Abstract
Several group I introns have been previously found in strains of the Bacillus cereus group at three different insertion sites in the nrdE gene of the essential nrdIEF operon coding for ribonucleotide reductase. Here, we identify an uncharacterized group IA intron in the nrdF gene in 12 strains of the B. cereus group and show that the pre-mRNA is efficiently spliced. The Bacillus thuringiensis ssp. pakistani nrdF intron encodes a homing endonuclease, denoted I-BthII, with an unconventional GIY-(X)8-YIG motif that cleaves an intronless nrdF gene 7 nt upstream of the intron insertion site, producing 2-nt 3′ extensions. We also found four additional occurrences of two of the previously reported group I introns in the nrdE gene of 25 sequenced B. thuringiensis and one B. cereus strains, and one non-annotated group I intron at a fourth nrdE insertion site in the B. thuringiensis ssp. Al Hakam sequenced genome. Two strains contain introns in both the nrdE and the nrdF genes. Phylogenetic studies of the nrdIEF operon from 39 strains of the B. cereus group suggest several events of horizontal gene transfer for two of the introns found in this operon.
INTRODUCTION
Self-splicing group I introns are RNA sequences that in a splicing event merge the two flanking sequences they interrupt into a contiguous RNA sequence (1). Bacterial group I introns are mainly found in tRNA genes (1–3). The first group I introns to be discovered in protein-coding genes were found in bacterial dsDNA viruses (4–6). Although group I introns rarely interrupt protein-coding genes in bacteria, several different group I introns have been recently found in DNA metabolizing genes among the Firmicutes family (7), in the nrdE gene coding for the essential class Ib ribonucleotide reductase which is required in the de novo production of deoxyribonucleotides for DNA synthesis (8–10) and in recA which is involved in recombination of DNA (11–14). The ribonucleotide reductase family consists of three major classes (I, II and III) with a common evolutionary relationship (9). The Bacillus cereus group strains (as most Bacillaceae) encode the subclass Ib and class III ribonucleotide reductases. The class Ib operon contains the nrdE and nrdF genes encoding the ribonucleotide reductase proper, which are preceded by the nrdI gene encoding a flavodoxin-like protein of unknown function (Figure 6A). The Ib ribonucleotide reductase is responsible for maintaining aerobic growth in these bacteria, whereas the class III ribonucleotide reductase is restricted to anaerobic growth.
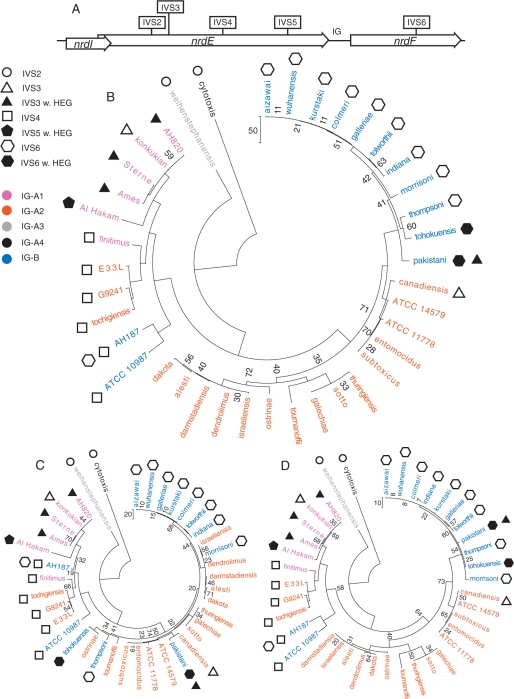
Figure 6.
The nrdEF genes in the B. cereus group. (A) Gene map showing the positioning of the genes and IG-sequence within the operon and the IVS insertion points in the operon. Phylogenetic trees based on the sequence of the (B) nrdIEF operon, (C) nrdE-coding sequences and (D) nrdF-coding sequences from 39 strains of the B. cereus group. Symbols represent the presence of IVS in the strain where filled and empty symbols represent a group I intron with and without HEG, respectively. Colours represent the IG-sequence groups each strain belongs to. The distance bars represent the number of nucleotide changes in the sequence alignments. Only bootstrap values below 75% are shown.
Not only do these nrdE genes contain group I introns but some of the introns are also found encoding homing endonuclease genes (HEGs). Homing endonucleases (HEases) cleave non-HEG-containing DNA close to their intronic insertion sites, leaving a single-strand break (nick) or a double-strand break (DSB) (15). The nick or DSB leads to recombination repair between the allele carrying the intron and the cleaved allele lacking the HEG. Via the recombination repair, the HEG is copied together with the intron and parts of the flanking genomic regions, to the non-HEG allele, and HEGs are therefore described as selfish genetic elements.
HEGs have been found encoded within intervening sequences (IVS) such as introns and inteins and at intergenic (IG) regions (15). Intron-encoded HEases almost exclusively cleave alleles lacking the intron, as the intron insertion with a few exceptions interrupts the recognition site of the HEase, preventing cleavage of the intron-containing allele. HEases typically promote non-Mendelian inheritance of the HEG-containing allele.
To make a horizontal gene transfer (HGT) possible the alleles must meet in a system where recombination events can take place. The most studied mechanism of HGT in bacteria is the transfer via phage infection, and IVSs have been found within prophage sequences in the genome of several members of the Bacillaceae family (16–18). The B. cereus group is a known host to IVSs of which some are not associated with any known phage or prophage sequence (7,8). This leaves the question whether they are transferred by other mechanisms. A different mechanism that has been discussed but not yet tested is the efficient uptake of environmental DNA involving the natural competence of bacteria, a mechanism that is common among the Firmicutes (13,14).
HEases are divided into several protein families based on conserved sequence motifs. These include the LAGLIDADG, HNH, His-box, GIY-YIG families and a single example of a PD-(D/E)-XK motif family (15,19,20). The HEases also contain DNA-binding motif(s) spanning 14–36 bp of the specific target site (8,15). This means that cleavage by HEase is rare, usually occurring only once per genome. In the B. cereus group, at least three different group I introns have been found at three different locations in the nrdE gene. At least one of these introns harbours a member of the GIY-YIG HEase family (8).
In this article, we report the identification of a group I intron in the nrdF gene found in several strains of closely related Bacillus thuringiensis. We show that the intron is spliced and encodes a fully functional HEase (denoted I-BthII) with a GIY-(X)8-YIG motif differing from all hitherto characterized GIY-YIG HEases having 9–11 amino acid residues separating the GIY-YIG triplets. We map the cleavage site and the recognition site of I-BthII, report the presence of several group I introns in the nrdE gene from 26 strains of the B. cereus group and also present phylogenetic indication for HGT of the group I introns in the nrdEF genes among these bacteria.
