Abstract
This work describes the novel use of tolC as a selectable/counter-selectable marker for the facile modification of DNA in Escherichia coli. Expression of TolC (an outer membrane protein) confers relative resistance to toxic small molecules, while its absence renders the cell tolerant to colicin E1. These features, coupled with the λredgam recombination system, allow for selection of tolC insertions/deletions anywhere on the E. coli chromosome or on plasmid DNA. This methodology obviates the need for minimal growth media, specialized wash protocols and the lengthy incubation times required by other published recombineering methods. As a rigorous test of the TolC selection system, six out of seven 23S rRNA genes were consecutively and seamlessly removed from the E. coli chromosome without affecting expression of neighboring genes within the complex rrn operons. The resulting plasmid-free strain retains one 23S rRNA gene (rrlC) in its natural location on the chromosome and is the first mutant of its kind. These new rRNA mutants will be useful in the study of rRNA gene regulation and ribosome function. Given its high efficiency, low background and facility in rich media, tolC selection is a broadly applicable method for the modification of DNA by recombineering.
INTRODUCTION
The tolC gene resides in an operon at 68.5 min on the Escherichia coli chromosome and precedes three non-essential open reading frames of unknown function (1,2). Sequence analysis and expression profiling postulate tolC to be transcribed from a tandem promoter (3). One study reports that tolC may fall under control of the mar-sox regulon (4), and other studies suggest the two-component system EvgAB regulates tolC (5,6). The gene itself encodes a 493 amino acid outer membrane protein with a signal sequence that allows SecB-dependent translocation (7,8). Once in the periplasm, mature TolC monomers undergo factor-independent assembly to form trimers subsequent to outer membrane insertion (9). Electron microscopy and X-ray crystallography have revealed a unique structure and topography for TolC (10) with the trimer existing as a trans-periplasmic tunnel, 140 Å in length (11). Upon interaction with various inner-membrane pumps and translocases, TolC plays an important role in hemolysin secretion (12,13), protein import (14,15) and antibiotic efflux (10,16). In E. coli for instance, the AcrB-TolC efflux system pumps a diverse set of small molecules from the cell that include novobiocin, erythromycin and sodium dodecyl sulfate (SDS).
In addition to its role in expelling toxic compounds from the cell, surface-exposed loops of the TolC channel provide for specific interactions with bacteriophage (17) and for the import of bacteriocins such as colicin E1 (18). Bacteriocins are bactericidal proteins that constitute part of an antimicrobial defense system and can be found in most bacterial genera and in the archaea as well (19). Most bacteriocins are large proteins and, in the case of colicins, the genes encoding these toxins reside typically on extrachromosomal elements (20). Group A colicins demonstrate diverse mechanisms of action and have a narrow killing spectrum. They bind to the E. coli vitamin B receptor BtuB and are dependent upon the Tol proteins for transport (15,21). Colicin E1 (522 amino acids in length) requires TolC exclusively for access to the cytoplasmic membrane where it forms voltage-gated ion channels to disrupt the bilayer and kill the cell (22).
The first tol mutants demonstrated a colicin tolerant phenotype (18). Such mutants lost their sensitivity to particular colicins but fully maintained the ability to adsorb toxin on the cell surface through BtuB. Mutations at the tolC locus differ from those at other tol loci, in that they make the cell tolerant only to colicin E1, while such mutants respond with full sensitivity to other group A colicins (23). It is precisely these features that form the basis for testing tolC's utility as a selectable/counter-selectable marker in genome engineering studies.
Recent years have witnessed remarkable advances in the application of new tools for the in vivo manipulation of DNA (24–28). Perhaps the most efficient of these tools is the bacteriophage λ-Red recombinase (29). This system has been used to ‘recombineer’ genomic and extra chromosomal DNA in E. coli through simple introduction of double- or single-stranded DNA substrates into the bacterial cell (30). The efficiency of the system is enhanced greatly by the incorporation of selectable genetic markers to assist in the identification of desired recombinants and a number of methods exist for constructing point mutations, deletions and in-frame fusions (31–36). Recent work has demonstrated continued improvements to these approaches such as the use of galK and thyA. These genes encode single open reading frames each able to function as both a selectable and counter-selectable marker (37,38), but some practical limitations remain. Current protocols for these markers involve extensive washing of cells and the use of specialized media during selection. In addition, the requirement of minimal media for cell growth often results in a delay of 2–3 days before new mutant colonies can be chosen for further manipulation.
The following report describes the novel use of tolC as a selectable/counter-selectable marker for recombination-mediated genetic engineering in E. coli. This new system maintains the flexibility and efficiency of the latest recombineering approaches while eliminating the need for specialized media and extended incubation times. To demonstrate the utility of the tolC single-gene marker system, deletions, point mutations and gene fusions were made to a variety of coding regions on the E. coli chromosome and on plasmid DNA. In addition, six of the seven 23S rRNA genes were seamlessly deleted from the E. coli genome using one universal recombination cassette. The resulting strain (containing a single functioning 23S rRNA gene) shows no polar effects on rRNA transcription and serves as a new tool for the study of rRNA gene regulation and ribosome function.
MATERIALS AND METHODS
Bacterial strains
Escherichia coli DY329 (W3110 ΔlacU169, nadA::Tn10, gal490, λ cI857 Δ[cro-bio]) was a generous gift of Don Court (28) and used with permission under US Public Health Service License Number L-159-2006/0. It contains a defective λ prophage expressing the gam, bet and exo recombination functions from the cI857 regulated pL promoter. All bacterial strains constructed during this work are derivatives of RS205 (a nad+ derivative of strain DY329) that was engineered according to the methods outlined in (39). A list of all bacterial strains can be found in Supplementary Data, Table S1.
Media and reagents
Bacterial strains were propagated on Luria Bertani broth and agar (Difco, BD Biosciences Franklin Lakes, NJ) according to (40). Agar plates for SDS (Fluka, Switzerland) selection were prepared according to standard procedures with an optimal concentration of 0.01% (w/v). Colicin E1 (SigmaAldrich St Louis, MO) selection was performed in 0.1 ml broth at a final concentration of 2 units/μl (see subsequently). DNA oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA) and a list describing their attributes can be found in Supplementary Data, Table S2. Linear dsDNA substrates were prepared using the GC-Rich PCR system (Roche Applied Science Indianapolis, IA) and purified following electrophoresis according to standard methods (41) using the WizardSV-Gel Clean-Up system (Promega Madison, WI). All PCR screening was performed with JumpStart REDTaq Ready Mix (SigmaAldrich) on purified, single bacterial colonies. Antibiotics and other bulk chemicals were purchased from SigmaAldrich.
Linear DNA substrate design, redgam induction and selection of recombinants
The tolC gene was removed from its location at 68.5 min on the E. coli chromosome by linear DNA transformation of redgam-induced cells with a 70-base single-stranded oligonucleotide (Rx-P15) of the following sequence: 5′-TTTCAGCGACGTTTGACTGCCGTTTGAGCAGTC-ATATGACGACGACGGGGCTTCGGCCCCGTCTGAACGT-3′ (Figure 1A). Colicin E1 selection was performed (see subsequently) and transformants were screened by PCR for the presence of a 310 base-pair (bp) product using Primers Rx-P19 (5′-GTTTCTCGTGCAATAATTTCTACATC-3′) and Rx-P20 (5′-CGTATGGATTTTGTCCGTTTCA-3′). A tolC − phenotype was confirmed by patching single colonies on 0.01% SDS agar and by cross streak against colicin E1 [plate receptor test- (23)].
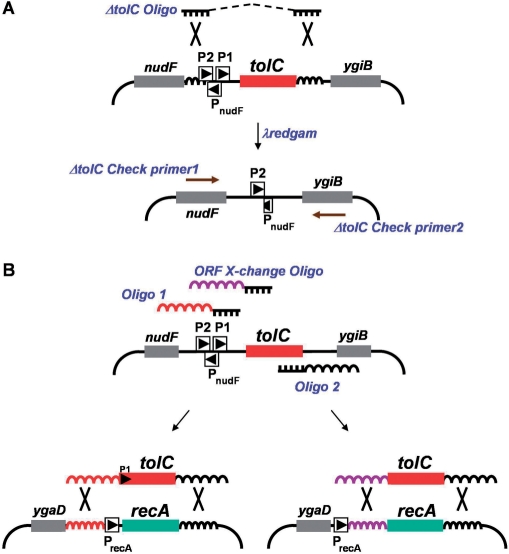
Figure 1.
