Abstract
This article presents the first evidence that the DNA base analogue 1,3-diaza-2-oxophenoxazine, tCO, is highly fluorescent, both as free nucleoside and incorporated in an arbitrary DNA structure. tCO is thoroughly characterized with respect to its photophysical properties and structural performance in single- and double-stranded oligonucleotides. The lowest energy absorption band at 360 nm (ε = 9000 M−1 cm−1) is dominated by a single in-plane polarized electronic transition and the fluorescence, centred at 465 nm, has a quantum yield of 0.3. When incorporated into double-stranded DNA, tCO shows only minor variations in fluorescence intensity and lifetime with neighbouring bases, and the average quantum yield is 0.22. These features make tCO, on average, the brightest DNA-incorporated base analogue so far reported. Furthermore, it base pairs exclusively with guanine and causes minimal perturbations to the native structure of DNA. These properties make tCO a promising base analogue that is perfectly suited for e.g. photophysical studies of DNA interacting with macromolecules (proteins) or for determining size and shape of DNA tertiary structures using techniques such as fluorescence anisotropy and fluorescence resonance energy transfer (FRET).
INTRODUCTION
The number of fluorescent DNA base analogues has increased considerably over the last decade and the search for new probes with improved photophysical and structural properties is continuing [for recent reviews on fluorescent DNA base analogues and advances in the field, see Wilsson and Kool (1), Asseline (2), Okamoto et al. (3), Hawkins (4), and Rist and Marino (5)]. The development of this class of fluorescent probes for studying biochemical reactions and interactions in DNA-containing systems is especially challenging due to the strict conditions set by the helical-stacking environment of DNA. Size, shape, charge, polarity and hydrogen-bonding motif are all important properties to be considered if this class of fluorophores is to cause as little perturbation to the native DNA structure as possible. Until recently all reported fluorescent DNA base analogues shared a common feature, namely, an emission intensity highly sensitive to its microenvironment [for example, see (6–20)]. Virtually all of them are quenched considerably when incorporated into DNA, the effect being strongly dependent on base sequence, position, and whether the DNA is single or double stranded. These features have been used extensively in DNA-containing systems to probe, for example, structure and stability of DNA constructs (21–26), DNA-macromolecular structures, interactions and activity (6,27–35), and DNA-mediated charge transport (36–39). In addition to DNA, fluorescent nucleobases have also been incorporated into RNA to study, for example, RNA-drug interaction (40–42). Utilizing this emission sensitivity to report on the microenvironment has proven to be extremely useful and has provided a range of important and valuable information. However, to expand the number of useful fluorescence applications for this class of DNA probes, new base analogues with new and different fluorescence properties are required.
Recently we reported on a novel fluorescent base analogue, tC, with unique fluorescence properties (43–47). In contrast to other base analogues, tC has a high fluorescence quantum yield (Φf = 0.2) that is virtually insensitive to the single- or double-strand nature of the DNA and the surrounding base sequence (46). We have also shown that tC works extremely well as a cytosine analogue and has only limited effects on the native conformation of DNA (45). Furthermore, tC is rigidly stacked within the duplex with no increased rate of base-flipping as compared to the native bases. Together with a single fluorescence lifetime in both single- and double-stranded DNA, these properties make tC unique as a fluorescent base analogue with great potential in applications where other fluorescent base analogues are less than ideal, e.g. in fluorescence resonance energy transfer (FRET) and fluorescence anisotropy experiments. Recently, tC was successfully used as a FRET donor to directly observe, for the first time, the inactive to active isomerization in the Klenow fragment (KF) polymerase (48). In the same study, fluorescence anisotropy of tC was successfully used to probe the effect of nucleotides on DNA-KF binding affinity.
Here we present the next fluorescent DNA base analogue in the tricyclic cytosine family, the tC homolog 1,3-diaza-2-oxophenoxazine (tCO, the superscript O indicates the change from sulphur in tC to oxygen in tCO, see Figure 1). Previously tCO has been shown to stabilize DNA-RNA- (49), DNA-DNA- (50) and PNA (peptide nucleic acid)-DNA-duplexes (51), and to discriminate well between A, T, C and G targets (49–51). Furthermore, the rigid structure of tCO has also provided the scaffold for derivatives with even greater stabilizing effects on nucleic acid duplexes (50–53) and very recently for a new spin label for the use in electron paramagnetic resonance experiments (54). We have now, for the first time, shown that tCO is fluorescent and performed an extensive photophysical and structural characterization of the tCO monomer and of tCO-containing single- and double-stranded oligonucleotides. We show, not surprisingly, that tCO structurally behaves very similarly to tC having a firm stacking within the DNA double helix and causing minimal perturbations to the native structure of the DNA. We also show that, unlike tC, the emission intensity of tCO is slightly affected by incorporation into DNA and the hybridization state of the DNA. However, once in double-stranded DNA the effect on emission intensity, for all possible combinations of neighbouring bases, is found to be negligible. Moreover, in terms of overall average brightness (= extinction coefficient × fluorescence quantum yield) tCO is unprecedented among base analogues when incorporated into DNA. Although tC has a high average brightness, it is still approximately 3 times weaker than tCO and other base analogues such as 2-aminopurine, pyrrolo-dC, 3-MI are one to several orders of magnitude weaker. Together with other properties presented herein, this makes tCO a very promising fluorescent DNA base analogue in a wide range of fluorescence applications especially FRET, fluorescence anisotropy and fluorescence melting experiments.
Figure 1.
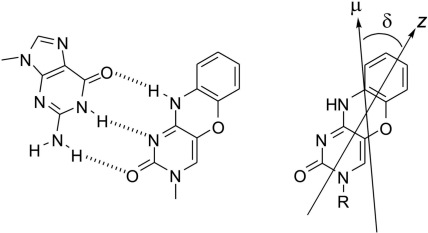
Structure of the G-tCO base-pair (left) and the derivatives of tCO used in the measurements (right) (R = CH2COO− K+ for the potassium salt of 1,3-diaza-2-oxophenoxazin-3-yl acetic acid (KtCO) and deoxyribose for the 2-deoxyribonucleoside of tCO). δ is an in-plane angle relative the molecular orientation axis (z) that is parallel to the molecular long axis. μ indicates the polarization of the lowest energy electronic transition moment.
MATERIALS AND METHODS
Chemicals
The tCO nucleoside, 1,3-diaza-2-oxophenoxazine-2′-deoxynucleoside, was synthesized following the procedure of Matteucci et al. (49) with the exception that toluoyl protection groups were used instead of acetyl. Preparation of the tCO nucleoside for standard automated solid-phase methods using β-cyanoethyl phosphoramidite was carried out by dimethoxytritylation at the 5′-OH group (49) and phosphitylation of the 3′-OH group (Supplementary Material) to afford the dimethoxytrityl-protected phosphoramidite monomer suitable for oligonucleotide synthesis. The potassium salt of 1,3-diaza-2-oxophenoxazin-3-yl acetic acid, KtCO, was obtained by saponification of the tert-butyl ester that was synthesized following the procedures of Rajeev et al. (51) and Ausín et al. (52).
The standard buffer in all measurements, unless stated otherwise, is a sodium phosphate buffer at pH 7.5 with a total Na+ concentration of 50 mM. Poly(vinyl alcohol) (PVA) was obtained as powder from E. I. du Pont de Nemours Co. (Elvanol). Quinine sulphate dihydrate, dissolved in 0.1 M H2SO4, was purchased from Molecular Probes. Oligonucleotides containing tCO were synthesized in the same manner as for tC, which is described elsewhere (46,47). Unmodified oligonucleotides, not containing tCO, were purchased from ATDBio Ltd.