MATERIALS AND METHODS
Bacterial strains and general methods
All strains of B. thuringiensis ssp. aizawai HD-131, alesti AUCT 10114, canadiensis HD-224, colmeri HD-847, dakota HD-932, darmstadiensis HD-146, dendrolimus HD-7, entomocidus HD-9, finitimus AUCT 10117, galechiae AUCT 2402, galleriae HD-8, indiana HD-521, kurstaki HD-1, morrisoni HD-12, ostrinae AUCT 10309, pakistani HD-395, sotto HD-770, subtoxicus HD-109, thompsoni NRRL 4060, thuringiensis AUCT 2507, tochigiensis HD-868, tohokuensis HD-866, tolworthii HD-537, toumanoffii HD-201, wuhanensis HD-525 and B. cereus ssp. ATCC 11778 sequenced in this study were obtained as kind gifts from Håkan Steiner at the Department of Genetics, Microbiology and Toxicology (GMT) at Stockholm University, Sweden. All strains were grown in Brain Heart Infusion medium (Becton Dickinson) at 30°C. Genomic DNA was extracted using the Genomic DNA extraction kit (Qiagen) according to manufacturer's recommendations. Target and template PCR products were amplified from genomic DNA using the High Fidelity DNA polymerase (Fermentas) according to manufacturer's recommendations and purified using QIAquick PCR purification kit (Qiagen). For sequencing, 600 fmol of PCR product were mixed with 10 pmol of respective sequencing primer, dried and sent to MWG-Biotech AG. Several primers were used for sequencing the nrdIEF operons (3500–6000 bases depending on appearance of IVSs). Primers used were complementary to sequences flanking the nrdIEF operon or to the operon itself. A full list of primers used in this article can be found in a Supplementary table.
In vivo splicing analysis
RNA isolation from B. thuringiensis ssp. colmeri and ssp. pakistani cells harvested at A640 = 1.0 was carried out using the RNeasy kit for total RNA isolation (Qiagen) according to the manufacturer's recommendations, including the on-column DNase I treatment. RT-PCR reactions were carried out using the ThermoScript RT-PCR system (Invitrogen) according to the manufacturer's recommendations. The RNA concentration used in the RT step was 76 µg/ml. After the RT step, presence of spliced and unspliced products was analysed by PCR using standard conditions with Platinum Taq DNA polymerase (Invitrogen) according to manufacturer's recommendations. Products were separated on agarose gel and visualized with ethidium bromide and UV light. The PCR product of the spliced RNA was sequenced with primers nrdF forw seq and nrdF rev2 seq.
Prediction of intron and I-BthII secondary structure
Prediction of the intron secondary structure was performed using Mfold default settings (http://frontend.bioinfo.rpi.edu/applications/mfold/cgi-bin/rna-form1-2.3.cgi) and corrected by hand according to the conventional folding suggested by Cech et al. (21). Prediction of I-BthII secondary structure was performed using Expresso (3DCoffee) (22,23) using I-TevI structure 1MK0A (24). Three-dimensional modelling was performed using the Swiss Model Workspace Automatic Modelling Mode (25) (http://swissmodel.expasy.org/workspace/) and the I-TevI 1MK0A structure (24) as template when the default settings could not find a fitting structure.
In vitro translation of endonuclease constructs
Template for in vitro translation was amplified from genomic DNA from B. thuringiensis ssp. pakistani and was used for in vitro translation using the kit TNT® T7 Quick for PCR DNA (Promega) according to manufacturer's recommendations. Radio labelled [35S]-Met was included in the reaction and the product was separated on a denaturing 15% polyacrylamide gel and analysed by PhosphoImager (Fujifilm FLA-3000).
In vitro endonuclease activity assays
Targets for HEase cleavage were amplified using B. thuringiensis ssp. pakistani spliced product from RT-PCR analysis and B. thuringiensis ssp. pakistani genomic DNA as templates. Cleavage reactions were performed using in vitro translated HEases directly in the reaction according to the method described previously (26). Reactions contained 1.8 pmol target DNA, 3 μl in vitro translation reaction and 0.1 μg RNaseA in 50 µl of 10 mM Tris–HCl (pH 8.5), 10 mM MgCl2, 100 mM KCl and 0.1 mg/ml BSA and were incubated at 37°C for 45 min. Incubations with mock in vitro translation without template DNA served as negative controls. All cleavage products were separated on agarose gels and stained with ethidiumbromide and visualized with UV light.
Cleavage site mapping
For mapping of the DSB one [32P] radiolabelled and one unlabelled primer were used in the target PCR for labelling of each strand separately. Primers [32P]-nrdF forw seq and nrdF rev2 seq were used for identifying the cut on the coding strand. Primers [32P]-nrdF rev2 seq and nrdF forw seq were used for identifying the cut on the template strand. The targets were cleaved with in vitro translated I-BthII. Cleavage products were purified and separated on 10% polyacrylamide gel with 7 M Urea, together with sequencing ladders produced with the fmol® DNA Cycle Sequencing System (Promega) according to the method described previously (26). The sequencing ladders were labelled using [32P]-labelled primers nrdF forw seq and nrdF rev2 seq. Template sequence for both cleaving and sequencing were IVS-less nrdF variant from B. thuringiensis ssp. pakistani RT-PCR splicing analysis. The gel was visualized and analysed by PhosphoImager (Fujifilm FLA-3000).
Targets used for fine mapping of the recognition site were amplified from IVS-less nrdF from B. thuringiensis ssp. pakistani RT-PCR splicing analysis using one fluorescein-labelled primer and unlabelled primers successively shortening the target sequence. The targets for the downstream part of the site were amplified using primers nrdF forw seq (F) and Ltd bind +22 to +25. The targets for the upstream part of the site were amplified using primers nrdF rev2 seq (F) and Ltd bind −3 to −6. Here, (F) denotes fluorescein labelling. Cleavage reactions were performed using in vitro translated HEases directly in the reaction according to the method described previously (26). Reactions contained 1 pmol target DNA, 2 μl in vitro translation reaction and 0.1 μg RNaseA in 20 µl of 10 mM Tris–HCl (pH 8.5), 10 mM MgCl2, 100 mM KCl and 0.1 mg/ml BSA and were incubated at 37°C for 45 min. Cleavage products were separated on a 10% polyacrylamide gel with 7 M Urea, and analysed by excitation of fluorescein-labelled targets and products at 473 nm and filtered at 520 nm (FujiFilm FLA-3000). Note that only one cleavage product was visualized, as only one strand was fluorescein labelled.
Sequence alignments and phylogenetic analysis
Sequences were aligned with Dialign (27). Phylogenetic analyses were performed with MEGA ver. 3.1 (28), using the Maximum Parsimony tool, and running the default setting of 1000 bootstraps. Recombination analyses were performed using the Recombination Analysis Tool (29) with minimum cut-off settings of 50–90% and default maximum cut-off, window and increment size settings.