Deletion of tolC from the E. coli genome and cassette design. Fragments of chromosomal DNA are depicted with bent lines. Target genes (tolC and recA) are denoted by colored boxes; neighboring genes that encode open reading frames are shown with shaded boxes. P2 and P1 refer to the promoters of tolC; PnudF and PrecA drive expression of the nudF and recA genes, respectively. (A) Strategy for deleting tolC from the chromosome. Recombination using the single-stranded DNA molecule ΔtolC Oligo (Rx-P15) and λ redgam is depicted. The Rx-P15 oligo contains 35 bases of DNA directly homologous to sequences flanking tolC on the chromosome (curvy lines). The dotted line in Rx-P15 is merely a representation demonstrating that the two halves of the 70-base oligo are contiguous. Large X's denote λ red-mediated cross-over between regions of homologous DNA. Following recombination and selection for colicin E1 resistance, the deletion of tolC from the genome was confirmed by PCR with primers shown as brown arrows (Rx-P19 and -P20). (B) Amplification of the tolC cassette and deletion of recA. Oligo 1 (RxP-43) and Oligo 2 (Rx-P44) PCR amplify a double-stranded tolC expression cassette. ORF X-change oligo (Rx-P45) is used with RX-P46 (data not shown) to amplify only the tolC open reading frame without its promoter. Curvy lines represent homologous DNA shared by the linear DNA substrate and the bacterial chromosome while large X's again denote λ red-mediated cross-over between regions of homologous DNA. In this example, the tolC expression cassette replaces the entire recA transcription unit (red) or only the recA ORF (purple) conferring SDS resistance. A similar strategy was used to delete the ara regulon, dicB and ygeX (data not shown). PCR primers internal to tolC and complementary to neighboring genes were used to confirm insertion of tolC and deletion of the target gene (see Figure 2 for an example). Removing tolC from its new location followed the methodology in (A) above. The sequence and description of all primers and oligos can be found in Table S2. Illustrations not drawn to scale.
A cassette containing the tolC gene (and upstream promoter sequence) was amplified by PCR from strain RS205 gDNA to serve directly as a linear DNA substrate. Primer design adhered to the following general scheme: 5′-N50-TTGAGGCACATTAACGCCCTAT-3′ and 5′-N50-CCCCGTCGTCGTCATCAG-3′, where ‘N’ represents nucleotides comprising the homology arms necessary for recombination. These tails share sequence identity with the regions of DNA flanking a target gene (Figure 1B). A 1.62 kb PCR product is generated using such primers and allows tolC expression when integrated into the chromosome (or plasmid DNA) via the 50-bp long tails. For experiments where the tolC ORF is used for exchange with a target open reading frame (i.e. no promoter), the following primer pair is used: 5′-N50-ATGAAGAAATTGCTCCCCATTCT-3′ and 5′-N50-TCAGTTACGGAAAGGGTTATGACC-3′. SDSR transformants are then screened for proper positioning of tolC using PCR with primers specific for regions flanking the target gene. Appropriate chromosomal rearrangements are confirmed by DNA sequencing. Removal of the tolC cassette followed the general strategy outlined in the previous paragraph.
General recombineering with tolC as a selectable and counter-selectable marker
RS205 and other derivatives were prepared for recombineering essentially as described (28,39). Briefly, cells were diluted 1/50 from a fresh overnight culture in LB broth at 35°C and grown to between 0.3 and 0.4 OD600. The pL promoter was de-repressed by incubation at 42°C for 15 min with optimal induction of the λredgam genes requiring longer incubation for some of the 23S rRNA mutants (see Results section for further detail). Induced cultures were chilled by constant swirling in an ice/water bath for 5–10 min and cells were pelleted by centrifugation. Cell pellets were washed in 2× their original volume of ice-cold water, pelleted and washed again in 1/20 their original volume with ice-cold water. After a final centrifugation step in a 1.5 ml microfuge tube, cells were resuspended to 1/100 their original volume in ice-cold water and used immediately for electroporation.
For linear DNA transformation, 50 μl of cell suspension was mixed with ∼100 ng DNA, transferred to a pre-chilled electroporation cuvette (1 mm), and pulsed at 1.8 kV in a Biorad Gene Pulser Xcell (Hercules, CA). Cells were transferred to a 15 ml tube (Falcon 2059) containing 1 ml LB and placed at 35°C with shaking for outgrowth.
Selection of tolC + recombinants was performed after a 2-h incubation followed by plating ≤ 0.1 ml onto LB agar containing 0.01% SDS. Most experiments yielded between 1.6 × 104 and 2.3 × 106 SDS-resistant transformants per 5 × 108 total cells. Selection of SDSR transformants for some of the 23S rRNA mutants required extended outgrowth times (see text).
To select for loss of the tolC marker, cells were incubated after electroporation for 5 h to allow for TolC turnover. Typically, 10 μl of cells was then transferred to a fresh tube containing 65 μl of LB and 200 units colicin E1 and incubated for 60 min at 35°C with shaking. Aliquots are plated on LB agar and grown overnight at 35°C. Most experiments yielded colicin-resistant recombinants at an ∼0.015 frequency. The colicin E1 tolerance of the resulting recombinants can be confirmed by a modified plate receptor test where, for instance, a 15 μl line of colicin E1 (8 units/μl) is pipetted down the center of an LB plate and each transformant is cross-streaked using a flat toothpick. Sensitivity was scored as a clear zone over the colicin E1 line.
Preliminary characterization of engineered rRNA mutants
Southern blots were performed as described (41) using standard procedures. Genomic DNA samples were prepared using the DNeasy Blood and Tissue kit (Qiagen Valencia, CA) and treated with restriction endonucleases BamHI and PstI (42). The 23S rDNA region from HpaI to SalI was PCR amplified from strain RS205 with the following primers 5′-ACGCTTCTCGCTCTCAACC-3′ (Rx-P200) and 5′-GAATAGGGGAGCCGAAGGG-3′ (Rx-P201) then labeled with 32P-NTP for use as probe.
Growth characteristics of the different rRNA mutants were determined by luminescence using BacTiter Glo (Promega) from 50 ml broth cultures in 250 ml Erlenmeyer flasks at 37°C using an orbital shaker set at 180 rpm. Doubling time was calculated according to the following: tgen = 1/k where the growth rate constant k = logXt-logX0/0.301t with X equal to cell number (lumens). Sensitivity to various antibiotics was assessed in LB broth according to CLSI procedures with minor modifications (43).
Ribosome isolation was performed according to published procedures (44). Briefly, cells were grown to mid/late log (OD600 ≤ 0.9) in LB medium at 37°C, concentrated by centrifugation, and frozen in liquid N2. One-gram cell pellets were resuspended in 10 ml of ice cold 1X TMK (10 mM Tris–Cl pH 7.5, 16 mM MgAc, 60 mM KAc, 1 mM DTT) and lysed at 20–25 000 psi using an EmulsiFlex C5 high pressure homogenizer (Avestin Inc. Ottawa, Canada). Cell lysate was cleared by centrifugation at 30 000g for 30 min at 4°C. Supernatant was mixed with sucrose to 220 mM final and centrifuged for 53 min at 90 000gave (4°C) to remove additional debris. Ribosomes were pelleted through a 20% v/v cushion of 1.1 M sucrose by centrifugation at 150 000gave for 18 h at 4°C, washed and resuspended in 1X TMK. A total of 10–30% sucrose gradients were prepared in 1X TMK using a Gradient Master (Biocomp Inc. Edenton, NJ) and ribosomes (2 OD260 nm units/0.1 ml) were analyzed following centrifugation in a SW60Ti rotor (Beckman Fullerton, CA) at 237 000g, 2 h, 7°C using a continuous flow UV monitor (Amersham Biosciences Piscataway, NJ).
RESULTS
Deleting the tolC promoter and coding region
As a first step in testing whether tolC could be used as both a positive and negative selectable marker, a strain lacking the tolC coding sequence was constructed. The strategy for removing tolC from its normal location at 68.5 min on the E. coli chromosome is shown in Figure 1A. Lambda recombineering was used to delete the gene, simultaneously demonstrating proof of principle for the strategy outlined earlier. Single-stranded DNA oligonucleotides were designed to have 35 bases of exact sequence homology to regions up and downstream of the tolC coding sequence. Recombination directed by this 70-base oligo (Rx-P15) would remove the tolC coding sequence in addition to its proximal promoter (P1), leaving the P2 promoter completely intact and slightly truncating the nudF promoter. When transformed to redgam-induced RS205 and selected for tolerance to colicin E1, Rx-P15 was able to remove the tolC gene with 25% efficiency (Table 1). Deletion of tolC was confirmed by PCR using primers directed against sequences within nudF and ygiB (Figure 1A) and the colicin E1 tolerance of the transformants was confirmed using the modified plate receptor test (data not shown). Oligonucleotide Rx-P17 (which leaves both P1tolC and PnudF intact) was just as efficient at removing tolC. When using DNA oligonucleotides exactly complementary to Rx-P15 and Rx-P17 (i.e. homologous to the lead replicating gDNA strand), tolC gene deletions were not detected under the same experimental conditions (data not shown). Thus, selection for tolC removal is possible through the use of colicin E1 and the resulting strain, RS206, can be used to test tolC's efficiency as a positive selectable marker.
Table 1.