Concentration determination
The concentrations of the oligonucleotides were determined by UV absorption measurements at 260 nm. The extinction coefficients for the modified oligonucleotides were calculated using a linear combination of the extinction coefficients of the natural nucleotides and the extinction coefficient of the tCO nucleoside at 260 nm. To account for the base stacking interactions, this linear combination was multiplied by 0.9 to give a final estimate of the extinction coefficients for the oligonucleotides. The individual extinction coefficients at 260 nm used in the calculation were εT = 9300 M−1 cm−1, εC = 7400 M−1 cm−1, εG = 11 800 M−1 cm−1, εA = 15 300 M−1 cm−1 (55) and εtCo = 11 000 M−1 cm−1. The total extinction coefficient of each sequence can be found in the Supplementary Material.
Characterization of the tCO monomer
The extinction coefficient of the tCO nucleoside was determined by measuring the absorption of samples of known concentration. Samples were prepared by weighing out small amounts of the tCO nucleoside, typically 1.5 mg, and dissolving them in known volumes of MQ water. Absorption spectra were recorded on a Varian Cary 4000 spectrophotometer at room temperature. The extinction coefficient was determined as an average of three measurements.
Linear dichroism (LD) measurements were performed in stretched PVA film. The film was made from a 12.5% (w/w) solution of PVA that was prepared by dissolving PVA in water under heating to 90°C. Portions of 5 mL were mixed with 3 mL of water solutions of KtCO, each containing ∼0.2 mg of substance. The mixtures were poured onto horizontal glass plates and left to dry in a dust-free environment for more than 48 h, where after the films were removed from the plates and mechanically stretched 4 times their original length under hot air from a hairdryer. The measurements were performed using a Varian Cary 4B spectrophotometer equipped with Glan air-space calcite polarizers in both sample and reference beam. LD is defined as
| 1 |
where AII(λ) and A⊥(λ) are the absorption of light polarized parallel and perpendicular to the macroscopic orientation axis (film-stretching direction), respectively. The reduced LD of a uniaxial sample is defined as
| 2 |
where Aiso is the absorption of the corresponding isotropic sample. The reduced LD for a pure electronic transition,  , for a molecule that has a rodlike orientation in a stretched PVA matrix is given by
, for a molecule that has a rodlike orientation in a stretched PVA matrix is given by
| 3 |
where δi is the angle between the molecular orientation axis, z (Figure 1), and the ith transition moment, and Szz is the Saupe orientation parameter for this axis.
The magnetic circular dichroism (MCD) was measured on KtCO in phosphate buffer, using a Jasco J-720 CD spectropolarimeter equipped with a permanent horseshoe magnet. MCD is measured exposing a molecule to a magnetic field parallel (by convention, the direction of the magnetic field is north to south) with the light propagation axis and defined as the difference in absorption of left- and right-circularly polarized light.
| 4 |
The spectra of KtCO were recorded with both NS (north–south) and SN (south–north) magnetic field orientation. The MCD was obtained by subtracting the SN spectrum from the NS spectrum and dividing by 2. For two electronic transitions close in energy but with different polarizations, the magnetic field would mix the two states giving them magnetic rotational strength of equal amplitude but of different signs that would result in a bisignate signal over the absorption band. For this reason MCD is very useful when determining whether an absorption band is due to one or several transitions.
Fluorescence quantum yields (Φf) were determined relative to the quantum yield of quinine sulphate in 0.5 M H2SO4 (Φf = 0.55 at 22°C) (56). The measurements were performed on a SPEX fluorolog 3 spectrofluorimeter (JY Horiba), using emission wavelengths between 370 and 700 nm and an excitation wavelength of 365 nm.
Fluorescence lifetimes were determined using time-correlated single photon counting. The samples were excited with a PicoQuant PLS-8-2-060 pulsed diode with an approximate centre wavelength of 370 nm (FWHM 50 nm). The diode was pulsed at a 10 MHz repetition rate with a pulse width of ∼1 ns and the emission was monitored at 460 nm. The photons were collected by a microchannel-plate photomultiplier tube (MCP-PMT R3809U-50; Hamamatsu) and fed into a multi-channel analyzer with 4096 channels. A minimum of 10 000 counts were recorded in the top channel. The intensity data were convoluted with the instrument response and evaluated with the software package FluoFit (PicoQuant). The experimental setup yielded a time resolution of ∼100 ps (FWHM).
The fluorescence excitation anisotropy spectra were measured on a SPEX fluorolog 3 spectrofluorimeter (JY Horiba) equipped with Glan polarizers both in the excitation and in the emission beam. The polarized excitation spectra of the tCO nucleoside in an H2O/ethylene glycol (1:2 mixture) glass at −100°C, were recorded from 250 to 405 nm, and the emission was set at the fluorescence maximum of the sample (433 nm). The fluorescence anisotropy (r) was calculated as (57)
| 5 |
where Ivh refers to excitation spectrum recorded with the excitation polarizer in the vertical and the emission polarizer in the horizontal position, and so forth, and G is the ratio Ihv/Ihh used for instrumental correction. For an immobile chromophore with an electronic transition (i), the fundamental anisotropy, r0i, is related to the angle αi between the absorbing and the emitting transition moments according to the following equation:
| 6 |
Quantum mechanical calculations of electronic absorption spectra of the tCO chromophore were performed with the semi-empirical ZINDO/S method as incorporated in the HyperChem program. All singly excited configurations using the 15 highest occupied and 15 lowest unoccupied orbitals were included in the configuration interaction (CI) calculation. The geometries used were obtained from AM1 optimizations as implemented in HyperChem.
Characterization of tCO-containing oligonucleotides
Double-stranded oligonucleotides were formed by mixing equimolar amounts of complementary single strands in phosphate buffer at room temperature (in fluorescence measurements an excess of 20% of the non-fluorescent complementary strand was used to ensure that all tCO strands were double stranded). The samples were heated to 85°C and thereafter annealed by slow cooling to 20°C. Melting temperatures were measured on a Varian Cary 4000 spectrophotometer equipped with a programmable multi-cell temperature block. The samples were heated from 10 to 85°C at a maximum rate of 0.5°C min−1 whereupon they were cooled to 10°C at the same rate. The absorption at 260 nm was measured with a temperature interval of 1°C.
Circular dichroism (CD) spectra were recorded on a Jasco J-720 spectropolarimeter at 25°C. The spectra of oligonucleotides containing tCO were recorded between 200 and 500 nm, and the spectra of oligonucleotides not containing tCO were recorded between 200 and 350 nm. All spectra were corrected for background contributions.
Fluorescence quantum yields and fluorescence lifetimes were determined as described for the monomer (see above).
Fluorescence excitation anisotropy of tC- and tCO-containing oligonucleotides
The steady-state anisotropy, r, for two tC-containing duplexes of different lengths (10 and 22 base pairs) and two tCO-containing duplexes of different lengths (10 and 21 base pairs) were measured (for sequences, see Supplementary Material). Fluorescence anisotropy spectra were measured as described for the monomer (see above). However, measurements were performed in phosphate buffer at room temperature. The anisotropy (r) for the duplexes was experimentally determined using Equation (5). The anisotropies at excitation wavelengths 375 and 395 nm were used for tCO- and tC-containing oligonucleotides, respectively, and the emission was monitored at 460 and 510 nm, respectively. In the theoretical calculation of the steady-state anisotropies, the DNA duplexes were modelled as cylinders with the rotational motion characterized by the diffusion coefficients D∥ and D⊥ for the rotation about the helical axis and about the axis perpendicular to the helical axis, respectively. D∥ and D⊥ were calculated using the equations of Tirado and Garcia de la Torre (58) to obtain the two rotational correlation times, θ1 and θ2 (Supplementary Material). The expected steady-state anisotropy, r, was calculated using the following equation:
| 7 |
where β1 and β2 are the amplitudes of θ1 and θ2, respectively. The fluorescence lifetime, τ, was experimentally determined for each sample. For details on the calculations and values used, see Supplementary Material.