Sequence data were obtained from the GenBank™ genome data [accession numbers NC_003997 for B. anthracis Ames, NC_005945 for B. anthracis Sterne, NZ_AAUF01000007 for B. cereus AH187, NZ_AAUE01000001 for B. cereus AH820, NC_003909 for B. cereus ATCC 10987, NC_004722 for B. cereus ATCC 14579, NZ_AALL01000009 for B. cereus cytotoxis, NC_006274 for B. cereus E33L, NZ_AAEK01000009 for B. cereus G9241, NC_008600 for B. thuringiensis Al Hakam (YP_894072 for the potential HEG), NZ_AAJM01000029 for B. thuringiensis israeliensis, NC_005957 for B. thuringiensis serovar konkukian str. 97–27 and NZ_AAOY01000003 for B. weihenstephanensis KBAB4].
Strains sequenced in this study have accession numbers: EU043128 for B. thuringiensis ssp. aizawai, EU043129 for B. thuringiensis ssp. canadiensis, EU043130 for B. thuringiensis ssp. finitimus, EU043131 for B. thuringiensis ssp. pakistani, EU043132 for B. thuringiensis ssp. tochigiensis, EU043133 for B. thuringiensis ssp. entomocidus, EU043134 for B. thuringiensis ssp. galleriae, EU043135 for B. thuringiensis ssp. kurstaki, EU043136 for B. thuringiensis ssp. thuringiensis, EU043137 for B. thuringiensis ssp. tolworthii, EU043138 for B. thuringiensis ssp. alesti, EU043139 for B. thuringiensis ssp. colmeri, EU043140 for B. thuringiensis ssp. dakota, EU043141 for B. thuringiensis ssp. darmstadiensis, EU043142 for B. thuringiensis ssp. dendrolimus, EU043143 for B. thuringiensis ssp. galechiae, EU043144 for B. thuringiensis ssp. indiana, EU043145 for B. thuringiensis ssp. morrisoni, EU043146 for B. thuringiensis ssp. ostrinae, EU043147 for B. thuringiensis ssp. sotto, EU043148 for B. thuringiensis ssp. subtoxicus, EU043149 for B. thuringiensis ssp. thompsoni, EU043150 for B. thuringiensis ssp. tohokuensis, EU043151 for B. thuringiensis ssp. toumanoffii, EU043152 for B. thuringiensis ssp. wuhanensis, EU043153 for B. cereus ATCC_11778.
RESULTS
A functional group IA intron in the nrdF gene of several members of the B. cereus group
Several IVSs have previously been found in the nrdE gene of the B. cereus group (7,8) but so far no IVSs have been reported in the nrdF gene in this group. By searching through sequenced genomes in GenBank, we found that the sequence of B. cereus ssp. AH187 contained a non-annotated group I intron with a remnant of a HEG (see below) in the non-annotated nrdF gene, and we named the group I intron IVS6 for intervening sequence at insertion site 6 in the nrdIEF operon.
In search for a functional HEG in the IVS6 intron in the nrdF gene in the B. cereus group, we sequenced 25 B. thuringiensis subspecies and one B. cereus subspecies. The IVS6 was found in 11 strains of which ssp. pakistani and tohokuensis contained a full-length HEG (Table 1). The HEases were found to differ at the 187th amino acid residue with an arginine in the ssp. pakistani HEase and a threonine in the ssp. tohokuensis HEase. The remnant HEG in ssp. AH187 would only give a HEase of 37 amino acids, where residues 1–3 aligns with the N-terminal part of ssp. pakistani and ssp. tohokuensis HEases and residues 5–37 aligns with their C-terminal part. The intronic sequences differed somewhat between the IVS6-containing strains with most differences in bulges or loops of the paired regions of the introns (Supplementary Figure 1). Most introns only differed by one to three bases (excluding the lack of full-length HEG in all but ssp. pakistani and ssp. tohokuensis) but the AH187 intronic sequence differed most from all other IVS6 with 14 nt difference to the ssp. pakistani intronic sequence (Supplementary Figure 1).
Table 1.
Occurrences of IVSs, HEGs and intergenic sequences in the nrdJEF operon in the B. cereus group
| Strains | ssp. | nrdE IVS | 1G1 | nrdF IVS | References |
|---|---|---|---|---|---|
| B. an ssp. | Ames | 1VS3 + (I-BanI)2 | Al | – | 4 |
| Sterne | 1VS3 + 1-Banl2 | Al | – | 5 | |
| B. ce ssp. | AH187 | 1VS4 | B | 1VS6 | 6, 7 |
| AH820 | lVS3+ (I-BanI)2 | Al | – | 8 | |
| ATCC 10987 | 1VS4 | B | – | 9 | |
| ATCC 11778 | – | A2 | – | 6 | |
| ATCC 14579 | – | A2 | – | 10 | |
| cytotoxis | 1VS2 | A4 | – | 11 | |
| E33L | 1VS4 | A2 | – | 12 | |
| G9241 | 1VS4 | A2 | – | 13 | |
| B. th ssp. | aizawai | – | B | IVS6 | 6 |
| alesti | – | A2 | – | 6 | |
| Al Hakam | IVS5 + HEG2 | Al | – | 6,14 | |
| canadensis | IVS3 | A2 | – | 6 | |
| colmeri | – | B | 1VS6 | 6 | |
| dakota | – | A2 | – | 6 | |
| darmstadiensis | – | A2 | – | 6 | |
| dendrolimus | – | A2 | – | 6 | |
| entomocidus | – | A2 | – | 6 | |
| finitimus | 1VS4 | Al | – | 6 | |
| galechiae | – | A2 | – | 6 | |
| galleriae | – | B | IVS6 | 6 | |
| indiana | – | B | IVS6 | 6 | |
| israeliensis3 | – | A2 | – | 15 | |
| konkukian | IVS3 | Al | – | 16 | |
| kurstaki | – | B | IVS6 | 6 | |
| morrisoni | – | B | IVS6 | 6 | |
| ostrinae | – | A2 | – | 6 | |
| pakistani | 1VS3 + (1-BanI)2 | B | 1VS6 +1-Bthll2 | 6 | |
| sotto | – | A2 | – | 6 | |
| subtoxicus | – | A2 | – | 6 | |
| thompsoni | – | B | 1VS6 | 6 | |
| thuringiensis | – | A2 | – | 6 | |
| tochigiensis | 1VS4 | A2 | – | 6 | |
| tohokuensis | – | B | 1VS6 +(1-BthII)2 | 6 | |
| tolworthii | – | B | 1VS6 | 6 | |
| toumanoffii | – | A2 | – | 6 | |
| wuhanensis | – | B | 1VS6 | 6 | |
| B. wei ssp. | KBAB4 | 1VS2 | A3 | – | 17 |
1Sequence lengths of IG-groups: A1 93 bp, A2: 81 bp, A3: 94 bp, A4: 152 bp, B: 154 bp.