Recombineering with tolC
| Target | Insertion method | Efficiency of tolC insertion | Deletion method | Efficiency of tolC removal |
|---|---|---|---|---|
| tolC | N/A | N/A | ss Oligo | 25% |
| recA | PtolC Cassette | 100% | ss Oligo | 86% |
| recA | PtolC Cassette | 100% | ds PCR product | 100% |
| araCBAD | PtolC Cassette | 100% | ss Oligo | 77% |
| dicB | PtolC Cassette | 100% | ss Oligo | 89% |
| ygeX | PtolC Cassette | 100% | ss Oligo | ND |
| rrIC | PtolC Cassette | 13% | ss Oligo | 100% |
| recA | tolC ORF-Xchange | 100% | ND | ND |
| dicB | tolC ORF-Xchange | 0% | ND | ND |
| ygeX | tolC ORF-Xchange | 0% | ND | ND |
A list of target genes for testing the efficiency of the tolC selectable/counter-selectable marker system is shown in column one. The table lists the methods used to insert tolC (either through a promoter-containing tolC cassette or through ORF-X change) and the methods used to remove tolC (either by using a single-stranded oligo or a double-stranded PCR product). Confirmation of tolC insertion or removal was performed by PCR (see Figure S2 for primers), phenotype analysis, and DNA sequencing when appropriate. The efficiencies for each method were calculated as follows: (number of recombinants testing positive for the insertion or deletion event/total number of resistant colonies tested) × 100. Deletion of tolC following ORF-X change was not performed. ND: not determined.
Design, construction and testing of a tolC expression cassette
With the tolC coding sequence and promoter deleted from the chromosome of RS206, a tolC expression cassette was amplified by PCR from the gDNA of RS205 (tolC +). Oligonucleotides were designed to amplify the region coding for the P1 tolC promoter and the entire tolC open reading frame with 50-base tails homologous to regions flanking an appropriate target gene (Figure 1B). The E. coli recA gene was selected as an appropriate candidate for testing if a target gene could be deleted using λ recombineering and tolC as a selectable marker. Upon introduction of the recA-tolC cassette into RS206 and selection of SDS resistance, resultant colonies were screened by PCR using primers located within the tolC cassette and those from neighboring genes (oraA and ygaD). The use of tolC as a selectable marker was highly efficient, as 100% of transformants tested (24 out of 24) demonstrated removal of recA and insertion of the tolC cassette.
TolC's facility as a selectable/counter selectable marker prompted a more detailed investigation of the selection parameters used during the proof of principle experiments. SDS selection was optimized by testing LB agar plates containing various concentrations of SDS or through direct spreading of SDS onto pre-poured LB agar plates (20 ml) before addition of cells. Both methods were successful as little to no background growth of SDS sensitive colonies was detected at SDS concentrations as low as 0.002%. Concentrations of SDS as high as 1% (10 mg/ml) were also tolerated (data not shown). Fixing the SDS concentration at 0.01% was sufficient to select for deletion of the entire arabinose operon from the E. coli genome, as well as the dicB and ygeX genes (Table 1) and for insertion of tolC into low- and medium-copy plasmids (data not shown).
A slightly different approach was used for colicin E1 counter-selection. As opposed to incorporating colicin E1 into agar (where background growth was problematic), the peptide was added to transformed cells in small volumes of broth as outlined in the Materials and Methods section. This simple method proved economical and successful as background growth was avoided. A variety of colicin E1 concentrations were tested in this fashion using the single-stranded DNA oligonucleotide strategy (Figure 1A) to remove tolC from the recA, ara and dicB loci (data not shown). A concentration of 2 units/μl (final) yielded reproducible results and removal of tolC by colicin E1 counter-selection was 86, 77 and 89% efficient, respectively (Table 1). Deletion of tolC from the recA locus was also performed with a double-stranded PCR product and 50 bp homology arms. This long, linear DNA (1197 bp) was 100% efficient at deleting tolC from the recA locus (Table 1).
The tolC marker system was also used to create in-frame fusions to specific genes in E. coli. A promoter-less tolC cassette was designed to replace the open reading frame of a specific target gene (coined ORF-Xchange in this work) leaving the promoter and other regulatory sequences of that gene intact (Figure1B). TolC ORF-Xchange would provide a preliminary indication of essentiality and expression level of that particular target gene. Simultaneous testing of that same target gene for replacement using the promoter-containing tolC cassette would help confirm or deny essentiality. Final demonstration of essentiality would rest on successful and efficient ORF-Xchange only when a complementing copy of the target gene was provided. To demonstrate that TolC can function in this capacity, ORF-Xchange experiments were performed on recA (a constitutively expressed, non-essential gene), dicB (a tightly regulated gene whose essentiality is controversial) and ygeX (a minimally expressed, non-essential gene). The recA coding sequence was replaced by tolC ORF-Xchange at 100% efficiency (Table 1). ORF-Xchange was not successful for dicB or ygeX (no SDS-resistant recombinants found compared to recA), but subsequent deletion of both genes by the promoter-containing tolC cassette was 100% efficient (Table 1). The essential genes metG and murA could not be replaced by either tolC cassette unless a complementing copy of those genes was provided on a plasmid [data not shown; (39)].
Remodeling the rRNA operons of Escherichia coli using tolC
As further proof of tolC's utility as a selectable/counter-selectable marker for λ recombineering, the genes encoding ribosomal RNAs were targeted for deletion. Escherichia coli contains seven different rRNA operons that map to various locations on the genome (Figure S3) (45). Deleting entire rRNA operons is not novel, as there is published work that used classically elegant genetic techniques to delete six and even all seven rRNA operons from the E. coli genome (46,47). Due to technical limitations, however, the strains containing a single rRNA operon require complementation by a plasmid that provides essential tRNA genes removed from the genome during the original genetic manipulations (46,47). So, a more precise manipulation of the E. coli rRNA operons was performed using the tolC marker system and λredgam with the intention of maintaining the structural fidelity of those complex operons and eliminating the requirement for extra-chromosomal elements.
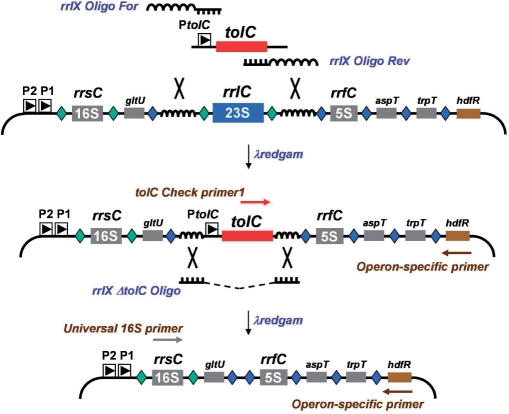
To that end, the gene encoding 23S rRNA was targeted exclusively for removal from all but one of the ribosomal RNA operons. Given the extraordinarily high degree of sequence homology among the seven rRNA operons, a universal tolC cassette (with promoter) was designed that would allow for deletion of any rrl gene regardless of its location on the E. coli genome. DNA oligonucleotides were synthesized for amplification of a tolC cassette that would contain 50-nucleotide tails homologous to regions just upstream and downstream of the rrl genes (see the rrlC example shown in Figure 2). The oligos were designed specifically so that tolC insertion would occur just after the last tRNA gene following 16S rDNA in all operons, deleting both RNAse III half-sites and the 23S rRNA gene itself. Following removal of the tolC cassette by a single-stranded DNA oligonucleotide, the proper arrangement of secondary RNase processing sites would be restored, permitting normal maturation of the neighboring 5S rRNA (rrf) and tRNA genes (Figure 2). Insertion and removal of the tolC cassette from specific rRNA operons was confirmed by PCR using a primer internal to tolC, a universal 16S rDNA primer common to all operons, and an operon-specific primer (Figure 2).
Figure 2.
Strategy for remodeling the Escherichia coli rRNA operons. A tolC cassette is shown amplified by rrlX oligos For and Rev (Rx-P50 and -P51, respectively) having universal homology arms that correspond to sequences flanking the 23S rRNA genes from all seven operons (Figure S3). In this example, removal of rrlC occurs following introduction of the tolC cassette and recombination. SDS-resistant colonies are screened by PCR using tolC Chk primer1 (Rx-P71) paired with an operon-specific primer (Px-P52 through P58). This screening can be multiplexed as primers were designed to amplify fragments of distinct sizes (Figure 3). Following confirmation of tolC insertion, the cassette is removed by introducing universal single-stranded oligos (RX-P59 or P60, depending on the direction of replication). After selecting for colicin E1 resistance, transformants are screened by plating on SDS and by PCR using a 16S rDNA primer with the operon specific primers mentioned before. Fragments of chromosomal DNA are depicted with bent lines. P2 and P1 refer to the promoters that drive expression of the rRNA operons and PtolC represents the P1 promoter for that gene. Neighboring genes are shown with shaded boxes: gltU, aspT and trpT encode tRNA; rrsC and rrfC encodes 16S and 5S rRNA, respectively. Green diamonds represent RNase III processing sites. Blue diamonds represent RNAse E processing sites for tRNA and 5S RNA. Primers used for PCR screening are shown with brown arrows. Curvy lines represent homologous DNA shared by the linear DNA substrate and the bacterial chromosome while large X's again denote λ red-mediated cross-over between regions of homologous DNA. Illustrations not drawn to scale.
