314. doi: 10.1261/rna.5850703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Afonyushkin T, Vecerek B, Moll I, Bläsi U, Kaberdin VR. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 2005;33:1678–1689. doi: 10.1093/nar/gki313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 15.Vecerek B, Moll I, Bläsi U. Control of Fur synthesis by the non-coding RNA RyhB and iron-responsive decoding. EMBO J. 2007;26:965–975. doi: 10.1038/sj.emboj.7601553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Møller T, Franch T, Hojrup P, Keene DR, Bachinger HP, Brennan RG, Valentin-Hansen P. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 17.Kawamoto H, Koide Y, Morita T, Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 2006;61:1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- 18.Robertson GT, Roop R.M., Jr The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol. Microbiol. 1999;34:690–700. doi: 10.1046/j.1365-2958.1999.01629.x. [DOI] [PubMed] [Google Scholar]
- 19.Sonnleitner E, Hagens S, Rosenau F, Wilhelm S, Habel A, Jäger KE, Bläsi U. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb. Pathog. 2003;35:217–228. doi: 10.1016/s0882-4010(03)00149-9. [DOI] [PubMed] [Google Scholar]
- 20.Christiansen JK, Larsen MH, Ingmer H, Søgaard-Andersen L, Kallipolitis BH. The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J. Bacteriol. 2004;186:3355–3362. doi: 10.1128/JB.186.11.3355-3362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding Y, Davis BM, Waldor MK. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol. 2004;53:345–354. doi: 10.1111/j.1365-2958.2004.04142.x. [DOI] [PubMed] [Google Scholar]
- 22.Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 23.Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 25.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogel J, Sharma CM. How to find small non-coding RNAs in bacteria. Biol. Chem. 2005;386:1219–1238. doi: 10.1515/BC.2005.140. [DOI] [PubMed] [Google Scholar]
- 27.Geissmann TA, Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Majdalani N, Vanderpool CK, Gottesman S. Bacterial small RNA regulators. Crit. Rev. Biochem. Mol. Biol. 2005;40:93–113. doi: 10.1080/10409230590918702. [DOI] [PubMed] [Google Scholar]
- 29.Lease RA, Woodson SA. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J. Mol. Biol. 2004;344:1211–1223. doi: 10.1016/j.jmb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 30.Brescia CC, Mikulecky PJ, Feig AL, Sledjeski DD. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA. 2003;9:33–43. doi: 10.1261/rna.2570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moll I, Leitsch D, Steinhauser T, Bläsi U. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep. 2003;4:284–289. doi: 10.1038/sj.embor.embor772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Udekwu KI, Darfeuille F, Vogel J, Reimegard J, Holmqvist E, Wagner EG. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 2005;19:2355–2366. doi: 10.1101/gad.354405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schumacher MA, Pearson RF, Møller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sauter C, Basquin J, Suck D. Sm-like proteins in Eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res. 2003;31:4091–4098. doi: 10.1093/nar/gkg480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikulin A, Stolboushkina E, Perederina A, Vassilieva I, Blaesi U, Moll I, Kachalova G, Yokoyama S, Vassylyev D, et al. Structure of Pseudomonas aeruginosa Hfq protein. Acta Crystallogr. D Biol. Crystallogr. 2005;61:141–146. doi: 10.1107/S0907444904030008. [DOI] [PubMed] [Google Scholar]
- 36.Achsel T, Stark H, Lührmann R. The Sm domain is an ancient RNA-binding motif with oligo(U) specificity. Proc. Natl Acad. Sci. USA. 2001;98:3685–3689. doi: 10.1073/pnas.071033998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Večerek B, Moll I, Bläsi U. Translational autocontrol of the Escherichia coli hfq RNA chaperone gene. RNA. 2005;11:976–984. doi: 10.1261/rna.2360205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat. Struct. Mol. Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun X, Wartell RM. Escherichia coli Hfq binds A18 and DsrA domain II with similar 2:1 Hfq6/RNA stoichiometry using different surface sites. Biochemistry. 2006;45:4875–4887. doi: 10.1021/bi0523613. [DOI] [PubMed] [Google Scholar]
- 40.Sun X, Zhulin I, Wartell RM. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 2002;30:3662–3671. doi: 10.1093/nar/gkf508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sonnleitner E, Moll I, Bläsi U. Functional replacement of the Escherichia coli hfq gene by the homologue of Pseudomonas aeruginosa. Microbiology. 2002;148:883–891. doi: 10.1099/00221287-148-3-883. [DOI] [PubMed] [Google Scholar]
- 42.Arluison V, Folichon M, Marco S, Derreumaux P, Pellegrini O, Seguin J, Hajnsdorf E, Regnier P. The C-terminal domain of Escherichia coli Hfq increases the stability of the hexamer. Eur. J. Biochem. 2004;271:1258–1265. doi: 10.1111/j.1432-1033.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- 43.Sonnleitner E, Napetschnig J, Afonyushkin T, Ecker K, Večerek B, Moll I, Kaberdin VR, Bläsi U. Functional effects of variants of the RNA chaperone Hfq. Biochem. Biophys. Res. Commun. 2004;323:1017–1023. doi: 10.1016/j.bbrc.2004.08.190. [DOI] [PubMed] [Google Scholar]
- 44.Steiner M, Lubitz W, Bläsi U. The missing link in phage lysis of gram-positive bacteria: gene 14 of Bacillus subtilis phage phi 29 encodes the functional homolog of lambda S protein. J. Bacteriol. 1993;175:1038–1042. doi: 10.1128/jb.175.4.1038-1042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller JH. Experiments in molecular genetics. Cold Spring Haror, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 46.Brückner R. A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene. 1992;122:187–192. doi: 10.1016/0378-1119(92)90048-t. [DOI] [PubMed] [Google Scholar]