2ORF found in respective intron, I-BanI and I-BthII indicate HEGs that have been tested for and show activity (Nord,D., Torrents,E. and Sjöberg,B.-M. A functional homing endonuclease in the Bacillus anthracis nrdE group I intron. J. Bacteriol. (2007) 189, 5293–5301, and this study), parenthesis indicate over 99% identity with I-BanI and I-BthII, respectively and HEG denotes no identified relative or activity tested.
3Sequence of nrdIEF1 operon, nrdE2 does not cluster close to B. cereus. group nrdE genes (Nord,D., Torrents,E. and Sjöberg,B.-M. A functional homing endonuclease in the Bacillus anthracis nrdE group I intron. J. Bacteriol. (2007) 189, 5293–5301).
4Acc no.: NC_003997 5Acc no.: NC_005945 6This study. 7Acc no.: NZ_AAUF01000007 8Acc no.: NZ_AAUE01000001 9Acc no.: NC_003909 10Acc no.: NC_004722 11Acc no.: NZ_AALL01000009 12Acc no.: NC_006274 13Acc no.: NZ_AAEK01000009 14Acc no.: NC_008600 15Acc no.: NZ_AAJM01000029 16Acc no.: NC_005957 17Acc no.: NZ_AAOY01000003.
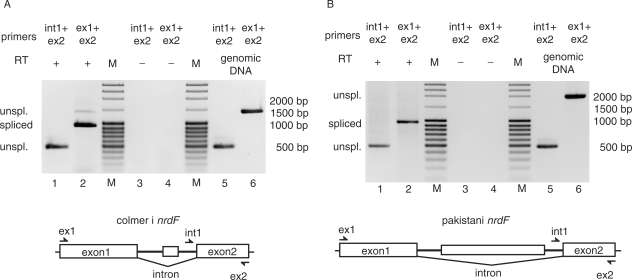
Figure 1.
Splicing of the group I self-splicing intron in B. thuringiensis ssp. colmeri and ssp. pakistani nrdF. (A) Agarose gel showing RT-PCR products of spliced and unspliced nrdF mRNA in total RNA extraction from B. thuringiensis ssp. colmeri. Intron and exon2-specific primers, int1 and ex2, respectively, in RT-PCR gave a product of 477 bp corresponding to unspliced nrdF mRNA (lane 1). Exon-specific primers ex1 and ex2 gave an RT-PCR product of 969 bp corresponding to spliced nrdF mRNA and also showing a weak band of unspliced mRNA of 1407 bp (lane 2). Primer pairs were used in control RT-PCR without reverse transcriptase (RT) and showed no DNA contamination in the RNA extraction (lanes 3 and 4). Primer pairs were also used in PCRs with genomic DNA as controls and showed products of 477 and 1407 bp, respectively (lanes 5 and 6). A molecular weight marker (M) was run for reference where bands 3–5 and 10 from the top correspond to 2000, 1500, 1000 and 500 bp, respectively. (B) Agarose gel showing RT-PCR products of spliced and unspliced nrdF mRNA in total RNA extraction from B. thuringiensis ssp. pakistani. Intron and exon2-specific primers, int1 and ex2 respectively, in RT-PCR gave a product of 477 bp corresponding to unspliced nrdF mRNA (lane 1). Exon-specific primers ex1 and ex2 gave an RT-PCR product of 969 bp corresponding to spliced nrdF mRNA (lane 2). Primer pairs were used in control RT-PCR without reverse transcriptase (RT) and showed no DNA contamination in the RNA extraction (lanes 3 and 4). Primer pairs were also used in PCRs with genomic DNA as controls and showed products of 477 and 2067 bp, respectively (lanes 5 and 6). The molecular weight marker (M) was as in (A).
The introns of the B. thuringiensis ssp. pakistani and ssp. colmeri nrdF genes, with and without the full-length HEG, respectively, were tested for in vivo splicing activity by non-quantitative RT-PCR (Figure 1A and B). RNA isolated from cells in the late log-phase contained both spliced and unspliced nrdF RNA species. Exon1 and exon2 specific primers gave a product of 969 bp corresponding to the size of the spliced product and a weak band of 1407 bp corresponding to the length of unspliced mRNA in the ssp. colmeri RNA but no detectable unspliced product in the ssp. pakistani RNA (2067 bp theoretical length) (Figure 1A and B, respectively, lane 2). Using intron and exon2 specific primers, it was possible to detect a product of 477 bp corresponding to unspliced mRNA from both strains (Figure 1A and B, respectively, lane 1). As controls, these primer pairs were used in PCR with genomic DNA, giving products of 477 bp (int1 and ex2 primers) and 1407 and 2067 bp for ssp. colmeri and ssp. pakistani, respectively (ex1 and ex2 primers) corresponding to the length of unspliced mRNA (Figure 1A and B, lanes 5 and 6). The primer pairs were also used in a RT-PCR reaction without RT giving no observed product, indicating that genomic contamination did not occur (Figure 1A and B, lanes 3 and 4). Sequencing of the spliced products confirmed that the splice sites were as predicted (see sequence in Figure 5A). These results show that the group I introns of B. thuringiensis ssp. colmeri and ssp. pakistani nrdF pre-mRNA are efficiently spliced in vivo.
Figure 5.
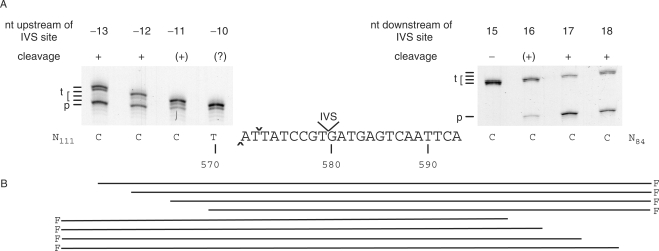
Mapping of minimum sequence required for I-BthII cleavage. (A) Polyacrylamide gels showing products from cleavage assays with fluorescein-labelled IVS-less targets with short flanking sequences either upstream of the IVS site (–13 to –10 bp, left gel) or downstream of the IVS site (+15 to +18 bp, right gel). Target (t) and product (p) is indicated for respective gel. Several target lengths are indicated since (t) is successively shortened by one to several base pairs for each target tested, as specified in (B). Cleavage is indicated with + for efficient cleavage, (+) for low or nearly no cleavage, (?) where no cleavage could be determined and (–) for no detected cleavage. Note that only one cleavage product was visualized, as only one strand was fluorescein labelled. The target and flanking sequences are indicated below the gels and the end positions are indicated below each corresponding lane. DSB is indicated with cap and inverted cap symbol for the cut on the template and coding strands, respectively. Numbers below sequence represent bases in the nrdF ORF. F denotes fluorescein label.