The rrlX-tolC cassette was electroporated to strain RS526 (RS206 ΔrecA Δara ΔtolC) following induction of λredgam at 42°C. Replacement of 23S rRNA genes with tolC in each operon was assessed by PCR using the tolC check primer and the specific primer from each rRNA operon. All SDS resistant recombinants showed deletion of 23S rRNA genes with no overt position effects as judged by the fact that tolC inserted into each of the seven operons with approximately equal frequency (data not shown). When a tolC cassette was designed to insert and establish transcription convergent to the strong tandem rRNA promoters P1P2, no recombinants were detected due presumably to antisense suppression of tolC (data not shown). Colicin E1 counter-selection was performed in the presence of the single stranded rrlX-ΔtolC oligo to remove the tolC cassette from rrnC (Figure 2). A 100% of the colicin E1-resistant colonies (48 out of 48) were SDS sensitive and had tolC removed as detected by PCR using primers Rx-P54 and Rx-P61 (Tables 1 and S2). These results demonstrate that the tolC marker system allows for seamless modification of the rRNA operons with no polar effects.
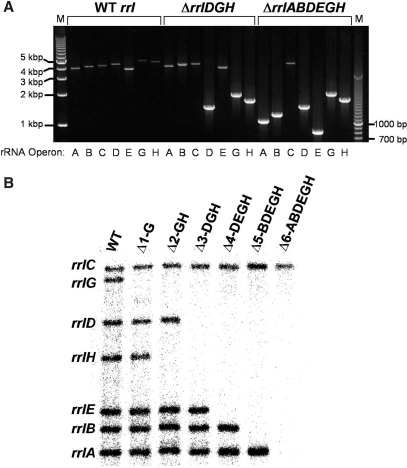
Starting with a tolC insert in rrlG, successive rounds of deletion/insertion were performed until only one rRNA operon contained the DNA encoding 23S rRNA (rrnC). Deletion of tolC using the single-stranded oligo strategy also demonstrated lagging strand bias specific to each rRNA operon (data not shown). PCR screening data from select strains is shown in Figure 3A. Using the 16S rDNA universal primer and primers specific for each operon, the wild-type strain gives large PCR products (4–5 kb) demonstrating the presence of the 23S rRNA gene in each operon. The tolC − strain deleted for rrlDGH (Figure 3A, middle section) shows smaller PCR products indicating the removal of the 23S RNA genes from those operons. The strain deleted for all but one 23S rRNA gene shows a WT PCR fragment only for rrlC. To confirm deletion of the appropriate 23S rRNA gene(s) from each strain, a Southern blot was performed using a probe directed against 23S rDNA (Figure 3B). The absence of the corresponding band in each of the respective strains verifies the deletion of the 23S rRNA genes from the E. coli genome. The restoration and proper alignment of RNase E processing sites was confirmed by DNA sequencing (data not shown).
Figure 3.
Confirmation of 23S rRNA deletions. (A) Agarose gel showing PCR products generated from gDNA of select E. coli strains. Strain RS526 (WT rrl) has all rRNA operons intact and yields large (∼4–5 kb) PCR products when the 16S and operon-specific primers are used (Figure 2). The ΔrrlDGH strain (RS547) shows smaller PCR fragments demonstrating the deletion of 23S rRNA genes from the respective operons. Lastly, strain RS676 (ΔrrlABDEGH) has only one copy of the gene for 23S rRNA located in the rrnC operon. Lanes 1 and 23: Marker DNA (1 kb ladder and 0.1 kb ladder, respectively). (B) Southern blot demonstrating the step-wise removal of 23S rRNA genes from six of the seven operons. The sizes of the DNA bands and corresponding operons follow: rrnC—16.9 kb; rrnG—15.5 kb; rrnE—11.2 kb; rrnH—9.6 kb; rrnD—8.1 kb; rrnB—7.2 kb and rrnA—6.6 kb. Strain designations: WT- RS526, Δ1- RS537, Δ2- RS544, Δ3- RS547, Δ4- RS550, Δ5- RS554 and Δ6-RS676.
Preliminary characterization of the new rrl mutant strains
During construction of these new E. coli rrl mutants, it became obvious that growth rate decreased in relation to the copy number of 23S rRNA genes present on the chromosome. Upon completion of the single-copy rrlC mutant (RS676), growth analysis was performed on the entire strain set according to the procedures outlined in Materials and Methods. The doubling time of each strain is reported in Table 2. RS526 (ΔtolC ΔrecA Δara) is the isogenic parent of all the rrl mutants (designated as wild-type-WT) and has a doubling time of 39 min. Deletion of rrlG and the double deletion ΔrrlGH increase generation time compared to WT by 21 and 47%, respectively. Interestingly, mutants that contain five, four, three and two functioning 23SrRNA gene copies all have doubling times between 57 and 60 min. Strain RS676 (single copy of a 23S rRNA structural gene in rrnC) has the longest generation time, 87 min.
Table 2.
Preliminary characterization of rrl mutants
| MIC (ng/ml) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Strain | rrl Genotype | tgen (min) | Amp | Nov | Rif | Cam | Ery | Kan | Tet |
| RS526 | WT | 39 | 2 | 0.5 | 4 | 1 | 2 | 4 | 0.5 |
| RS537 | Δ1-rrIG | 48 | 2 | 0.5 | 4 | 1 | 2 | 4 | 0.5 |
| RS544 | Δ2-rrlGH | 58 | 2 | 0.5 | 4 | 1 | 2 | 8 | 0.5 |
| RS547 | Δ3-rrlDGH | 59 | 2 | 0.5 | 4 | 1 | 2 | 8 | 1 |
| RS550 | Δ4-MDEGH | 57 | 2 | 0.5 | 4 | 1 | 4 | 4 | 1 |
| RS554 | Δ5-MBDEGH | 60 | 1 | 0.5 | 4 | 1 | 4 | 4 | 1 |
| RS676 | Δ6-rrlABEDGH | 87 | 1 | 0.5 | 4 | 0.5 | 4 | 4 | 0.5 |
Bacterial strain numbers are listed in column one and their genotypes with respect to 23S rRNA genes are shown in column two (please refer to Table S1 for complete genotype). Column three shows doubling time in LB media at 37°C (tgen in minutes). Minimum inhibitory concentration (µg/ml) of known antibacterial agents are shown for each strain tested in microtiter plates in LB broth at 35°C. Amp: ampicillin; Nov: novobiocin; Rif: rifampicin; Cam: chloramphenicol; Ery: erythromycin; Kan: kanamycin; Tet: tetracycline. Other agents tested but not shown: Linezolid, Puromycin, Azithromycin, Kasugamycin, Spectinomycin, Fusidic Acid, and Kirromycin.
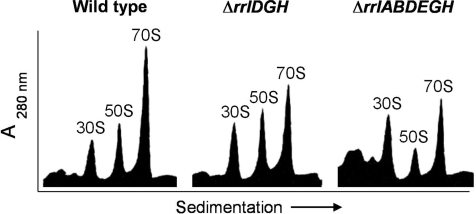
Given that RS676 has only one copy of the gene encoding 23S rRNA and seven intact copies of the 16S rRNA gene, it was tempting to reason that such a mutant would be unable to maintain the proper ratio of 30S/50S ribosomal subunits. In order to address this point, ribosomes were purified from RS526 (WT), RS547 (ΔrrlDGH) and RS676 (single-copy rrlC). Figure 4 shows the sedimentation profiles of ribosomes from these strains after analysis by sucrose density gradient centrifugation. The ribosome profile differs among the three strains, with the amount of 70S ribosomes clearly decreasing as the number of 23S rRNA genes drop. The Δ6 strain (containing only rrlC) has the least amount of free 50S ribosomal subunits and an excess of free 30S subunits compared to both WT and the Δ3 mutant.
Figure 4.
Ribosome sedimentation profiles from rrl mutants. Ribosomes were purified and analyzed by sucrose density gradient sedimentation according to Materials and Methods. The positions and size (S = Svedberg units) are shown: 70S = mature ribosomes, 50S = large ribosomal subunits, 30S = small ribosomal subunits. WT- strain RS526 (rrl+), ΔrrlDGH- strain RS547 and ΔrrlABDEGH- strain RS676.
To determine whether the differences in ribosome profiles might confer upon the rrl mutant strains an easily testable phenotype, their sensitivity to a number of well-characterized antibiotics was measured (Table 2). The minimum inhibitory concentration (MIC) of the cell wall-targeting antibiotic ampicillin did not change for any of the mutants tested. Similarly, there is no difference in the MIC values for rifampin (targets RNA polymerase and transcription) and novobiocin (targets DNA gyrase and replication). These results suggest there is little, if any, detectable effect of the rrl mutants on essential cellular processes not relating to protein synthesis. The rrl mutants were also tested with drugs and antibiotics that block protein synthesis through binding to either the 30S or 50S subunits of the ribosome. Table 2 shows that all rrl mutants are comparable to WT in their sensitivity to ribosome targeting compounds under the conditions tested.
DISCUSSION
Genome engineering has undergone remarkable advances due to the development of efficient systems for recombination in model organisms like E. coli (25,27,28,48,49). It is now routine to create deletions and point mutations in target genes (on the chromosome or on plasmids) using DNA oligonucleotides and PCR (26,32,50,51). This study describes the development of novel methodology that uses the gene coding for a single open reading frame, tolC, as both a selectable and counter-selectable marker for genome engineering studies. TolC selection obviates the need for minimal media, specialized wash protocols, extended incubation times and the traditional antibiotic selections required by other techniques.