RESULTS
Spectroscopic characterization of the tCO monomer
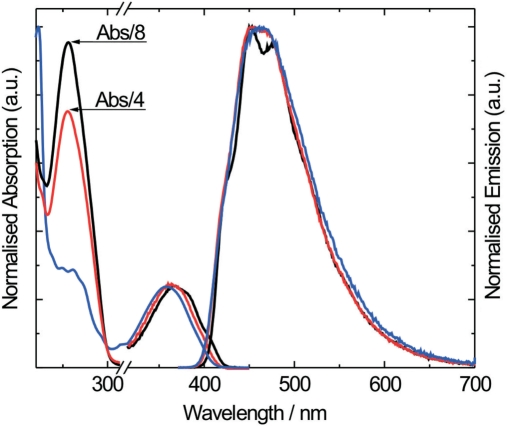
The absorption and emission spectra of the tCO nucleoside are presented in Figure 2. The low energy absorption band is centred at 360 nm (ε = 9000 M−1cm−1) and the emission maximum is found at 465 nm. The fluorescence quantum yield, Φf, and lifetime, τ, in aqueous buffer at pH 7.5, 22°C is 0.30 and 3.4 ns, respectively. Neither the absorption nor the emission spectrum shows significant changes in the pH range 5 to 9 and at Na+-concentrations from 50 mM to 1 M (data not shown). Furthermore, the emission intensity decreases linearly by approximately 30% when increasing the temperature from 10 to 90°C (data not shown).
Figure 2.
Isotropic absorption and emission spectra of the tCO nucleoside (blue), tCO-containing single strand (AA, red) and tCO-containing double strand (AA, black). The spectra have been normalized at the maxima of the lowest energy absorption band (left) and of the emission (right), respectively.
A detailed characterization of the lowest energy absorption band was performed using LD, fluorescence anisotropy, MCD, and quantum chemical calculations. Figure 3a shows the isotropic absorption, reduced LD (LDr) and fluorescence anisotropy spectra. The LDr value for the lowest energy absorption band is found to be 1.31 (taken as the plateau value at long wavelengths). Using Equation (3) and assuming that tCO, like tC, has the same orientation factor, Szz, as methylene blue (previously shown to be ∼0.78) (59) the angle between the molecular orientation axis, coinciding with the molecular long axis, z, as defined in Figure 1, and the lowest electronic transition moment was calculated to be 33°. The assumption that methylene blue and tCO orient in a similar fashion in the stretched PVA matrix is reasonable since the two molecules are very similar in shape. The fundamental fluorescence anisotropy of tCO immobilized in a solid H2O/ethylene glycol (1:2 mixture) matrix has a constant value (0.37) close to the maximum theoretical value (0.4) over the whole absorption band. This suggests both that the band is a result of a single absorption transition moment, since multiple transitions would give rise to a variation with wavelength, and also that the emission and absorption transition moments are virtually parallel. A single transition moment in the low energy absorption band is also evidenced by the MCD (Figure 3b). In this region, the MCD signal for tCO has the same sign and appears as the mirror image of the isotropic absorption spectrum. Both these observations are strong indications that this absorption band is due to a single electronic transition moment. Figure 3c shows the electronic spectrum of tCO calculated using the ZINDO/S method. The geometry of the neutral tCO was predicted from AM1 calculations to be slightly bent at an angle of 166° over the middle ring oxygen–nitrogen axis. A similar slight deviation from planarity has previously been found for tC (44). The calculation shows that the first two strong electronic transitions are well separated by ∼35 nm, again suggesting that only a single transition moment is responsible for the lowest energy absorption band of tCO. From the calculation, the S0 → S1 transition (358 nm) was determined to be a virtually in-plane transition with an angle of ∼41° relative the long axis, z (as defined in Figure 1), of tCO. This agrees fairly well with the experimentally estimated value (∼33°, see above).
Figure 3.

(a) Isotropic absorption (Aiso, black) and excitation anisotropy spectra (r, red) of the tCO nucleoside and reduced linear dichroism spectra (LDr, blue) of KtCO. Aiso was measured in a phosphate buffer (50 mM Na+, pH 7.5), r in a solid H2O/ethylene glycol (1:2 mixture) matrix at −100°C and LDr in a stretched PVA film. (b) Magnetic circular dichroism (MCD) of KtCO in a phosphate buffer (50 mM Na+, pH 7.5). (c) Electronic transitions of the tCO chromophore and their oscillator strengths calculated using the ZINDO/S method on an AM1 geometry optimized structure. Note that the calculated wavenumbers have been multiplied by 0.9 to facilitate comparison.
Incorporation of tCO into DNA oligonucleotides
Since emission properties of fluorescent base analogues are notoriously sensitive to base surroundings, we incorporated tCO into a series of decamers positioning it between every possible combination of nearest neighbours while keeping all the other bases the same. The sequences are named using the closest neighbours to tCO, e.g. in sequence GA the tCO has a G on the 5′ side and an A on the 3′ side (for full sequences, see Table 1). In addition to these 16 oligonucleotides one more sequence was included, GAAG (GA on the 5′ side of the tCO and AG on the 3′ side, for full sequence, see Table 1), to investigate possible effects from guanines positioned one base away from tCO. The series also provided for a very thorough characterization of the effects on DNA conformation and stability upon tCO incorporation.
Table 1.
Melting temperatures of tCO-containing duplexes (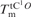 ), the corresponding unmodified duplexes (
), the corresponding unmodified duplexes (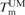 ) and the difference in melting temperature as a result of changing one C for tCO (ΔTm)
) and the difference in melting temperature as a result of changing one C for tCO (ΔTm)
| Sequencea | Sequence name |
 (°C)b (°C)b
|
 (°C)b (°C)b
|
Δ Tm(°C) |
|---|---|---|---|---|
| 5′-CGCA-GtCOA-TCG-3′ | GA | 42 | 45 | −3 |
| 5′-CGCA-GtCOG-TCG-3′ | GG | 53 | 50 | 3 |
| 5′-CGCA-GtCOC-TCG-3′ | GC | 48 | 49 | −1 |
| 5′-CGCA-GtCOT-TCG-3′ | GT | 46 | 46 | 0 |
| 5′-CGCA-AtCOA-TCG-3′ | AA | 41 | 41 | 0 |
| 5′-CGCA-AtCOG-TCG-3′ | AG | 45 | 43 | 2 |
| 5′-CGCA-AtCOC-TCG-3′ | AC | 43 | 43 | 0 |
| 5′-CGCA-AtCOT-TCG-3′ | AT | 39 | 40 | −1 |
| 5′-CGCA-CtCOA-TCG-3′ | CA | 51 | 46 | 5 |
| 5′-CGCA-CtCOG-TCG-3′ | CG | 53 | 48 | 5 |
| 5′-CGCA-CtCOC-TCG-3′ | CC | 55 | 48 | 7 |
| 5′-CGCA-CtCOT-TCG-3′ | CT | 49 | 44 | 5 |
| 5′-CGCA-TtCOA-TCG-3′ | TA | 45 | 41 | 4 |
| 5′-CGCA-TtCOG-TCG-3′ | TG | 50 | 45 | 5 |
| 5′-CGCA-TtCOC-TCG-3′ | TC | 45 | 41 | 4 |
| 5′-CGCA-TtCOT-TCG-3′ | TT | 44 | 39 | 5 |
| 5′-GCC-GAtCOAG-CG-3′ | GAAG | 49 | 49 | 0 |
aIn unmodified sequences, tCO is replaced by cytosine.
bTm values were determined from the derivative of the melting curves. Measurements were performed in a phosphate buffer (50 mM Na+, pH 7.5) at a duplex concentration of 2.5 μM.
Structure of tCO-modified duplex DNA
Effects on DNA secondary structure and duplex stability were investigated by CD and UV-melting measurements. The CD spectra (Supplementary Material) show for all tCO-modified decamers typical characteristics of B-form DNA confirming that incorporation of tCO has no effect on the overall conformation of the DNA. Surprisingly, in contrast to tC, no significant CD signal was found for the long wavelength absorption band of tCO. The reason for this is still not fully understood.
The thermal stability of the tCO-DNA duplexes is summarized in Table 1. Incorporation of tCO results in an increase in melting temperature by on average 2.7°C. Furthermore, as for tC, it should be noted that a purine at the 5′ side of the base analogue has modest effects on the melting temperature (−3°C < ΔTm<+3°C; average 0°C) while a 5′-pyrimidine increases the Tm by ∼4–7°C (average 5°C).