- 47.Takayanagi Y, Tanaka K, Takahashi H. Structure of the 5′ upstream region and the regulation of the rpoS gene of Escherichia coli. Mol. Gen. Genet. 1994;243:525–531. doi: 10.1007/BF00284200. [DOI] [PubMed] [Google Scholar]
- 48.Lin-Chao S, Bremer H. Effect of the bacterial growth rate on replication control of plasmid pBR322 in Escherichia coli. Mol. Gen. Genet. 1986;203:143–149. doi: 10.1007/BF00330395. [DOI] [PubMed] [Google Scholar]
- 49.Sonnleitner E, Schuster M, Sorger-Domenigg T, Greenberg EP, Bläsi U. Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol. Microbiol. 2006;59:1542–1558. doi: 10.1111/j.1365-2958.2006.05032.x. [DOI] [PubMed] [Google Scholar]
- 50.Mayer O, Rajkowitsch L, Lorenz C, Konrat R, Schroeder R. RNA chaperone activity and RNA-binding properties of the E. coli protein StpA. Nucleic Acids Res. 2007;35:1257–1269. doi: 10.1093/nar/gkl1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gottesman S, McCullen CA, Guillier M, Vanderpool CK, Majdalani N, Benhammou J, Thompson KM, FitzGerald PC, Sowa NA, et al. Small RNA regulators and the bacterial response to stress. Cold Spring Harb. Symp. Quant Biol. 2006;71:1–11. doi: 10.1101/sqb.2006.71.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morita T, Mochizuki Y, Aiba H. Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc. Natl Acad. Sci. USA. 2006;103:4858–4863. doi: 10.1073/pnas.0509638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lease RA, Cusick ME, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl Acad. Sci. USA. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl Acad. Sci. USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sledjeski DD, Gupta A, Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 56.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arluison V, Hohng S, Roy R, Pellegrini O, Regnier P, Ha T. Spectroscopic observation of RNA chaperone activities of Hfq in post-transcriptional regulation by a small non-coding RNA. Nucleic Acids Res. 2007;35:999–1006. doi: 10.1093/nar/gkl1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tolstorukov MY, Virnik KM, Adhya S, Zhurkin VB. A-tract clusters may facilitate DNA packaging in bacterial nucleoid. Nucleic Acids Res. 2005;33:3907–3918. doi: 10.1093/nar/gki699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Azam TA, Hiraga S, Ishihama A. Two types of localization of the DNA-binding proteins within the Escherichia coli nucleoid. Genes Cells. 2000;5:613–626. doi: 10.1046/j.1365-2443.2000.00350.x. [DOI] [PubMed] [Google Scholar]
- 60.Zambrano MM, Siegele DA, Almiron M, Tormo A, Kolter R. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]
- 61.Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18:1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- 62.Sukhodolets MV, Garges S. Interaction of Escherichia coli RNA polymerase with the ribosomal protein S1 and the Sm-like ATPase Hfq. Biochemistry. 2003;42:8022–8034. doi: 10.1021/bi020638i. [DOI] [PubMed] [Google Scholar]
- 63.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mohanty BK, Maples VF, Kushner SR. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol. Microbiol. 2004;54:905–920. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- 65.Bossi L, Figueroa-Bossi N. A small RNA downregulates LamB maltoporin in Salmonella. Mol. Microbiol. 2007;65:799–810. doi: 10.1111/j.1365-2958.2007.05829.x. [DOI] [PubMed] [Google Scholar]
- 66.Antal M, Bordeau V, Douchin V, Felden B. A small bacterial RNA regulates a putative ABC transporter. J. Biol. Chem. 2005;280:7901–7908. doi: 10.1074/jbc.M413071200. [DOI] [PubMed] [Google Scholar]
- 67.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 68.Heidrich N, Chinali A, Gerth U, Brantl S. The small untranslated RNA SR1 from the Bacillus subtilis genome is involved in the regulation of arginine catabolism. Mol. Microbiol. 2006;62:520–536. doi: 10.1111/j.1365-2958.2006.05384.x. [DOI] [PubMed] [Google Scholar]
- 69.Heidrich N, Moll I, Brantl S. In vitro analysis of the interaction between the small RNA SR1 and its primary target ahrC mRNA. Nucleic Acids Res. 2007;35:4331–4346. doi: 10.1093/nar/gkm439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huntzinger E, Boisset S, Saveanu C, Benito Y, Geissmann T, Namane A, Lina G, Etienne J, Ehresmann B, et al. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005;24:824–835. doi: 10.1038/sj.emboj.7600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bohn C, Rigoulay C, Bouloc P. No detectable effect of RNA-binding protein Hfq absence in Staphylococcus aureus. BMC Microbiol. 2007;7:10. doi: 10.1186/1471-2180-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Christiansen JK, Nielsen JS, Ebersbach T, Valentin-Hansen P, Søgaard-Andersen L, Kallipolitis BH. Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA. 2006;12:1383–1396. doi: 10.1261/rna.49706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Romero P, Obradovic Z, Dunker AK. Sequence data analysis for long disordered regions prediction in the calcineurin family. Genome Inform. 1997;8:110–124. [PubMed] [Google Scholar]
- 76.Li X, Romero P, Rani M, Dunker AK, Obradovic Z. Predicting protein disorder for N-, C-, and internal regions. Genome Inform. 1999;10:30–40. [PubMed] [Google Scholar]
- 77.Romero P, Obradovic Z, Li X, Garner E, Brown C, Dunker AK. Proteins: sequence complexity of disordered protein. Struct. Funct. Gen. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]