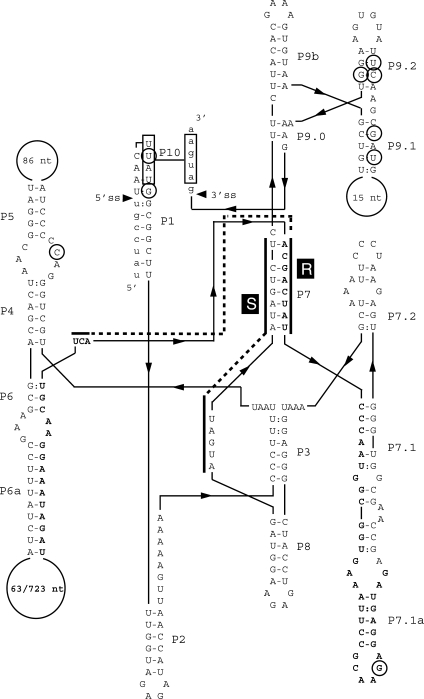
Prediction of the secondary structure of the ssp. pakistani intron sequence showed that it contains all conserved regions of pairing (P1–P10) and the conserved sequence elements R and S and shares characteristics with the subgroup IA introns (30). The ORF is predicted to start in the loop of the P6a region and spanning over the P7 region with the conserved R element (Figure 2). The ssp. colmeri intron differs from the ssp. pakistani intron by a deletion of a major part of the HEG and by one base in the loop in P5 (see Figure 2 and Supplementary Figure 1). The introns showed low general similarity to the predicted secondary structure of the group I intron in the nrdE gene of B. anthracis (8) with 18 identities in 23 nt over the conserved R and S sequence elements.
Figure 2.
Secondary structure of the group I self-splicing intron in B. thuringiensis ssp. pakistani and ssp. colmeri nrdF. Predicted secondary structure of the nrdF intron. Small letters indicate coding sequence of the nrdF gene. Capital letters indicate intronic sequence. Bold capital letters indicate the ORF in the intron with the numbers in the loop in P6.1 representing the sequence of B. thuringiensis ssp. colmeri and ssp. pakistani, respectively. Conserved sequence elements (R and S), conserved base-paired regions (P1 to P9) and additional pairings (P6a, P7.1, P7.2, P9a, P9.1 and P9.2) are shown. Alignment between the 5′ and 3′ splice site can be promoted by the boxed nucleotides, UUAU, in the P1 loop and the AUGA, in exon 2, making pairing P10. Circled letters indicate differences from the other IVS6-containing strains.
The HEG in the B. thuringiensis ssp. pakistani nrdF intron is an unconventional GIY-YIG endonuclease
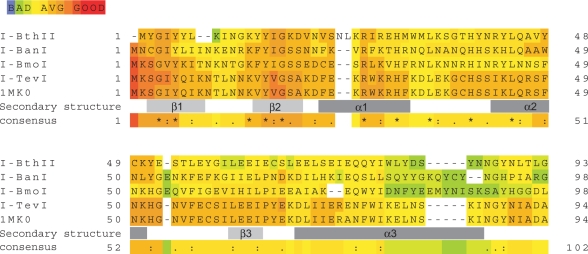
The nrdF intron in ssp. pakistani contains an ORF encoding a 257 amino acid residues HEase. We named this ORF I-BthII, according to the suggested nomenclature for HEases (31). I-BthII was not identified as a member of any of the four known HEase families using the protein blast tool at NCBI (32) with default parameters but with further analysis and secondary structure prediction of the I-BthII sequence a GIY-(X)8-YIG sequence motif was found. The motif in I-BthII thus differs from the hitherto reported GIY-(X)9-11-YIG motifs (33). Prediction of the secondary structure of I-BthII showed a similar structure to the I-TevI 3D structure (24,34) with the GIY and the YIG triplets in the sequence motif included in two β-strands followed by an α-helix (Figure 3) as typical for the GIY-YIG HEase family catalytic domain (32,33). I-BthII also displays an insert of two amino acid residues N-terminal to the first lysine residue and the conserved arginine residue in helix α1. Using the I-TevI 3D structure (24) as a template modelling of I-BthII showed a similar structure for the α-helices and the third β-sheet but the GIY-YIG triplet motif was not modelled into the structure. Only by expanding the GIY-YIG motif by two amino acid residues (GIY-YLK)-NT-(INGKY-YIG) thereby mimicking the I-TevI (GIY-QIK)-NT-(LNNKV-YVG) motif could the structure of the expanded I-BthII be modelled in full with the GIY and YIG sequences predicted in β-strands.
Figure 3.
The B. thuringiensis ssp. pakistani nrdF intron encodes a GIY-YIG-like homing endonuclease. Alignments made with Expresso (3DCoffee) showing I-BthII GIY-YIG domain aligned with I-BanI, I-BmoI and I-TevI. Light and dark grey boxes below the aligned sequences show β-strands and α-helices of I-TevI catalytic domain based on the 3D structure of the N-terminal domain (24).
In vitro translation of the ssp. pakistani I-BthII gave rise to ∼30 kDa product (data not shown) which is in agreement with the predicted molecular mass for this ORF. The product was tested for DSB cleaving activity on target sequences from ssp. pakistani nrdF. No cleavage activity was detected when the nrdF sequence containing the intron sequence was incubated with I-BthII in vitro translation or with mock translation (no DNA template in the in vitro translation reaction), (Figure 4A, lanes 4 and 6). The IVS-less target showed cleavage products when incubated with I-BthII, indicating that the target site was created by deletion of the intron (Figure 4A, lane 5). No cleavage product was detected when IVS-less target was incubated with mock translation (Figure 4A, lane 3).
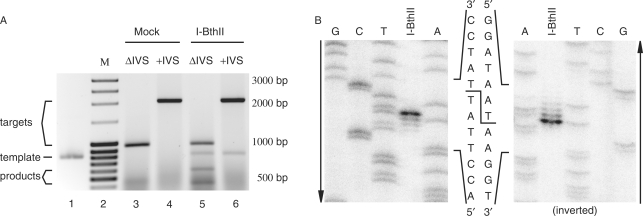
Figure 4.
Mapping of the cleavage site of the I-BthII homing endonuclease. (A) Agarose gel showing products from cleavage assay, ssp. pakistani IVS-less nrdF target (969 bp, lanes 3 and 5), ssp. pakistani wild-type nrdF target (2067 bp, lanes 4 and 6), incubated with mock translation and I-BthII. No cleavage products were found for targets incubated with mock translation (lanes 3 and 4). Cleavage products of 572 and 424 bp were found for IVS-less nrdF target incubated with I-BthII (lane 5). No cleavage product was found for pakistani wild-type nrdF target incubated with I-BthII (lane 6). Template for I-BthII in vitro translation was run for reference in lane 1 (815 bp). A molecular weight marker (M, lane 2) was run for reference where bands 3–5 and 10 from the top correspond to 2000, 1500, 1000 and 500 bp, respectively. (B) Polyacrylamide gel showing sequencing reactions run alongside [32P] single-strand labelled cleavage reactions to identify I-BthII cleavage site and overhang. The second gel is inverted to show the cleavage sites on both strands more clearly. Arrows indicate the direction of the gels.