Photophysics of tCO in single-stranded DNA
Figure 2 shows an absorption and an emission spectrum of a tCO-containing single strand and Table 2 lists the photophysical properties of tCO when incorporated into single-stranded DNA. The long wavelength absorption maximum is slightly red shifted (363–370 nm) upon incorporation into single-stranded oligonucleotides as compared to the free monomer (360 nm). In contrast, the emission of tCO is not significantly shifted upon incorporation into single-stranded oligonucleotides, however, the spectra become slightly more structured. The emission peak is split into two closely lying vibronic peaks that cannot always be resolved making the peak appear truncated. The relative intensities of the two peaks vary and, thus, for comparison, the peak at shorter wavelengths is listed in Table 2. Although these values might give the impression that the emission is blue-shifted upon single-strand incorporation, this is, as mentioned above, not the case. Interestingly, the emission spectra of GG, GA, GT and GC are more structured and slightly red-shifted compared to the rest of the single strands (Supplementary Material). We will return to this in the discussion.
Table 2.
Lowest energy absorption and emission maxima, fluorescence quantum yield, fluorescence lifetimes and intensity averaged lifetimes of tCO-containing single strands
| Sequencea | Absmax (nm)b | Emmax (nm)b | Φfb, c | τ1(α1) (ns)b, d | τ2(α2) (ns)b, d | <τ> (ns)e |
|---|---|---|---|---|---|---|
| GA | 370 | 461 | 0.14 | 4.7 (0.13) | 2.3 (0.87) | 2.6 |
| GG | 369 | 458 | 0.14 | 4.1 (0.23) | 2.0 (0.77) | 2.5 |
| GC | 369 | 456 | 0.17 | 4.0 (0.18) | 2.6 (0.82) | 2.9 |
| GT | 368 | 456 | 0.15 | 3.6 (0.19) | 2.5 (0.81) | 2.7 |
| AA | 365 | 450 | 0.38 | 5.6 (1.00) | f | 5.6 |
| AG | 365 | 448 | 0.41 | 5.8 (0.98) | 2.9 (0.02) | 5.7 |
| AC | 365 | 454 | 0.33 | 5.4 (0.69) | 3.3 (0.31) | 4.7 |
| AT | 365 | 453 | 0.36 | 5.2 (0.67) | 3.9 (0.33) | 4.7 |
| TA | 365 | 449 | 0.34 | 5.5 (0.81) | 3.1 (0.19) | 5.0 |
| TG | 365 | 446 | 0.36 | 5.5 (0.90) | 2.9 (0.10) | 5.2 |
| TC | 364 | 448 | 0.29 | 5.1 (0.66) | 2.7 (0.34) | 4.3 |
| TT | 365 | 448 | 0.30 | 4.7 (0.80) | 2.5 (0.20) | 4.3 |
| CA | 365 | 452 | 0.34 | 5.4 (0.84) | 3.3 (0.16) | 5.1 |
| CG | 365 | 447 | 0.41 | 5.8 (0.91) | 3.7 (0.09) | 5.7 |
| CC | 366 | 450 | 0.31 | 5.2 (0.63) | 3.0 (0.37) | 4.4 |
| CT | 366 | 451 | 0.30 | 5.1 (0.61) | 3.3 (0.39) | 4.4 |
| GAAG | 363 | 454 | 0.40 | 5.6 (0.97) | 3.3 (0.03) | 5.5 |
aSequence names indicate the nearest neighbours to tCO. For full sequences, see Table 1.
bMeasurements were performed in phosphate buffer (50 mM Na+, pH 7.5) at room temperature.
cFluorescence quantum yields are measured relative to quinine sulphate in 0.5 M H2SO4 (Φf = 0.55) (56).
dThe amplitudes are indicated in parenthesis.
eMean fluorescence lifetimes <τ > = ∑αiτi2/∑αiτi.
fWhen fitting with two lifetimes, The second lifetime gets a negative amplitude.
Table 2 also lists the fluorescence quantum yields, Φf, and mean fluorescence lifetimes, <τ >, for tCO incorporated into single-stranded DNA. The fluorescence decays were fitted with two fluorescence lifetimes in all cases even though in a few cases, where the sequence has a high fluorescence quantum yield, a single fluorescence lifetime would have been sufficient (Table 2). When comparing the mean fluorescence lifetimes of all sequences (Table 2), GG, GA, GT and GC once again stand out with lower values (2.8–3.0 ns) than both the free monomer (3.4 ns) and the other single strands (4.4–5.8 ns). These four sequences also have fluorescence quantum yield of about half (Φf ∼ 0.15) that of the free monomer (Φf = 0.30) and the other sequences (Φf = 0.29–0.41). Apparently the guanine on the 5′ side of tCO has a fluorescence-quenching effect in these sequences. The other single strands all have high fluorescence quantum yields and interestingly, the single strands with a guanine on the 3′ side of tCO are among the highest. The sequence GAAG, which has two adenines as closest neighbours to tCO and guanines as next neighbours, shows no significant difference in either the fluorescence quantum yield or the lifetime as compared to the sequence AA indicating that the quenching effect of guanine has a steep distance dependence.
Photophysics of tCO in double-stranded DNA
Also included in Figure 2 are an absorption and an emission spectrum of tCO in double-stranded DNA and the photophysical properties of tCO when incorporated into double-stranded DNA are listed in Table 3. The long wavelength absorption maximum (363–369 nm) is in the same range as for the single strands. However, there is very weak vibrational structure on the long wavelength side of the absorption spectra and the emission is significantly more structured for the double strands as compared to both the single strands and the free nucleoside. In contrast to the single strands, all emission spectra are very similar and the first of the two vibronic peaks being the strongest (Figure 2). The emission is not significantly shifted compared to either the single strands or the free nucleoside. As in the case for the single-stranded samples, the values for the emission maximum listed in Table 3 might give the impression that the emission is blue-shifted compared to the free nucleoside. However, as discussed above, this is an effect of the vibronic splitting and not an actual shift of the spectra.
Table 3.
Lowest energy absorption and emission maxima, fluorescence quantum yield, fluorescence lifetimes, radiative rate constants and non-radiative rate constants of tCO-containing double strands and the tCO nucleoside
| Sequencea | Absmax (nm)b | Emmax (nm)b | Φfb, c | τ (ns)b | kf (107 s−1)d | knr (108 s−1)e |
|---|---|---|---|---|---|---|
| GA | 367 | 453 | 0.17 | 3.5 | 4.8 | 2.3 |
| GG | 365 | 452 | 0.18 | 4.0 | 4.6 | 2.1 |
| GC | 367 | 453 | 0.20 | 4.0 | 5.0 | 2.0 |
| GT | 367 | 454 | 0.17 | 3.4 | 5.0 | 2.4 |
| AA | 366 | 451 | 0.23 | 4.5 | 5.1 | 1.7 |
| AG | 367 | 453 | 0.24 | 4.6 | 5.2 | 1.6 |
| AC | 369 | 453 | 0.27 | 4.8 | 5.6 | 1.5 |
| AT | 369 | 454 | 0.27 | 4.8 | 5.6 | 1.5 |
| TA | 366 | 448 | 0.21 | 4.0 | 5.3 | 2.0 |
| TG | 363 | 448 | 0.19 | 3.8 | 5.0 | 2.1 |
| TC | 366 | 450 | 0.23 | 4.2 | 5.5 | 1.8 |
| TT | 367 | 450 | 0.22 | 4.4 | 5.0 | 1.8 |
| CA | 366 | 447 | 0.18 | 3.6 | 5.0 | 2.3 |
| CG | 364 | 448 | 0.19 | 3.5 | 5.4 | 2.3 |
| CC | 366 | 448 | 0.21 | 3.9 | 5.4 | 2.0 |
| CT | 364 | 450 | 0.23 | 4.3 | 5.3 | 1.8 |
| GAAG | 367 | 453 | 0.25 | 4.7 | 5.3 | 1.6 |
| tCOnucleoside | 359 | 461 | 0.30 | 3.4 | 8.8 | 2.1 |
aSequence names indicate the nearest neighbours to tCO. For full sequences, see Table 1.
bMeasurements were performed in phosphate buffer (50 mM Na+, pH 7.5) at room temperature.
cFluorescence quantum yields are measured relative to quinine sulphate in 0.5 M H2SO4 (Φf = 0.55) (56).
dRadiative rate constant kf = Φf/τ.
eNon-radiative rate constant knr = kf/Φf − kf.