Historically, selectable markers used in recombinant DNA work have been small genetic elements that express cytoplasmic enzymes or proteins conferring resistance to antibiotics (52). Use of tolC as a genetic marker is somewhat unorthodox in that the gene encodes an outer membrane protein that forms a transperiplasmic channel through which various efflux pumps extrude toxic molecules from the bacterial cell (10). TolC's properties, however, meet perfectly the requirements for use as a tool in genome engineering especially when coupled with the highly efficient λredgam recombination system. In an E. coli cell deleted for tolC, SDS and other small toxic molecules are unable to be pumped efficiently from the cell. Introduction of the tolC ORF confers relative resistance to these compounds but for some molecules (e.g. novobiocin) spontaneous resistance mutants may also arise (53). Consequently, SDS was used in this work as the reagent to select for tolC insertions as it causes cell lysis and the frequency of spontaneous SDS-resistant mutants is extremely low (<5 × 10−10) in a tolC null background [data not shown, (54)]. Indeed, the data in Table 1 show that SDS selection results in 100% recombination efficiency at any unique targeted site and gives no background at SDS concentrations as low as 0.002% (data not shown). The efficiencies reported in Table 1 only reflect proper placement of the tolC cassette at the targeted site. The high frequency of recombination seen with tolC is governed by the λredgam recombination system itself (28,32,55) and is not influenced by SDS selection. SDS selection is, however, economical and allows for facile incorporation into molten agar without the need for cooling to intermediate temperatures as is necessary with heat labile antibiotics. Furthermore, positive selection for tolC using SDS is performed on rich media (LB) commonly used in many laboratories. Therefore, recombinants often arise after only 16 h (overnight) incubation at 35°C. This incubation period is similar to other gene cassettes that select for chloramphenicol resistance or gentamycin resistance (30,36) but those markers are incapable of providing counter-selection. Finally, when creating mutants to study gene regulation or other aspects of E. coli physiology, the tolC strategy does not require antibiotics or specialized media that might interfere with facile experimental design or data interpretation (see subsequently).
The key feature of TolC as a tool for genetic engineering in E. coli is the ability to select for its absence from the cell. A number of counter-selectable markers have been developed to perform genetic manipulation with varying degrees of success (31,32,34,35,52,56,57). Unlike tolC, these counter-selectable markers must be used in tandem with a gene that allows for positive selection. Though widely applied, the sacB-neo fusion cassette involves antibiotic selection and sucrose toxicity (24) which can result in high background after negative selection. Alternatively, the rpsL gene has been coupled with neo to provide concerted antibiotic selection (32). In a bacterial strain containing a mutant rpsL gene on the chromosome, loss of a second copy of wild-type rpsL converts the cell from streptomycin-sensitive to streptomycin-resistant (57). Two recent efforts coupled the λredgam recombineering system with either galK or thyA selection in E. coli (37,38). These and other auxotrophic markers can be used quite readily in a positive selection mode, allowing recombinants having these markers integrated into the chromosome to grow on minimal media. Clever counter-selection methods were utilized to permit efficient removal of galK and thyA by incorporating 2-deoxygalactose and trimethoprim/thymine, respectively, into agar plates. Such methods, however, require extensive washing of cells to reduce background during positive selection (since minimal media is required) and recombinants appear only after 3-days growth (35,37,38).
Like the galK and thyA techniques, the tolC system also provides for efficient counter-selection. Through its surfaced exposed residues, the TolC protein allows import of colicin E1, a bactericidal protein that normally functions as part of an antimicrobial defense system (14). Therefore, cells containing tolC are lysed when exposed to colicin E1. This feature of the tolC counter-selection strategy helps to reduce background without the need for specialized wash protocols (35,37,38). Furthermore, the procedure can be performed in rich media, allowing recombinants to grow after only 16 h incubation. The short incubation time is advantageous as it permits PCR screening of colonies and phenotypic testing for both SDS sensitivity and colicin E1 resistance/tolerance all within one workday following electroporation.
The efficiency of colicin E1 counter-selection against the tolC cassette using single-stranded oligonucleotides is quite high, generally 80–100% for targets that map to various locations on the genome (Table 1 and data not shown). Removal of tolC from its normal location (using ss DNA) on the E. coli chromosome was, however, less efficient with only 25% of the E1-resistant recombinants demonstrating tolC deletion. The lower efficiency in this case could be due simply to sub-optimal recombineering conditions. Alternatively, genome position effects or the minimal length of the single stranded oligonucleotides used as a recombination substrate may have also contributed to the reduced efficiency. Increasing the efficiency of tolC deletion at this locus might be achieved by lengthening the ss-oligo to provide more than 35 bases of homologous sequence flanking the tolC target or by using double stranded DNA to increase homology further. Ellis et al. (55) showed that when using linear DNA in this way, recombination efficiency can improve with increased length, a fact confirmed by the removal of tolC from the recA locus at 100% efficiency using a double-stranded PCR product (Table 1). Such experiments were not performed on the tolC locus as additional manipulation was not required to demonstrate proof of principle.
An alternative to colicin E1 for counter-selection of tolC was tried but did not yield favorable results. TLS phage uses TolC as a receptor for attachment to E. coli and following infection and replication inside tolC+ strains, TLS kills the cell (17). Removal of tolC from the ara locus was attempted using TLS phage counter-selection on agar plates and in broth. Individual recombinants often demonstrated weak phage resistance (upon purification and phage cross-streak) as well as the ability to retain resistance to SDS (data not shown). As the colicin E1 counter-selection provided favorable results and is not affected by unforeseen changes in surface lipopolysaccharides, further optimization of TLS phage for selection was not pursued.
Colicin E1 selection is not without drawbacks as E1-resistant (or tolerant) mutants can arise through spontaneous mutation of btuB or tolC. The involvement of btuB mutants can be identified easily by screening colonies in the plate receptor test (23) and patching on SDS-containing media. Mutations in btuB would appear colicin-resistant, but SDS-resistant as well. In addition, point mutations in tolC that interfere directly with colicin E1 binding and/or translocation but remain efflux competent (15) would also be identified through this counter-screen. In fact, most of the infrequent E1-resistant recombinants shown not to have tolC deletions by PCR fell into this category (data not shown). Particular mutations in tolC (L3S, ΔL3, L412P, and C-terminal deletions) confer both novobiocin sensitivity and colicin E1 resistance (58–60), but none have been specifically reported for SDS and colicin E1. Nevertheless, such spontaneous mutations could interfere with efficient counter-selection using the method described in this work. Mutations at multiple loci (e.g. mutations in both tolC and an efflux pump) could also confound selection as such cells might be both colicin E1-resistant and SDS-sensitive. Given the extremely high recombination rates catalyzed by λ redgam (28), these potential false positives were not problematic and efficient selection against tolC was possible (Table 1).
Along the same lines, removal of the tolC cassette may occur through recombination at repetitive sequences flanking the intended deletion junctions. Such events occur irrespective of the counter-selectable marker used and the tolC strategy cannot overcome this inherent drawback of the system. Recombinants that have lost tolC expression through recombination at flanking repeats would also be colicin E1-resistant and would appear as false positives during selection. PCR testing with primers that bracket the proper deletion junction (as shown in Figure 2) would exclude these false positives. Thus, Table 1 reports efficiencies that reflect only proper removal of tolC from the targeted site. Interestingly, the highly homologous ribosomal operons of E. coli represent a situation where deletion through flanking repeats may occur (see subsequently).
As mentioned previously, the tolC marker system does not require specialized media or small molecule antibiotics and can be used economically in laboratories familiar with the growth and manipulation of E. coli strains in the course of normal molecular biology protocols. A potential confounding factor of the technique is that antibiotic resistant markers are not needed. While this can be advantageous when studying physiological processes that may be altered by the presence of antibiotics, lack of antibiotics does permit the growth of contaminants in the absence of proper sterile technique. Should contamination be problematic, a straightforward conversion of the ΔtolC parent strain to streptomycin resistance by creating a point mutation in rpsL using λ redgam may offer a solution.
The strategy of colicin selection may have broader application to other model organisms where a dearth of genetic tools has hampered progress toward understanding molecular processes. For instance, some archaebacteria synthesize and respond to peptide toxins called halocins (19). The receptors and molecular targets of these toxins have not been fully characterized, but they may provide a similar set of tools for genome engineering in the archaea. In fact, an outer membrane protein with weak sequence homology but functional similarity to E. coli tolC has been identified in Chromohalobacter marismortui (61) Given that archaea are known to take up and recombine linear DNA (62,63), a gene disruption strategy similar to that described in this work may have potential.