Mapping of the I-BthII cleavage site
Based on fragment size from cleavage assays on IVS-less target (Figure 4A) a DSB was estimated to occur close to the intron insertion site in the ssp. pakistani DNA. In separate experiments, the DSB was identified by isotopic labelling of one or the other strand of the IVS-less target incubated with I-BthII. The cleavage products were run alongside sequencing reactions using the same primers as for target construction. Figure 4B shows that the DSB occurs 7 and 9 nt upstream of the intron insertion site with 2-nt 3′ extensions, i.e. 572 and 570 nt from the start of the ssp. pakistani nrdF on the coding and template strand, respectively.
To map the approximate size of the recognition site, targets were amplified from the IVS-less (spliced) ssp. pakistani RT-PCR product using primers successively shortening either the upstream or the downstream sequences flanking the cleavage site as described in Materials and Methods section. Figure 5 shows that when the target sequence was limited to 12 and 13 bp upstream (and 102 bp downstream) of the IVS site, it was readily cleaved by I-BthII. Targets limited to 11 bp upstream of the IVS site showed less cleavage efficiency. Due to appearance of double bands of target DNA in the experiment shown in Figure 5A, we could not exclude cleavage for the target limited to only 10 bp upstream of the IVS site. Figure 5 also shows that limiting the target sequence to 15 bp downstream (and 124 bp upstream) of the IVS site abolished cleavage of I-BthII. A very small amount of cleavage product was detected with 16 bp downstream of the IVS site, whereas a target sequence limited to 17 and 18 bp downstream of the IVS site showed full activity of I-BthII cleavage. These experiments suggest that the I-BthII recognition site is 27–29 bp with the DSB cleavage site at the 5′-end of the top strand (i.e. the recognition site covers 4–5 bp upstream and 23–24 bp downstream of the cut on the coding strand), and with the IVS insertion site approximately in the middle of the recognition site.
The B. cereus group nrdIEF operons contain IVSs at five different locations
Encouraged by the finding of several IVS6 introns in the sequenced B. cereus group of strains of which two contained a full-length HEG, we continued to screen and sequence the entire nrdIEF operon in the 26 strains to investigate the total presence of IVSs in the operon. We have earlier identified several different insertion sites (IVS2–IVS5) in the nrdE gene among the Firmicutes and of these, all sites were found to harbour group I introns (8) (Figure 6A and Table 1). In the current study, the ssp. canadiensis and ssp. pakistani nrdE genes were found to contain the IVS3 (Table 1). The ssp. canadiensis IVS3 had a similar deletion of the HEG as found in B. thuringensis ssp. konkukian differing by 2 nt, while ssp. pakistani harboured a full-length HEG differing by 10 nt from the I-BanI HEG in ssp. Ames, Sterne and AH820. Except for the differences in the HEGs, all the IVS3 introns were found to be identical with the exception of ssp. konkukian differing by 1 nt in the bulge in the predicted P5.1 (8,35). An IVS4 group I intron was found in ssp. finitimus and tochigiensis both without a HEG. A comparison of all IVS4 introns showed that the ssp. tochigiensis intron was identical to ssp. E33l and G9241 introns and to the non-annotated AH187 intron in nrdE. The ssp. finitimus intron differed in sequence from these four introns by 1 nt and the ATCC 10987 intron is 2 nt longer than the ssp. finitimus intron. The B. thuringiensis ssp. Al Hakam was shown to contain a non-annotated group I intron with a HEG at the IVS5 position in the nrdE gene. The B. thuringiensis ssp. alesti, dakota, darmstadiensis, dendrolimus, entomocidus, galechiae, ostrinae, sotto, subtoxicus, thuringiensis, toumanoffi and B. cereus ssp. ATCC 11778 did not show any presence of IVS in the nrdIEF operon. Of all strains in this study, only the ssp. AH187 and ssp. pakistani were shown to harbour two IVSs, one in nrdE and one in nrdF. Interestingly, sequence analysis of the operon also revealed a distinct difference in the IG sequences between nrdE and nrdF, dividing the strains in two IG groups based on sequence, IG-A and IG-B. The IG-A group was further divided into four subgroups based on the length of the IG sequence, IG-A1 (93 bp), IG-A2 (81 bp), IG-A3 (94 bp) and IG-A4 (152 bp) (Table 1 and Supplementary Figure 2). The sequences within the IG-groups differ very little, as do the coding sequence of the operons.
Phylogenetic studies of the nrdIEF operon showed a clustering of the different subspecies of the B. cereus group into groups harbouring different IVSs and also some general correlation with the IG-groups (Figure 6B). The IVS2-containing B. weihenstephanensis and B. cereus ssp. cytotoxis differ the most from the other species and both are single members of IG-A subgroups, IG-A3 and IG-A4, respectively. The introns at IVS2 are similar, but the B. weiheistephanensis intron is shorter (158 nt) than the ssp. cytotoxic intron. The IVS4-containing strains ssp. finitimus (IG-A1) and ssp. E33L, G9241, tochigiensis (all with IG-A2) cluster together with the two IVS4-containing strains, ssp. ATCC 10987 and AH187, both belonging to IG-B. These two IG-B strains (of which ssp. AH187 also contains IVS6) were found separate from all other IG-B strains of which all were IVS6-containing (ssp. aizawai, colmeri, galleriae, indiana, kurstaki, morrisoni, pakistani, thompsoni, tohokuensis, tolworthii and wuhanensis). The IVS3-containing ssp. Ames, Sterne, konkukian and AH820 (all with IG-A1) and the IVS5-containing ssp. Al Hakam (IG-A1) all clustered closely together, but apart from the IVS3-containing strains ssp. canadiensis (IG-A2) and ssp. pakistani (IG-B). Instead, the ssp. canadiensis clusters with the majority of the IG-A2 strains without IVSs and ssp. pakistani clusters with the majority of the IG-B strains.
Phylogenetic studies of an operon tree where the IG-regions had been manually deleted gave the same clustering of strains as the full operon tree. Also, phylogenetic analyses of the nrdE and nrdF sequences separately (Figure 6C and D, respectively) showed in general the same distribution of strains as for the operon sequences with an exception for the main cluster of IVS6-containing strains. In the nrdE tree ssp. pakistani containing both IVS6 and IVS3 clusters with IVS3-containing ssp. canadiensis, and also ssp. AH187, tohokuensis, thompsoni and morrisoni showed a scattered distribution to the rest of the IVS6-containing strains. In the nrdF tree, the IVS6-containing strains and IG-groups cluster as in the operon tree indicating that the nrdF and IG-B sequences are related to the distribution of IVS6 (Figure 6 and Table 1).