Table 3 also lists the fluorescence quantum yields and fluorescence lifetimes for the double strands. Both the fluorescence quantum yields and fluorescence lifetimes show significantly less variation between different double-strand sequences compared to the single-stranded case. The fluorescence quantum yield is relatively insensitive to base sequence and is in the range 0.22 ± 0.05 for all sequences. All emission decay curves could be fitted to a single fluorescence lifetime (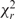 ) ranging between 3.4 and 4.8 ns. The calculated radiative rate constants, kf, and non-radiative rate constants, knr, for both the free tCO nucleoside and the tCO-containing double strands can also be found in Table 3. The kf values range between 4.6 × 107 and 5.6 × 107 s−1 for the double strands. This can be compared to 8.8 × 107 s−1 for the free monomer. The knr of tCO is less affected upon incorporation into double strands and ranges between 1.5 × 108 and 2.3 × 108 s−1 compared to 2.1 × 108 s−1 for the free monomer.
) ranging between 3.4 and 4.8 ns. The calculated radiative rate constants, kf, and non-radiative rate constants, knr, for both the free tCO nucleoside and the tCO-containing double strands can also be found in Table 3. The kf values range between 4.6 × 107 and 5.6 × 107 s−1 for the double strands. This can be compared to 8.8 × 107 s−1 for the free monomer. The knr of tCO is less affected upon incorporation into double strands and ranges between 1.5 × 108 and 2.3 × 108 s−1 compared to 2.1 × 108 s−1 for the free monomer.
Fluorescence anisotropy experiments on DNA using rigidly stacked base analogues
To demonstrate the advantage of using rigidly stacked base analogues in fluorescence anisotropy experiments, we performed steady-state measurements on four different DNA duplexes labelled with tC or tCO. Table 4 shows the measured fluorescence lifetimes and steady-state anisotropy as well as the calculated steady-state anisotropy of two tC-labelled oligonucleotides and two tCO-labelled oligonucleotides. The expected steady-state anisotropy is calculated from Equation (7), using measured lifetimes and calculated rotational correlation times. The rotational correlation times, θ1 and θ2 together with their amplitudes, β1 and β2, were calculated using the equations in Supplementary Material, S3. In the calculation the DNA duplex is modelled as a rotating cylinder with a length, L = Nbp × 3.4 Å and diameter, d = 20 Å (60–62). The calculated anisotropies for the tC oligonucleotides, 0.12 and 0.21 agree very well with the experimental values 0.12 and 0.20 for the 10-mer and 22-mer, respectively. Also for the tCO oligonucleotides, the calculated values, 0.15 and 0.23 agree very well with the experimental values 0.14 and 0.22 for the 10-mer and 21-mer, respectively.
Table 4.
Fluorescence lifetimes, experimental and calculated steady-state fluorescence anisotropy of tC- and tCO-containing double-stranded oligonucleotides
| Sequencea | Base pairs | τ (ns)b | rexp b | rcalc c |
|---|---|---|---|---|
| tC10 | 10 | 6.8 | 0.12 | 0.12 |
| tC22 | 22 | 7.0 | 0.20 | 0.20 |
| tCO10 | 10 | 4.5 | 0.14 | 0.15 |
| tCO21 | 21 | 4.5 | 0.22 | 0.23 |
aSequence name indicates the probe and the length of the oligonucleotide in number of base pairs (for full sequences, see Supplementary Material).
bMeasurements were performed in phosphate buffer (50 mM Na+, pH 7.5) at 20°C.
cCalculated steady-state anisotropies of DNA duplexes with the length, L = Nbp × 3.4 Å and the hydrodynamic diameter, d = 20 Å (60–62). For more details, see Supplementary Material.
DISCUSSION
In light of our previous findings regarding the highly fluorescent and promising DNA base analogue 1,3-diaza-2-oxophenothiazine, tC, we undertook the task of investigating the tC homolog 1,3-diaza-2-oxophenoxazine, tCO (Figure 1). It had previously been shown that tCO has good potential as a cytosine analogue, forming selective base pairs with guanine (49–51) and stabilizing DNA-RNA- (49), DNA-DNA- (50) and PNA-DNA-duplexes (51). In this study we have not only performed a more thorough investigation of effects on DNA conformation and stability upon incorporation of tCO, but more importantly, we show the first evidence that this base analogue is highly fluorescent and has very interesting and useful fluorescent properties.
Characterization of the tCO monomer shows that pH, salt, and temperature have very limited effects on tCO and its photophysics under biological conditions. This is very important for the intended use of tCO as a probe in biochemical and biophysical experiments. The nearly constant fluorescence anisotropy and LDr, the purely negative MCD, and quantum chemical calculations all support that the lowest energy absorption band of tCO at ∼365 nm is due to a single electronic transition. This will simplify both experimental work and analysis of data and, thus, yield more accurate results, especially when using techniques such as fluorescence anisotropy and FRET. Furthermore, the LD measurements and the quantum chemical calculations show that this transition is at an angle of ∼35° to the long axis of tCO and virtually in the plane of the molecule. In addition, the high fundamental fluorescence anisotropy of the matrix immobilized tCO suggests that the emission and absorption transitions are nearly parallel. The extinction coefficient for tCO at the long wavelength absorption maxima (ε360nm = 9000 M−1cm−1) is more than twice as high as for tC (ε375nm = 4000 M−1cm−1). Since the intensity of emission is proportional to the extinction coefficient and fluorescence quantum yield a high value of ε is obviously a desirable property. The characterization of the fluorophore itself is important for both experimental design and data analysis. However, even more important for future use of tCO as a fluorescent DNA base analogue is the characterization of its effect on DNA and also the effect of DNA on tCO itself, which will be discussed below.
tCO-dynamics in duplex DNA and effects on DNA structure and stability
Results from both CD and UV-melting experiments show that tCO works excellently as a cytosine analogue. The CD indicates only limited effects on the overall secondary structure of DNA and, more importantly, the spectrum is typically that of B-form DNA for all sequences. Duplex stability is on average slightly increased by incorporation of tCO (∼2.7°C). However, with a purine on the 5′ side of tCO the melting temperature is, on average, not significantly changed whereas with a pyrimidine it is increased 5°C. Hence, with an intelligent choice of the 5′ neighbour of tCO the stability of the duplex can be increased or left unchanged, thus, offering a potentially very important control. The behaviour is very similar to tC and, as suggested in that case, the enhanced stability is most likely a result of increased π–π overlap between the extended ring system of tCO and the neighbouring bases.
Further insight into the geometry and dynamics of tCO in the DNA double helix can be obtained from the absorption and emission spectra as well as from the fluorescence lifetimes. The observed single fluorescence lifetime is a strong indication of a single conformation of the fluorophore, whereas multiple lifetimes would have indicated several conformations normally suggesting fast base flipping of the fluorophore. The latter is often accompanied by a decrease in duplex stability. Additional indications of tCO being situated in a well-defined and rigid environment come from the more structured absorption spectra and especially from the emission spectra that show significant vibrational structure. Similar vibrational structure was also found for tCO in a rigid low-temperature H2O/ethylene glycol matrix (data not shown). These results suggest that tCO is firmly stacked and has a very well-defined position and geometry at least on the time scale of fluorescence. This means that any emission-monitored motion of tCO will be strongly coupled to the motion of the DNA. This property is extremely useful in, for example, fluorescence anisotropy experiments (see below).
Combining these findings with previous reports on the base-pairing selectivity, it is evident that tCO is an excellent cytosine analogue and that exchanging C for tCO will only introduce minute perturbations to the native structure of DNA.