As shown in the Results section, the tolC marker system can also be used to assess the essentiality and expression levels of various target genes through ORF-Xchange. A similar strategy was recently described in detail using antibiotic resistant markers (64). For a constitutively expressed, non-essential gene like recA, ORF-Xchange with tolC occurred at high frequency and was 100% efficient (Table 1). The product of the dicB gene stimulates MinC function and interferes with normal cell division (65,66). DicB synthesis is tightly regulated (67), and is one of the least abundant mRNAs/proteins found in the E. coli cell (∼ 60-fold less than recA) (68). Its essentiality has been questioned, however, as some groups have reported conflicting results using various experimental techniques (69,70). Table 1 shows that the promoter-containing tolC cassette is able to delete the dicB gene with 100% efficiency, confirming its non-essentiality. In addition, dicB's low expression level was confirmed by unsuccessful ORF-Xchange under standard SDS selection. Similar results were obtained for ygeX, a gene of unknown function whose expression levels are just slightly below that of dicB (68). In agreement with previous results (39), deletion of the known essential genes murA and metG could not be achieved with high frequency by either tolC cassette unless a complementing copy of either gene was provided. These data demonstrate that the tolC marker system can be used successfully to screen for gene essentiality and expression in the same manner as described by Bubunenko et al. (64).
Perhaps the best demonstration of the utility and reproducibility of tolC selection/counter-selection is the seamless modifications made in six of the seven 23S rRNA genes on the E. coli chromosome (Figure 3). These genes occupy various positions on the genomic map and reside in complex operons interspersed between other essential structural rRNA and tRNA genes (Figure S3) (1,71). The intergenic sequences of the rRNA operons contain RNAse cleavage sites necessary for the correct processing and maturation of the individual cistrons (Figures 2 and S3) (72). In addition, there is a high degree of sequence homology among the RNA coding sequences and among the intergenic regions (73). Such features make directed genetic modification of the individual rRNA genes on the chromosome a challenging task. Previously published work was relegated to creating large deletions in the rRNA operons (46,74).
As this work describes, the tolC marker system provides the tools necessary to precisely remodel the rRNA operons in E. coli. The keys to this advancement are the use of bacteriophage λ's redgam recombination system combined with tolC's selection properties: namely the ability to use rich media without traditional antibiotic markers. Successful deletion of the rrlC gene alone (Figure 2, Table 1) demonstrates that tolC modification of the ribosomal operons can be achieved without conferring polar effects. RS1443 (ΔrrlC strain) has a growth phenotype indistinguishable from RS537 (ΔrrnG), with no observable defect in protein synthesis, as maximal λ redgam induction occurs within 15 min as seen with the parent strain (data not shown). Since the viability of RS1443 depends upon proper expression and processing of the essential tryptophan tRNA (trpT) gene located downstream, these results establish that trpT expression is not disturbed by the genetic manipulations performed and that the experimental design shown in Figure 2 is sound.
Using this strategy, deletion of six 23S rRNA genes was straightforward (Figure 3). Mutations in any rRNA operon were maintained by preventing homogenotization through the use of the recA deleted, RS526 strain background. The decision to leave rrnC as the only intact operon was based on its proximity to the origin of DNA replication, with the intention that gene dosage effects might provide a more favorable growth rate to a single-copy mutant (74,75). In fact, the ability to perform these genetic manipulations in rich media was crucial as the growth rate of cells with decreased 23S rRNA gene copies was significantly reduced (Table 2). Interestingly, mutants with two to five copies of 23S rRNA genes had very similar growth rates (2.2% SD between strains) but were collectively ∼34% slower than their isogenic parent.
In addition to the effect on growth rate, deleting rrl genes from the chromosome of E. coli has a pronounced effect on protein synthesis. This was observed indirectly through assessment of the time required for sufficient λredgam induction to yield recombinants at non-essential loci. For example, where a 15 min induction at 42°C yielded thousands of recombinants in the WT strain, a minimum of 60 min was required for strains having only one or two copies of the 23S rRNA gene (data not shown). The Δ5 strain (RS554) required 2.5× more time for induction than the Δ3 strain (RS547) while the single-copy strain (RS676), despite its even slower growth rate, was comparable to RS554 for redgam induction (data not shown). Given the unique construction of these mutants, decreases in 23S rRNA gene copy number may not yield predictable changes in growth rate and ribosome efficiency. Although the increase in doubling time is similar to that seen with other rRNA deletion strains reported in the literature (46,75), a direct comparison is difficult as the strains constructed here are not deleted for entire rRNA operons nor are plasmids used to provide essential functions to the cell. Therefore, a more thorough investigation of the physiological properties of these new mutants is required.
As a first step towards understanding the molecular basis of the mutant phenotypes, the ribosome profiles from select mutant strains were determined (Figure 4). Reducing the number of 23S rRNA genes affected the ribosome profiles of the mutants constructed in this work. The Δ3 mutant (having four copies of 23S rRNA genes), shows an increase in free 30S and 50S ribosomal subunits compared to WT but a reduced amount of 70S ribosomes. The effect is more pronounced in the single-copy rrlC strain RS676 where 70S ribosomes and free 50S subunits levels are further reduced compared to WT, but an excess of 30S subunits is present. This imbalance in ribosome content could account for the observations concerning growth rate and protein synthesis discussed in the previous paragraph. A similar imbalance has been detected in ribosomes from normal cells grown at 16°C (76), from cells over-expressing incomplete rRNA genes from plasmids (77), and from cdsA mutants when grown at low temperatures (78). The cdsA mutants, however, lead to accumulation of a 40S particle not yet seen with the mutants described in this work. In the studies where a 3′ segment of 23S rRNA was expressed from a plasmid, it was determined that the material sedimenting at 30S was misfolded 23S rRNA bound to an incomplete complement of ribosomal proteins (78).
The unbalanced ribosome profile of RS676 prompted a test of whether this strain (or any of the other mutants) showed altered sensitivity towards antibiotics that bind to the E. coli ribosome given the potential for excess free 30S ribosomal subunits to act as a sink for inhibitor binding. Coupled with reduced 50S and 70S ribosomes levels, this could make RS676 relatively resistant to antibiotics that target the 30S ribosomal subunit. Upon testing all mutant strains against a number of different drugs and antibiotics (Table 2), no clear difference was detected under the conditions used. Interestingly, antibiotics that target normal cellular processes not related to protein synthesis (e.g. DNA replication, transcription, cell wall biosynthesis) also showed no change in potency compared to wild-type. These data suggest that the rRNA mutants constructed do not suffer gross abnormalities in essential cellular pathways.
The control of ribosome synthesis occurs at many levels and it has been the focus of many comprehensive studies [reviewed in (79,80)]. Ribosomal RNA synthesis is regulated at both the initiation and termination steps of transcription. The P1P2 promoters (shared by all rRNA operons) are subject to the action of a transcriptional activator (Fis), the alarmone ppGpp and the concentration of initiating nucleotide triphosphates (81). To prevent premature termination of the untranslated rRNA transcripts, antitermination mechanisms (similar to those used by phage λ), are also important (82). These and other potential control mechanisms work together to ensure that rRNA synthesis is proportional to the steady-state growth rate of the E. coli cell (growth-rate-dependent control) (83).
The synthesis of ribosomal proteins is regulated by negative feedback inhibition of translation by free, unassembled r-proteins (84). Ribosomal protein operons are multi-cistronic and contain the genes encoding both small and large r-proteins. In most cases, one r-protein within an operon functions as an autogenous translational repressor, binding to a site on the message and blocking synthesis of all proteins in the operon (84). Coupled with the growth-rate-dependent control of rRNA production, these mechanisms help balance the synthesis of r-proteins with rRNA and ensure the cooperative assembly of ribosomes in E. coli. Given the imbalance of ribosomal subunits noted for the single copy 23S rRNA mutant RS676 (Figure 4), it is tempting to speculate that remodeling of the rRNA operons in the manner described here has lead to a short-circuiting of the regulatory pathways that control ribosome synthesis. Further investigation of this and other rRNA mutants constructed with the tolC marker system will guide additional studies on the regulation of ribosome synthesis in E. coli.