DISCUSSION
The many occurrences of different self-splicing group I introns with and without HEGs in the nrdIEF operon in several strains of the B. cereus group indicate a high frequency of HGT in this group of gram-positive bacteria. In this study, we identify a new group I intron in the nrdF gene in 11 B. thuringiensis strains and one B. cereus strain. We show that it is spliced in vivo and that two of the introns encode a HEase, named I-BthII, with a GIY-YIG-like motif. The motif in I-BthII has only eight amino acid residues between the GIY-YIG triplets compared to the previously characterized HEases belonging to the GIY-YIG family that have 9–11 residues separating the GIY-YIG triplets (33). From the results in Figure 5, we suggest that the recognition site of I-BthII covers 4–5 bp upstream and 23–24 bp downstream of the cut on the coding strand. This correlates with what is expected of a HEase of the GIY-YIG family with the produced DSB close to one end of the recognition site. Alignments of I-TevI, I-BmoI and I-BanI and comparison to the 3D structure of I-TevI show that I-BthII is a member of the GIY-YIG family. The GIY and YIG motifs are modelled in β-strands, and the subsequent α-helix seems shorter, but would still allow for the conserved and functionally critical arginine residue to have a similar position in I-BthII as in I-TevI. The GIY-YIG family contains four GIY-(X)9-YIG HEases, nine GIY-(X)10-YIG HEases and one GIY-(X)11-YIG HEase (33). We propose that HEases of the GIY-YIG family should also include the I-BthII HEases making it a GIY-(X)8-11-YIG motif.
Interestingly, the nrdIEF operon in bacteria is host to 10 different group I introns at five different locations (IVS2–IVS6) of which at least one representative for each site has been found in the B. cereus group (8,16–18,this study). Phylogenetic studies of the screened strains in this study and previously sequenced strains of this group's nrdIEF operons show that these strains are closely related despite the variable occurrences of different IVSs. Interestingly, the distributions of IVSs in the different trees (Figure 6) suggest that several rounds of HGT are likely to have occurred in the B. cereus group bacteria.
When HGT occurs it is dependent on recombination of flanking sequences of the insertion site of the IVS. This leads to a transfer of sequences from donor to recipient making recipient flanking sequences identical to the donor. Phylogenetic studies of sequences closely flanking IVSs are therefore less reliable when a HGT event has occurred in one strain but not the other. On the other hand, if sequences further away from the IVS insertion site differ between strains with and without the IVS, it may indicate that HGT has occurred. Unfortunately, the coding regions of the nrdIEF operons in this study are too similar to allow a direct dissection of the recombination events. Based on our phylogenetic analyses we suggest that several HGT events have happened for IVS3 and IVS6, see discussion below.
Interestingly the ssp. AH187 was found to contain both the IVS4 intron in nrdE and the IVS6 in nrdF. With the exception of ssp. AH187, the strains containing the IVS6 intron all have the IG-B sequence and cluster in the same clade in the operon tree (Figure 6B). They also group together in the nrdF tree (Figure 6D) again with the exception for ssp. AH187. This suggests that the IVS6 has been subject to HGT at least twice in the B. cereus group, once in the ssp. AH187 clade and once in the clade with the rest of the IVS6-containing strains. Further, in the nrdE tree (Figure 6C) the IVS6-containing strains show a scattered distribution in several clades suggesting that the HGT of IVS6 may have happened more than twice.
The IG-B, which is strongly associated with the IVS6 shows up in ssp. ATCC 10987 as an exception as ssp. ATCC 10987 does not harbour the IVS6. Ssp. ATCC 10987 does contain IVS4 differing in sequence from the IVS4 in ssp. AH187. The divergence between the nrdE sequence (including IVS4) in the two strains suggest that the IVS6 may have been lost in ssp. ATCC 10987 leaving an IG-region trace and a similarity in nrdF sequence to the ssp. AH187. Alternatively, the ssp. ATCC 10987 may have inherited the IG-B region by vertical transfer or recombination with another strain of the IG-B group without IVS6. Unfortunately as ssp. ATCC 10987 is the only strain of the IG-B group without IVS6, these alternatives cannot be tested without finding close relatives to ssp. ATCC 10987 belonging to the IG-B group and containing IVS6.
The ssp. pakistani contains two IVSs in the nrdIEF operon, IVS3 in nrdE and IVS6 in nrdF. Ssp. pakistani clusters with the majority of the IVS6- and IG-B-containing strains in the operon and nrdF trees but shows a different clustering in the nrdE tree with a close relation to ssp. canadiensis which contains the IVS3 but belongs to the IG-A2 group and does not have the IVS6 intron. Considering the differing IG-groups for these two strains and the diverse grouping in the nrdE versus the nrdF trees, it is difficult to say which IVS preceded the other in ssp. pakistani. Hypothetically, if IVS3 insertion preceded IVS6 and the IVS6 was transferred only once to the major IVS6 clade, we would expect to find IVS3 in the species belonging to the closely related IVS6 clade of ssp. pakistani nrdF, which we do not (Figure 6D). If, on the other hand, IVS6 insertion preceded IVS3, the close relation of ssp. canadiensis and ssp. pakistani nrdE would make us expect to see remnants of the IVS6 transfer, i.e. IG-B and nrdF clustering after a subsequent loss of IVS6 in ssp. canadiensis, which we do not (Figure 6C). The topology of the distribution of IVS3-containing strains was found to be similar in all trees with the IG-A1 strains ssp. AH820, Ames, konkukian and Sterne grouping apart from the ssp. pakistani (IG-B) and ssp. canadiensis (IG-A2). A comparison of the deletion in the ORF in IVS3 of ssp. canadiensis versus ssp. konkukian shows a difference in sequence and suggests that these have been two separate events of deletion.