Photophysics of tCO in single-stranded DNA
The fluorescence quantum yield of tCO in single-stranded oligonucleotides shows some sensitivity to base sequence. Four of the sequences, GG, GA, GT and GC, all containing a guanine 5′ to tCO, have 50% lower fluorescence quantum yield than the nucleoside free in solution. On the contrary, for sequences with A, T or C 5′ to tCO, it is virtually the same or slightly higher. In addition, the mean fluorescence lifetime is significantly shorter (2.8–3.0 ns) for the GG, GA, GT and GC strands compared to the other single strands (4.4–5.8 ns) and also shorter than for the free nucleoside (3.4 ns). This fluorescence quenching might be due to electron transfer from the nearby guanine, the natural DNA base with the lowest oxidation potential. In sequences AA and GAAG, tCO is surrounded by adenines, however, the next to nearest neighbours are changed from adenine and thymine in AA to two guanines in GAAG. Since both the fluorescence quantum yield and average fluorescence lifetime are virtually the same for those sequences, the effects of guanines further away from tCO seem to be negligible. Interestingly, sequences with a guanine at the 3′ side are among the ones with highest fluorescence quantum yield and longest fluorescence lifetimes. Thus, not only proximity to a guanine is important for the quenching but also subtle structural differences. Most likely, parameters such as relative orientation and stacking between tCO and neighbouring guanines play an important role.
In a preliminary study including the sequence GT-2 (see Supplementary Material for full sequence) interesting aspects of the importance of orientation for the quenching become clear. In this sequence only the order of the two bases at the 5′-end is reversed while keeping the rest of the sequence the same as for the GT. The GT-2 has a quantum yield only slightly lower (Φf = 0.27) than the free nucleoside and approximately twice as high as for GT. The average fluorescence lifetime for GT-2 is slightly lower than for the other single strands (3.9 ns) but almost 50% longer than for GT. As shown above, a change in sequence that distant from tCO should have no direct effects on the fluorescence, so the explanation has to lie elsewhere. Combining information obtained from CD and the emission spectra allowed for a deeper understanding of the structural features of the single strands. As will be discussed below, the results support our initial statement that minute structural differences play a very important role in the quenching mechanism. The CD spectra of the single strands show similar structural features for all strands, suggesting a certain degree of secondary structure in all cases. However, the four sequences GG, GA, GT and GC all have a CD signal at ∼280 nm of approximately twice the intensity of that of all other single strands including GT-2 (Supplementary Material). This suggests that these four sequences have a more ordered secondary structure compared to the other single strands. Interestingly, from the lower CD signal for GT-2 we can conclude that the small change in the sequence for GT-2 compared to GT is enough to disrupt the more ordered structure. Furthermore, the more ordered secondary structure is not a result of incorporating tCO since the same high CD signal was observed also for the unmodified single strands (Supplementary Material). This indicates that the more ordered secondary structure is an intrinsic property of these sequences. In addition, the emission spectra of GG, GA, GT and GC are more red-shifted (Table 2) and have more pronounced vibrational structure compared to the other single strands (Supplementary Material). Vibrational structure in the spectra is an indication that the fluorophore is in a rigid environment and, thus, suggests that tCO is better stacked and has a more well-defined position in GG, GA, GT and GC compared to all the other single strands. Since these four sequences are the only ones showing any significant quenching (∼50%) it is obvious, especially when comparing with GT-2, that a specific stacking interaction between the tCO and the guanine is also required for significant quenching to occur. That the quenching of tCO is so strongly dependent on relative orientation and interaction between tCO and guanine indicate that the driving force for quenching, possibly an effect of electron transfer, is relatively small.
Photophysics of tCO in double-stranded DNA
The behaviour of tCO in duplex DNA is much less complex than in single strands. Here all sequences behave virtually the same having a decreased fluorescence quantum yield compared to the nucleoside and a single fluorescence lifetime. The single fluorescence lifetime is a great advantage in techniques such as fluorescence anisotropy and FRET and has previously only been reported for one fluorescent DNA base analogue, namely tC. Despite that the fluorescence quantum yield is reduced upon duplex formation it is still 0.22 ± 0.05 in all possible combinations of nearest neighbours which in combination with ε = 9000 M−1cm−1 result in a DNA-incorporated fluorescent base analogue that is unprecedented in terms of overall average brightness (∝ ε × Φf). We would like to point out that there are other base analogues with similar quantum yields in specific sequences (7,12). However, in those cases the same base analogue can have quantum yields less then a few percent in other sequences. In contrast to the single strands, the reduced fluorescence quantum yield is not a result of quenching since the fluorescence lifetime is longer and the non-radiative rate constant is approximately the same as compared to the free tCO nucleoside (Table 3). Instead, the major change is found in the radiative rate constant, which is lowered by 35–50%. This is most likely a consequence of the electronic interaction between tCO and the other bases in the DNA stack leading to a hypochromic effect in the tCO chromophore. Thus, since the radiative rate constant is proportional to the extinction coefficient as shown by the Strickler–Berg equation (63), the radiative rate constant will be lowered by this hypochromicity. The hypochromicity of double-stranded DNA as compared to a free nucleoside is typically <40% (64) which is, within error margins, similar to the observed change in the radiative rate constant.
The sequences GG, GA, GT and GC that were quenched as single strands now behaves as all the other double strands, i.e. they do not exhibit any fluorescence quenching. This suggests that the formation of duplex effectively turns off the quenching pathway present in the single strands, supporting the notion of a quenching with a low driving force and, thus, a high sensitivity to minute structural changes.
Fluorescence anisotropy experiments on DNA using rigidly stacked base analogues
As shown in previous publications and here both tC and tCO are rigidly stacked within the DNA duplex. This suggests that these probes will report primarily on the motion of the DNA rather than any independent motion of the probe. Indeed this is what we find when comparing the experimental results of the tC and tCO sequences with the theoretically estimated values (Table 4). The experimental steady-state anisotropy values agree very well with those obtained from calculations proving that any motional freedom of tC or tCO not related to the mobility of the whole DNA is of very small amplitude. This shows the superiority of these probes compared to many other existing fluorescent DNA probes when using fluorescence anisotropy to study DNA–macromolecule interactions or in the determination of size and shape of DNA tertiary structures as well as DNA-macromolecule complexes.
CONCLUSIONS
We have shown the first evidence that the base analogue tCO is highly fluorescent and presented a thorough characterization of it both as free monomer as well as incorporated into DNA. In terms of average overall brightness (∝ ε × Φf) and, thus, high sensitivity when incorporated into DNA, tCO is unprecedented among fluorescent DNA base analogues. Only tC is close to its average brightness, but it is still approximately 3 times weaker, while other base analogues such as 2-aminopurine, pyrrolo-dC, 3-MI are one to several orders of magnitude weaker. Although tCO has many promising properties in common with tC, there is one significant difference, apart from the fact that tCO is a different colour that expands the wavelength range of highly fluorescent DNA base analogues, namely that the emission from tCO is sensitive to its DNA micro-environment. This sensitivity is something that tCO shares with virtually all other base analogues but other properties make tCO superior. Most obvious is the significantly higher brightness of tCO mentioned above and also that tCO has no significant effects on the native DNA conformation and stability. Furthermore, tCO is rigidly stacked within the double helix and it has a single fluorescence lifetime in double-stranded DNA. This makes tCO a highly interesting contributor to the growing numbers of fluorescent base analogues. The vast majority of the current analogues work well if one is only interested in monitoring emission intensity change. tCO, on the other hand, in addition to that is also very well suited in techniques such as FRET and fluorescence anisotropy.
In summary, tCO combines some of the favourable properties that make tC unique with the useful property shared by the currently exploited base analogues. Thus, tCO is suitable for use in a wide range of fluorescence applications, especially FRET, fluorescence anisotropy and fluorescence melting experiments, monitoring segmental melting of complex nucleic acid structures (Börjesson and coworkers and Sandin and coworkers; manuscripts in preparation).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGMENTS
Tomio Takahashi, Oscar Janson and Daniel Farkas are gratefully acknowledged for their preliminary initial studies for this paper. European Commission's Sixth Framework Programme (Project reference AMNA, contract no. 013575); Swedish Research Council (VR). Funding to pay the Open Access publication charges for this article was provided by the Swedish Research Council (VR).