Finally, the TolC selection method reported in this work can be broadly applied to genetic manipulation of any DNA substrate capable of amplification within E. coli. The construction of new plasmid vectors, point mutations, deletions and insertions is possible using the tolC marker system and λredgam. The type and specific nature of the modifications are not limited by TolC selection. The method is highly reproducible and may serve as sufficient replacement for other current selection systems.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
I am grateful to Anjana Agarwal, Dan Stoughton and Maria Grazia Cappiello for help in performing the first experiments with tolC. I thank Don Court for strains and Rajeev Misra for phage. Special thanks to Eugene Skripkin and Francois Franceschi for their expert advice on ribosome preparation, to Paul Danese for critical reading of the manuscript, and to the rest of my colleagues at Rib-X Pharmaceuticals for their support and encouragement. Funding to pay the Open Access publication charges for this article was provided by Rib-X Pharmaceuticals Inc.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bachmann BJ. Linkage map of Escherichia coli K-12, edition 8. Microbiol. Rev. 1990;54:130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Che D, Li G, Mao F, Wu H, Xu Y. Detecting uber-operons in prokaryotic genomes. Nucleic Acids Res. 2006;34:2418–2427. doi: 10.1093/nar/gkl294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salgado H, Gama-Castro S, Peralta-Gil M, Diaz-Peredo E, Sanchez-Solano F, Santos-Zavaleta A, Martinez-Flores I, Jimenez-Jacinto V, Bonavides-Martinez C, et al. RegulonDB (version 5.0): Escherichia coli K-12 transcriptional regulatory network, operon organization, and growth conditions. Nucleic Acids Res. 2006;34:D394–397. doi: 10.1093/nar/gkj156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aono R, Tsukagoshi N, Yamamoto M. Involvement of outer membrane protein TolC, a possible member of the mar-sox regulon, in maintenance and improvement of organic solvent tolerance of Escherichia coli K-12. J. Bacteriol. 1998;180:938–944. doi: 10.1128/jb.180.4.938-944.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masuda N, Church GM. Escherichia coli gene expression responsive to levels of the response regulator EvgA. J. Bacteriol. 2002;184:6225–6234. doi: 10.1128/JB.184.22.6225-6234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eguchi Y, Oshima T, Mori H, Aono R, Yamamoto K, Ishihama A, Utsumi R. Transcriptional regulation of drug efflux genes by EvgAS, a two-component system in Escherichia coli. Microbiology. 2003;149:2819–2828. doi: 10.1099/mic.0.26460-0. [DOI] [PubMed] [Google Scholar]
- 7.Hackett J, Reeves P. Primary structure of the tolC gene that codes for an outer membrane protein of Escherichia coli K12. Nucleic Acids Res. 1983;11:6487–6495. doi: 10.1093/nar/11.18.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hackett J, Misra R, Reeves P. The TolC protein of Escherichia coli K12 is synthesised in a precursor form. FEBS Lett. 1983;156:307–310. doi: 10.1016/0014-5793(83)80518-3. [DOI] [PubMed] [Google Scholar]
- 9.Werner J, Augustus AM, Misra R. Assembly of TolC, a structurally unique and multifunctional outer membrane protein of Escherichia coli K-12. J. Bacteriol. 2003;185:6540–6547. doi: 10.1128/JB.185.22.6540-6547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koronakis V, Eswaran J, Hughes C. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu. Rev. Biochem. 2004;73:467–489. doi: 10.1146/annurev.biochem.73.011303.074104. [DOI] [PubMed] [Google Scholar]
- 11.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 12.Wandersman C, Delepelaire P. TolC, an Escherichia coli outer membrane protein required for hemolysin secretion. Proc. Natl Acad. Sci. USA. 1990;87:4776–4780. doi: 10.1073/pnas.87.12.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vakharia H, German GJ, Misra R. Isolation and characterization of Escherichia coli tolC mutants defective in secreting enzymatically active alpha-hemolysin. J. Bacteriol. 2001;183:6908–6916. doi: 10.1128/JB.183.23.6908-6916.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao Z, Klebba PE. Mechanisms of colicin binding and transport through outer membrane porins. Biochimie. 2002;84:399–412. doi: 10.1016/s0300-9084(02)01455-4. [DOI] [PubMed] [Google Scholar]
- 15.Masi M, Vuong P, Humbard M, Malone K, Misra R. Initial steps of colicin E1 import across the outer membrane of Escherichia coli. J. Bacteriol. 2007;189:2667–2676. doi: 10.1128/JB.01448-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fralick JA. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 1996;178:5803–5805. doi: 10.1128/jb.178.19.5803-5805.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.German GJ, Misra R. The TolC protein of Escherichia coli serves as a cell-surface receptor for the newly characterized TLS bacteriophage. J. Mol. Biol. 2001;308:579–585. doi: 10.1006/jmbi.2001.4578. [DOI] [PubMed] [Google Scholar]
- 18.Nagel de Zwaig R, Luria SE. Genetics and physiology of colicin-tolerant mutants of Escherichia coli. J. Bacteriol. 1967;94:1112–1123. doi: 10.1128/jb.94.4.1112-1123.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riley MA, Wertz JE. Bacteriocins: evolution, ecology, and application. Annu. Rev. Microbiol. 2002;56:117–137. doi: 10.1146/annurev.micro.56.012302.161024. [DOI] [PubMed] [Google Scholar]
- 20.Pugsley AP. The ins and outs of colicins. Part I: production, and translocation across membranes. Microbiol. Sci. 1984;1:168–175. [PubMed] [Google Scholar]
- 21.Di Masi DR, White JC, Schnaitman CA, Bradbeer C. Transport of vitamin B12 in Escherichia coli: common receptor sites for vitamin B12 and the E colicins on the outer membrane of the cell envelope. J. Bacteriol. 1973;115:506–513. doi: 10.1128/jb.115.2.506-513.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cramer WA, Dankert JR, Uratani Y. The membrane channel-forming bacteriocidal protein, colicin El. Biochim. Biophys. Acta. 1983;737:173–193. doi: 10.1016/0304-4157(83)90016-3. [DOI] [PubMed] [Google Scholar]
- 23.Davies JK, Reeves P. Genetics of resistance to colicins in Escherichia coli K-12: cross-resistance among colicins of group A. J. Bacteriol. 1975;123:102–117. doi: 10.1128/jb.123.1.102-117.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 25.Murphy KC. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu D, Court DL. A new system to place single copies of genes, sites and lacZ fusions on the Escherichia coli chromosome. Gene. 1998;223:77–81. doi: 10.1016/s0378-1119(98)00163-2. [DOI] [PubMed] [Google Scholar]
- 27.Muyrers JP, Zhang Y, Testa G, Stewart AF. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costantino N, Court DL. Enhanced levels of lambda Red-mediated recombinants in mismatch repair mutants. Proc. Natl Acad. Sci. USA. 2003;100:15748–15753. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawitzke JA, Thomason LC, Costantino N, Bubunenko M, Datta S, Court DL. Recombineering: in vivo genetic engineering in E. coli, S. enterica, and beyond. Methods Enzymol. 2007;421:171–199. doi: 10.1016/S0076-6879(06)21015-2. [DOI] [PubMed] [Google Scholar]
- 31.Fabret C, Ehrlich SD, Noirot P. A new mutation delivery system for genome-scale approaches in Bacillus subtilis. Mol. Microbiol. 2002;46:25–36. doi: 10.1046/j.1365-2958.2002.03140.x. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Muyrers JP, Rientjes J, Stewart AF. Phage annealing proteins promote oligonucleotide-directed mutagenesis in Escherichia coli and mouse ES cells. BMC Mol. Biol. 2003;4:1. doi: 10.1186/1471-2199-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li MZ, Elledge SJ. MAGIC, an in vivo genetic method for the rapid construction of recombinant DNA molecules. Nat. Genet. 2005;37:311–319. doi: 10.1038/ng1505. [DOI] [PubMed] [Google Scholar]
- 34.Brans A, Filee P, Chevigne A, Claessens A, Joris B. New integrative method to generate Bacillus subtilis recombinant strains free of selection markers. Appl. Environ. Microbiol. 2004;70:7241–7250. doi: 10.1128/AEM.70.12.7241-7250.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura GS, Bratt DS, Yim HH, Nittayajarn A. Use of glnQ as a counterselectable marker for creation of allelic exchange mutations in group B streptococci. Appl. Environ. Microbiol. 2005;71:587–590. doi: 10.1128/AEM.71.1.587-590.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poteete AR, Rosadini C, St Pierre C. Gentamicin and other cassettes for chromosomal gene replacement in Escherichia coli. Biotechniques. 2006;41:261–262, 264. doi: 10.2144/000112242. [DOI] [PubMed] [Google Scholar]
- 37.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong QN, Ng VC, Lin MC, Kung HF, Chan D, Huang JD. Efficient and seamless DNA recombineering using a thymidylate synthase A selection system in Escherichia coli. Nucleic Acids Res. 2005;33:e59. doi: 10.1093/nar/gni059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeVito JA, Mills JA, Liu VG, Agarwal A, Sizemore CF, Yao Z, Stoughton DM, Cappiello MG, Barbosa MD, et al. An array of target-specific screening strains for antibacterial discovery. Nat. Biotechnol. 2002;20:478–483. doi: 10.1038/nbt0502-478. [DOI] [PubMed] [Google Scholar]
- 40.Miller JH. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 41.Sambrook J, Russel DW, Sambrook J. Molecular Cloning: A Laboratory Manual. 3rd. Plainview, NY: Cold Spring Harbor Laboratories; 2001. [Google Scholar]