We have shown that screening of a closely related group of bacteria is a useful method to find a missing HEG in a group I intron. Our identification of IVSs in 25 of 39 strains in the B. cereus group, including two strains with IVSs in both the nrdE and the nrdF gene, make the B. cereus group a most interesting candidate for study of HGT in bacteria. A screening expanded to other closely related groups and genes where IVSs have been found might prove a very efficient way of revealing the relations between the group I introns in bacteria.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by a grant from the Swedish Research Council. We thank Daniel Lundin and Mark Hoeppner for supporting tips on how to build phylogenetic trees, Steinar Johanssen for help with secondary folding of IVS6 and David Edgell for an interesting discussion on how IVSs might be lost from a genome. Funding to pay the Open Access publication charges for this article was provided by the Swedish Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cech TR. Self-splicing of group I introns. Annu. Rev. Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- 2.Haugen P, Simon DM, Bhattacharya D. The natural history of group I introns. Trends Genet. 2005;21:111–119. doi: 10.1016/j.tig.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Lambowitz AM, Belfort M. Introns as mobile genetic elements. Annu. Rev. Biochem. 1993;62:587–622. doi: 10.1146/annurev.bi.62.070193.003103. [DOI] [PubMed] [Google Scholar]
- 4.Chu FK, Maley GF, Maley F, Belfort M. Intervening sequence in the thymidylate synthase gene of bacteriophage T4. Proc. Natl Acad. Sci. USA. 1984;81:3049–3053. doi: 10.1073/pnas.81.10.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sjöberg B-M, Hahne S, Mathews CZ, Mathews CK, Rand KN, Gait MJ. The bacteriophage T4 gene for the small subunit of ribonucleotide reductase contains an intron. EMBO J. 1986;5:2031–2036. doi: 10.1002/j.1460-2075.1986.tb04460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gott JM, Shub DA, Belfort M. Multiple self-splicing introns in bacteriophage T4: evidence from autocatalytic GTP labeling of RNA in vitro. Cell. 1986;47:81–87. doi: 10.1016/0092-8674(86)90368-5. [DOI] [PubMed] [Google Scholar]
- 7.Read TD, Peterson SN, Tourasse N, Baillie LW, Paulsen IT, Nelson KE, Tettelin H, Fouts DE, Eisen JA, et al. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature. 2003;423:81–86. doi: 10.1038/nature01586. [DOI] [PubMed] [Google Scholar]
- 8.Nord D, Torrents E, Sjöberg B-M. A functional homing endonuclease in the Bacillus anthracis nrdE group I intron. J. Bacteriol. 2007;189:5293–5301. doi: 10.1128/JB.00234-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nordlund P, Reichard P. Ribonucleotide reductases. Annu. Rev. Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 10.Torrents E, Sahlin M, Biglino D, Gräslund A, Sjöberg B-M. Efficient growth inhibition of Bacillus anthracis by knocking out the ribonucleotide reductase tyrosyl radical. Proc. Natl Acad. Sci. USA. 2005;102:17946–17951. doi: 10.1073/pnas.0506410102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ko M, Choi H, Park C. Group I self-splicing intron in the recA gene of Bacillus anthracis. J. Bacteriol. 2002;184:3917–3922. doi: 10.1128/JB.184.14.3917-3922.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen I, Dubnau D. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2004;2:241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 13.Claverys JP, Prudhomme M, Martin B. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu. Rev. Microbiol. 2006;60:451–475. doi: 10.1146/annurev.micro.60.080805.142139. [DOI] [PubMed] [Google Scholar]
- 14.Chen I, Christie PJ, Dubnau D. The ins and outs of DNA transfer in bacteria. Science. 2005;310:1456–1460. doi: 10.1126/science.1114021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belfort M, Roberts RJ. Homing endonucleases: keeping the house in order. Nucleic Acids Res. 1997;25:3379–3388. doi: 10.1093/nar/25.17.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lazarevic V. Ribonucleotide reductase genes of Bacillus prophages: a refuge to introns and intein coding sequences. Nucleic Acids Res. 2001;29:3212–3218. doi: 10.1093/nar/29.15.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazarevic V, Soldo B, Dusterhoft A, Hilbert H, Mauel C, Karamata D. Introns and intein coding sequence in the ribonucleotide reductase genes of Bacillus subtilis temperate bacteriophage SPβ. Proc. Natl Acad. Sci. USA. 1998;95:1692–1697. doi: 10.1073/pnas.95.4.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stankovic S, Soldo B, Beric-Bjedov T, Knezevic-Vukcevic J, Simic D, Lazarevic V. Subspecies-specific distribution of intervening sequences in the Bacillus subtilis prophage ribonucleotide reductase genes. Syst. Appl. Microbiol. 2007;30:8–15. doi: 10.1016/j.syapm.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 19.Stoddard BL. Homing endonuclease structure and function. Q. Rev. Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 20.Zhao L, Bonocora RP, Shub DA, Stoddard BL. The restriction fold turns to the dark side: a bacterial homing endonuclease with a PD-(D/E)-XK motif. EMBO J. 2007;26:2432–2442. doi: 10.1038/sj.emboj.7601672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cech TR, Damberger SH, Gutell RR. Representation of the secondary and tertiary structure of group I introns. Nat. Struct. Biol. 1994;1:273–280. doi: 10.1038/nsb0594-273. [DOI] [PubMed] [Google Scholar]
- 22.Armougom F, Moretti S, Poirot O, Audic S, Dumas P, Schaeli B, Keduas V, Notredame C. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 2006;34:W604–W608. doi: 10.1093/nar/gkl092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 24.Van Roey P, Meehan L, Kowalski JC, Belfort M, Derbyshire V. Catalytic domain structure and hypothesis for function of GIY-YIG intron endonuclease I-TevI. Nat. Struct. Biol. 2002;9:806–811. doi: 10.1038/nsb853. [DOI] [PubMed] [Google Scholar]
- 25.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandegren L, Nord D, Sjöberg B-M. SegH and Hef: two novel homing endonucleases whose genes replace the mobC and mobE genes in several T4-related phages. Nucleic Acids Res. 2005;33:6203–6213. doi: 10.1093/nar/gki932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgenstern B. DIALIGN: multiple DNA and protein sequence alignment at BiBiServ. Nucleic Acids Res. 2004;32:W33–W36. doi: 10.1093/nar/gkh373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 29.Etherington GJ, Dicks J, Roberts IN. Recombination Analysis Tool (RAT): a program for the high-throughput detection of recombination. Bioinformatics. 2005;21:278–281. doi: 10.1093/bioinformatics/bth500. [DOI] [PubMed] [Google Scholar]
- 30.Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 31.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev S, Dryden DT, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, et al. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunin-Horkawicz S, Feder M, Bujnicki JM. Phylogenomic analysis of the GIY-YIG nuclease superfamily. BMC Genomics. 2006;7:98. doi: 10.1186/1471-2164-7-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bujnicki JM, Rotkiewicz P, Kolinski A, Rychlewski L. Three-dimensional modeling of the I-TevI homing endonuclease catalytic domain, a GIY-YIG superfamily member, using NMR restraints and Monte Carlo dynamics. Protein Eng. 2001;14:717–721. doi: 10.1093/protein/14.10.717. [DOI] [PubMed] [Google Scholar]
- 35.Tourasse NJ, Helgason E, Økstad OA, Hegna IK, Kolstø AB. The Bacillus cereus group: novel aspects of population structure and genome dynamics. J. Appl. Microbiol. 2006;101:579–593. doi: 10.1111/j.1365-2672.2006.03087.x. [DOI] [PubMed] [Google Scholar]