Conflict of interest statement. None declared.
REFERENCES
- 1.Wilson JN, Kool ET. Fluorescent DNA base replacements: reporters and sensors for biological systems. Org. Biomol. Chem. 2006;4:4265–4274. doi: 10.1039/b612284c. [DOI] [PubMed] [Google Scholar]
- 2.Asseline U. Development and applications of fluorescent oligonucleotides. Curr. Org. Chem. 2006;10:491–518. [Google Scholar]
- 3.Okamoto A, Saito Y, Saito I. Design of base-discriminating fluorescent nucleosides. Photochem. Photobiol. C: Photochem. Rev. 2005;6:108–122. [Google Scholar]
- 4.Hawkins ME. Fluorescent nucleoside analogues as DNA probes. In: Lakowicz JR, editor. Topics in Fluorescence Spectroscopy: DNA Technology. Vol. 7. New York: Kluwer Academic/Plenum Publishers; 2003. pp. 151–175. [Google Scholar]
- 5.Rist MJ, Marino JP. Fluorescent nucleotide base analogs as probes of nucleic acid structure, dynamics and interactions. Curr. Org. Chem. 2002;6:775–793. [Google Scholar]
- 6.Liu CH, Martin CT. Fluorescence characterization of the transcription bubble in elongation complexes of T7 RNA polymerase. J. Mol. Biol. 2001;308:465–475. doi: 10.1006/jmbi.2001.4601. [DOI] [PubMed] [Google Scholar]
- 7.Driscoll SL, Hawkins ME, Balis FM, Pfleiderer W, Laws WR. Fluorescence properties of a new guanosine analog incorporated into small oligonucleotides. Biophys. J. 1997;73:3277–3286. doi: 10.1016/S0006-3495(97)78352-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hawkins ME, Pfleiderer W, Balis FM, Porter D, Knutson JR. Fluorescence properties of pteridine nucleoside analogs as monomers and incorporated into oligonucleotides. Anal. Biochem. 1997;244:86–95. doi: 10.1006/abio.1996.9879. [DOI] [PubMed] [Google Scholar]
- 9.Hawkins ME, Pfleiderer W, Jungmann O, Balis FM. Synthesis and fluorescence characterization of pteridine adenosine nucleoside analogs for DNA incorporation. Anal. Biochem. 2001;298:231–240. doi: 10.1006/abio.2001.5399. [DOI] [PubMed] [Google Scholar]
- 10.Godde F. A fluorescent base analog for probing triple helix formation. Antisense Nucleic Acid Drug Dev. 1998;8:469–476. doi: 10.1089/oli.1.1998.8.469. [DOI] [PubMed] [Google Scholar]
- 11.Godde F, Toulme JJ, Moreau S. 4-amino-1H-benzo[g]quinazoline-2-one: A fluorescent analog of cytosine to probe protonation sites in triplex forming oligonucleotides. Nucleic Acids Res. 2000;28:2977–2985. doi: 10.1093/nar/28.15.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Godde F, Toulme JJ, Moreau S. Benzoquinazoline derivatives as substitutes for thymine in nucleic acid complexes. Use of fluorescence emission of benzo[g]quinazoline-2,4-(1H,3H)-dione in probing duplex and triplex formation. Biochemistry. 1998;37:13765–13775. doi: 10.1021/bi9811967. [DOI] [PubMed] [Google Scholar]
- 13.Okamoto A, Tainaka K, Saito I. Synthesis and properties of a novel fluorescent nucleobase, naphthopyridopyrimidine. Tetrahedron Lett. 2003;44:6871–6874. [Google Scholar]
- 14.Okamoto A, Tainaka K, Saito I. Clear distinction of purine bases on the complementary strand by a fluorescence change of a novel fluorescent nucleoside. J. Am. Chem. Soc. 2003;125:4972–4973. doi: 10.1021/ja034090u. [DOI] [PubMed] [Google Scholar]
- 15.Okamoto A, Tanaka K, Fukuta T, Saito I. Design of base-discriminating fluorescent nucleoside and its application to T/C SNP typing. J. Am. Chem. Soc. 2003;125:9296–9297. doi: 10.1021/ja035408l. [DOI] [PubMed] [Google Scholar]
- 16.Secrist JA, Barrio JR, Leonard NJ. Fluorescent modification of adenosine-triphosphate with activity in enzyme-systems – 1,N6-ethenoadenosine triphosphate. Science. 1972;175:646–647. doi: 10.1126/science.175.4022.646. [DOI] [PubMed] [Google Scholar]
- 17.Ward DC, Reich E, Stryer L. Fluorescence studies of nucleotides and polynucleotides. I. Formycin 2-aminopurine riboside 2,6-diaminopurine riboside and their derivatives. J. Biol. Chem. 1969;244:1228–1237. [PubMed] [Google Scholar]
- 18.Coleman RS, Madaras ML. Synthesis of a novel coumarin C-riboside as a photophysical probe of oligonucleotide dynamics. J. Org. Chem. 1998;63:5700–5703. [Google Scholar]
- 19.Greco NJ, Tor Y. Simple fluorescent pyrimidine analogues detect the presence of DNA abasic sites. J. Am. Chem. Soc. 2005;127:10784–10785. doi: 10.1021/ja052000a. [DOI] [PubMed] [Google Scholar]
- 20.Kim SJ, Kool ET. Sensing metal ions with DNA building blocks: Fluorescent pyridobenzimidazole nucleosides. J. Am. Chem. Soc. 2006;128:6164–6171. doi: 10.1021/ja0581806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Augustyn KE, Wojtuszewski K, Hawkins ME, Knutson JR, Mukerji I. Examination of the premelting transition of DNA A-tracts using a fluorescent adenosine analogue. Biochemistry. 2006;45:5039–5047. doi: 10.1021/bi0518343. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Not so crystal clear: the structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andreatta D, Lustres JLP, Kovalenko SA, Ernsting NP, Murphy CJ, Coleman RS, Berg MA. Power-law solvation dynamics in DNA over six decades in time. J. Am. Chem. Soc. 2005;127:7270–7271. doi: 10.1021/ja044177v. [DOI] [PubMed] [Google Scholar]
- 24.Andreatta D, Sen S, Lustres JLP, Kovalenko SA, Ernsting NP, Murphy CJ, Coleman RS, Berg MA. Ultrafast dynamics in DNA: “fraying” at the end of the helix. J. Am. Chem. Soc. 2006;128:6885–6892. doi: 10.1021/ja0582105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu D, Evans KO, Nordlund TM. Melting and premelting transitions of an oligomer measured by DNA base fluorescence and absorption. Biochemistry. 1994;33:9592–9599. doi: 10.1021/bi00198a027. [DOI] [PubMed] [Google Scholar]
- 26.Nordlund TM, Andersson S, Nilsson L, Rigler R, Gräslund A, McLaughlin LW. Structure and dynamics of a fluorescent DNA oligomer containing the EcoRI recognition sequence: fluorescence, molecular dynamics, and NMR studies. Biochemistry. 1989;28:9095–9103. doi: 10.1021/bi00449a021. [DOI] [PubMed] [Google Scholar]
- 27.Lenz T, Bonnist EYM, Pljevaljcic G, Neely RK, Dryden DTF, Scheidig AJ, Jones AC, Weinhold E. 2-aminopurine flipped into the active site of the adenine-specific DNA methyltransferase M.TaqI: Crystal structures and time-resolved fluorescence. J. Am. Chem. Soc. 2007;129:6240–6248. doi: 10.1021/ja069366n. [DOI] [PubMed] [Google Scholar]
- 28.Arzumanov A, Godde F, Moreau S, Toulme JJ, Weeds A, Gait MJ. Use of the fluorescent nucleoside analogue benzo[g]quinazoline 2′-O-methyl-beta-D-ribofuranoside to monitor the binding of the HIV-1 Tat protein or of antisense oligonucleotides to the TAR RNA stem-loop. Helv. Chim. Acta. 2000;83:1424–1436. [Google Scholar]