- 42.Condon C, Philips J, Fu ZY, Squires C, Squires CL. Comparison of the expression of the seven ribosomal RNA operons in Escherichia coli. Embo. J. 1992;11:4175–4185. doi: 10.1002/j.1460-2075.1992.tb05511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clinical Laboratory Standards Institute. Wayne, PA: Clinical and Laboratory Standards Institute; 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. CLSI publication number M7-A7. [Google Scholar]
- 44.Bommer UA, Burkhardt N, Junemann R, Spann CMT, Triana-Alonso FJ, Nierhaus KH. Subcellular Fractionation: A Practical Approach. Oxford: Oxford University Press; 1996. [Google Scholar]
- 45.Kiss A, Sain B, Venetianer P. The number of rRNA genes in Escherichia coli. FEBS Lett. 1977;79:77–79. doi: 10.1016/0014-5793(77)80354-2. [DOI] [PubMed] [Google Scholar]
- 46.Asai T, Condon C, Voulgaris J, Zaporojets D, Shen B, Al-Omar M, Squires C, Squires CL. Construction and initial characterization of Escherichia coli strains with few or no intact chromosomal rRNA operons. J. Bacteriol. 1999;181:3803–3809. doi: 10.1128/jb.181.12.3803-3809.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Asai T, Zaporojets D, Squires C, Squires CL. An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc. Natl Acad. Sci. USA. 1999;96:1971–1976. doi: 10.1073/pnas.96.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muyrers JP, Zhang Y, Stewart AF. Techniques: Recombinogenic engineering–new options for cloning and manipulating DNA. Trends Biochem. Sci. 2001;26:325–331. doi: 10.1016/s0968-0004(00)01757-6. [DOI] [PubMed] [Google Scholar]
- 49.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murphy KC, Campellone KG, Poteete AR. PCR-mediated gene replacement in Escherichia coli. Gene. 2000;246:321–330. doi: 10.1016/s0378-1119(00)00071-8. [DOI] [PubMed] [Google Scholar]
- 51.Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 52.Reyrat JM, Pelicic V, Gicquel B, Rappuoli R. Counterselectable markers: untapped tools for bacterial genetics and pathogenesis. Infect. Immun. 1998;66:4011–4017. doi: 10.1128/iai.66.9.4011-4017.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jovanovic M, Lilic M, Janjusevic R, Jovanovic G, Savic DJ. tRNA synthetase mutants of Escherichia coli K-12 are resistant to the gyrase inhibitor novobiocin. J. Bacteriol. 1999;181:2979–2983. doi: 10.1128/jb.181.9.2979-2983.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whitney EN. The tolC locus in Escherichia coli K12. Genetics. 1971;67:39–53. doi: 10.1093/genetics/67.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ellis HM, Yu D, DiTizio T, Court DL. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc. Natl Acad. Sci. USA. 2001;98:6742–6746. doi: 10.1073/pnas.121164898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tischer BK, von Einem J, Kaufer B, Osterrieder N. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques. 2006;40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 57.Sander P, Meier A, Bottger EC. rpsL+: a dominant selectable marker for gene replacement in mycobacteria. Mol. Microbiol. 1995;16:991–1000. doi: 10.1111/j.1365-2958.1995.tb02324.x. [DOI] [PubMed] [Google Scholar]
- 58.Yamanaka H, Izawa H, Okamoto K. Carboxy-terminal region involved in activity of Escherichia coli TolC. J. Bacteriol. 2001;183:6961–6964. doi: 10.1128/JB.183.23.6961-6964.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamanaka H, Nomura T, Morisada N, Shinoda S, Okamoto K. Site-directed mutagenesis studies of the amino acid residue at position 412 of Escherichia coli TolC which is required for the activity. Microb. Pathog. 2002;33:81–89. doi: 10.1006/mpat.2002.0519. [DOI] [PubMed] [Google Scholar]
- 60.Yamanaka H, Morisada N, Miyano M, Tsuge H, Shinoda S, Takahashi E, Okamoto K. Amino-acid residues involved in the expression of the activity of Escherichia coli TolC. Microbiol. Immunol. 2004;48:713–722. doi: 10.1111/j.1348-0421.2004.tb03593.x. [DOI] [PubMed] [Google Scholar]
- 61.Tokunaga H, Mitsuo K, Ichinose S, Omori A, Ventosa A, Nakae T, Tokunaga M. Salt-inducible multidrug efflux pump protein in the moderately halophilic bacterium Chromohalobacter sp. Appl. Environ. Microbiol. 2004;70:4424–4431. doi: 10.1128/AEM.70.8.4424-4431.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sato T, Fukui T, Atomi H, Imanaka T. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 2003;185:210–220. doi: 10.1128/JB.185.1.210-220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cline SW, Schalkwyk LC, Doolittle WF. Transformation of the archaebacterium Halobacterium volcanii with genomic DNA. J. Bacteriol. 1989;171:4987–4991. doi: 10.1128/jb.171.9.4987-4991.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bubunenko M, Baker T, Court DL. Essentiality of ribosomal and transcription antitermination proteins analyzed by systematic gene replacement in Escherichia coli. J. Bacteriol. 2007;189:2844–2853. doi: 10.1128/JB.01713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson JE, Lackner LL, de Boer PA. Targeting of (D)MinC/MinD and (D)MinC/DicB complexes to septal rings in Escherichia coli suggests a multistep mechanism for MinC-mediated destruction of nascent FtsZ rings. J. Bacteriol. 2002;184:2951–2962. doi: 10.1128/JB.184.11.2951-2962.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnson JE, Lackner LL, Hale CA, de Boer PA. ZipA is required for targeting of DMinC/DicB, but not DMinC/MinD, complexes to septal ring assemblies in Escherichia coli. J. Bacteriol. 2004;186:2418–2429. doi: 10.1128/JB.186.8.2418-2429.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bejar S, Bouche F, Bouche JP. Cell division inhibition gene dicB is regulated by a locus similar to lambdoid bacteriophage immunity loci. Mol. Gen. Genet. 1988;212:11–19. doi: 10.1007/BF00322439. [DOI] [PubMed] [Google Scholar]
- 68.Wei Y, Lee JM, Richmond C, Blattner FR, Rafalski JA, LaRossa RA. High-density microarray-mediated gene expression profiling of Escherichia coli. J. Bacteriol. 2001;183:545–556. doi: 10.1128/JB.183.2.545-556.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kang Y, Durfee T, Glasner JD, Qiu Y, Frisch D, Winterberg KM, Blattner FR. Systematic mutagenesis of the Escherichia coli genome. J. Bacteriol. 2004;186:4921–4930. doi: 10.1128/JB.186.15.4921-4930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gerdes SY, Scholle MD, Campbell JW, Balazsi G, Ravasz E, Daugherty MD, Somera AL, Kyrpides NC, Anderson I, et al. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 2003;185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Komine Y, Adachi T, Inokuchi H, Ozeki H. Genomic organization and physical mapping of the transfer RNA genes in Escherichia coli K12. J. Mol. Biol. 1990;212:579–598. doi: 10.1016/0022-2836(90)90224-A. [DOI] [PubMed] [Google Scholar]
- 72.King TC, Sirdeskmukh R, Schlessinger D. Nucleolytic processing of ribonucleic acid transcripts in procaryotes. Microbiol. Rev. 1986;50:428–451. doi: 10.1128/mr.50.4.428-451.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Condon C, Squires C, Squires CL. Control of rRNA transcription in Escherichia coli. Microbiol. Rev. 1995;59:623–645. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Condon C, French S, Squires C, Squires CL. Depletion of functional ribosomal RNA operons in Escherichia coli causes increased expression of the remaining intact copies. EMBO J. 1993;12:4305–4315. doi: 10.1002/j.1460-2075.1993.tb06115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Condon C, Liveris D, Squires C, Schwartz I, Squires CL. rRNA operon multiplicity in Escherichia coli and the physiological implications of rrn inactivation. J. Bacteriol. 1995;177:4152–4156. doi: 10.1128/jb.177.14.4152-4156.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jiang M, Sullivan SM, Walker AK, Strahler JR, Andrews PC, Maddock JR. Identification of novel Escherichia coli ribosome-associated proteins using isobaric tags and multidimensional protein identification techniques. J. Bacteriol. 2007;189:3434–3444. doi: 10.1128/JB.00090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Siehnel RJ, Morgan EA. Unbalanced rRNA gene dosage and its effects on rRNA and ribosomal-protein synthesis. J. Bacteriol. 1985;163:476–486. doi: 10.1128/jb.163.2.476-486.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Charollais J, Dreyfus M, Iost I. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 2004;32:2751–2759. doi: 10.1093/nar/gkh603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nomura M. Regulation of ribosome biosynthesis in Escherichia coli and Saccharomyces cerevisiae: diversity and common principles. J. Bacteriol. 1999;181:6857–6864. doi: 10.1128/jb.181.22.6857-6864.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Keener J, Nomura M. In: Escherichia Coli and Salmonella Typhimurium: Cellular and Molecular Biology. 2nd. Neidhardt FC, editor. Vol. 1. Washington DC: American Society for Microbiology; 1996. pp. 1417–1431. [Google Scholar]
- 81.Schneider DA, Ross W, Gourse RL. Control of rRNA expression in Escherichia coli. Curr. Opin. Microbiol. 2003;6:151–156. doi: 10.1016/s1369-5274(03)00038-9. [DOI] [PubMed] [Google Scholar]
- 82.Squires CL, Greenblatt J, Li J, Condon C, Squires CL. Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc. Natl Acad. Sci. USA. 1993;90:970–974. doi: 10.1073/pnas.90.3.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gourse RL, Gaal T, Bartlett MS, Appleman JA, Ross W. rRNA transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Annu. Rev. Microbiol. 1996;50:645–677. doi: 10.1146/annurev.micro.50.1.645. [DOI] [PubMed] [Google Scholar]
- 84.Zengel JM, Lindahl L. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog. Nucleic Acid Res. Mol. Biol. 1994;47:331–370. doi: 10.1016/s0079-6603(08)60256-1. [DOI] [PubMed] [Google Scholar]