- 29.Deprez E, Tauc P, Leh H, Mouscadet JF, Auclair C, Hawkins ME, Brochon JC. DNA binding induces dissociation of the multimeric form of HIV-1 integrase: A time-resolved fluorescence anisotropy study. Proc. Natl Acad. Sci. USA. 2001;98:10090–10095. doi: 10.1073/pnas.181024498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tleugabulova D, Reha-Krantz LJ. Probing DNA polymerase-DNA interactions: Examining the template strand in exonuclease complexes using 2-aminopurine fluorescence and acrylamide quenching. Biochemistry. 2007;46:6559–6569. doi: 10.1021/bi700380a. [DOI] [PubMed] [Google Scholar]
- 31.Singleton SF, Shan F, Kanan MW, McIntosh CM, Stearman CJ, Helm JS, Webb KJ. Facile synthesis of a fluorescent deoxycytidine analogue suitable for probing the RecA nucleoprotein filament. Org. Lett. 2001;3:3919–3922. doi: 10.1021/ol0167863. [DOI] [PubMed] [Google Scholar]
- 32.Hawkins ME, Pfleiderer W, Mazumder A, Pommier YG, Falls FM. Incorporation of a fluorescent guanosine analog into oligonucleotides and its application to a real-time assay for the HIV-1 integrase 3′-processing reaction. Nucleic Acids Res. 1995;23:2872–2880. doi: 10.1093/nar/23.15.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wojtuszewski K, Hawkins ME, Cole JL, Mukerji I. HU binding to DNA: Evidence for multiple complex formation and DNA bending. Biochemistry. 2001;40:2588–2598. doi: 10.1021/bi002382r. [DOI] [PubMed] [Google Scholar]
- 34.Hariharan C, Bloom LB, Helquist SA, Kool ET, Reha-Krantz LJ. Dynamics of nucleotide incorporation: Snapshots revealed by 2-aminopurine fluorescence studies. Biochemistry. 2006;45:2836–2844. doi: 10.1021/bi051644s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neely RK, Daujotyte D, Grazulis S, Magennis SW, Dryden DTF, Klimasauskas S, Jones AC. Time-resolved fluorescence of 2-aminopurine as a probe of base flipping in M.HhaI-DNA complexes. Nucleic Acids Res. 2005;33:6953–6960. doi: 10.1093/nar/gki995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Neill MA, Barton JK. DNA-mediated charge transport requires conformational motion of the DNA bases: Elimination of charge transport in rigid glasses at 77 K. J. Am. Chem. Soc. 2004;126:13234–13235. doi: 10.1021/ja0455897. [DOI] [PubMed] [Google Scholar]
- 37.O'Neill MA, Barton JK. DNA charge transport: conformationally gated hopping through stacked domains. J. Am. Chem. Soc. 2004;126:11471–11483. doi: 10.1021/ja048956n. [DOI] [PubMed] [Google Scholar]
- 38.O'Neill MA, Barton JK. 2-aminopurine: A probe of structural dynamics and charge transfer in DNA and DNA: RNA hybrids. J. Am. Chem. Soc. 2002;124:13053–13066. doi: 10.1021/ja0208198. [DOI] [PubMed] [Google Scholar]
- 39.Kelley SO, Barton JK. Electron transfer between bases in double helical DNA. Science. 1999;283:375–381. doi: 10.1126/science.283.5400.375. [DOI] [PubMed] [Google Scholar]
- 40.Srivatsan SG, Tor Y. Fluorescent pyrimidine ribonucleotide: synthesis, enzymatic incorporation, and utilization. J. Am. Chem. Soc. 2007;129:2044–2053. doi: 10.1021/ja066455r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tam VK, Kwong D, Tor Y. Fluorescent HIV-1 dimerization initiation site: design, properties, and use for ligand discovery. J. Am. Chem. Soc. 2007;129:3257–3266. doi: 10.1021/ja0675797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaul M, Barbieri CM, Pilch DS. Fluorescence-based approach for detecting and characterizing antibiotic-induced conformational changes in ribosomal RNA: comparing aminoglycoside binding to prokaryotic and eukaryotic ribosomal RNA sequences. J. Am. Chem. Soc. 2004;126:3447–3453. doi: 10.1021/ja030568i. [DOI] [PubMed] [Google Scholar]
- 43.Wilhelmsson LM, Holmén A, Lincoln P, Nielsen PE, Nordén B. A highly fluorescent DNA base analogue that forms Watson-Crick base pairs with guanine. J. Am. Chem. Soc. 2001;123:2434–2435. doi: 10.1021/ja0025797. [DOI] [PubMed] [Google Scholar]
- 44.Wilhelmsson LM, Sandin P, Holmén A, Albinsson B, Lincoln P, Nordén B. Photophysical characterization of fluorescent DNA base analogue, tC. J. Phys. Chem. B. 2003;107:9094–9101. [Google Scholar]
- 45.Engman KC, Sandin P, Osborne S, Brown T, Billeter M, Lincoln P, Nordén B, Albinsson B, Wilhelmsson LM. DNA adopts normal B-form upon incorporation of highly fluorescent DNA base analogue tC: NMR structure and UV-Vis spectroscopy characterization. Nucleic Acids Res. 2004;32:5087–5095. doi: 10.1093/nar/gkh844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sandin P, Wilhelmsson LM, Lincoln P, Powers VEC, Brown T, Albinsson B. Fluorescent properties of DNA base analogue tC upon incorporation into DNA – negligible influence of neighbouring bases on fluorescence quantum yield. Nucleic Acids Res. 2005;33:5019–5025. doi: 10.1093/nar/gki790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sandin P, Lincoln P, Brown T, Wilhelmsson LM. Synthesis and oligonucleotide incorporation of fluorescent cytosine analogue tC: a promising nucleic acid probe. Nat. Protocols. 2007;2:615–623. doi: 10.1038/nprot.2007.80. [DOI] [PubMed] [Google Scholar]
- 48.Stengel G, Gill JP, Sandin P, Wilhelmsson LM, Albinsson B, Nordén B, Millar DP. Conformational dynamics of DNA polymerase probed with a novel fluorescent DNA base analog. Biochemistry. 2007;46:12289–12297. doi: 10.1021/bi700755m. [DOI] [PubMed] [Google Scholar]
- 49.Lin KY, Jones RJ, Matteucci M. Tricyclic 2′-deoxycytidine analogs – syntheses and incorporation into oligodeoxynucleotides which have enhanced binding to complementary RNA. J. Am. Chem. Soc. 1995;117:3873–3874. [Google Scholar]
- 50.Lin KY, Matteucci MD. A cytosine analogue capable of clamp-like binding to a guanine in helical nucleic acids. J. Am. Chem. Soc. 1998;120:8531–8532. [Google Scholar]
- 51.Rajeev KG, Maier MA, Lesnik EA, Manoharan M. High-affinity peptide nucleic acid oligomers containing tricyclic cytosine analogues. Org. Lett. 2002;4:4395–4398. doi: 10.1021/ol027026a. [DOI] [PubMed] [Google Scholar]
- 52.Ausin C, Ortega JA, Robles J, Grandas A, Pedroso E. Synthesis of amino- and guanidino-G-clamp PNA monomers. Org. Lett. 2002;4:4073–4075. doi: 10.1021/ol026815p. [DOI] [PubMed] [Google Scholar]
- 53.Flanagan WM, Wolf JJ, Olson P, Grant D, Lin KY, Wagner RW, Matteucci MD. A cytosine analog that confers enhanced potency to antisense oligonucleotides. Proc. Natl Acad. Sci. USA. 1999;96:3513–3518. doi: 10.1073/pnas.96.7.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barhate N, Cekan P, Massey AP, Sigurdsson ST. A nucleoside that contains a rigid nitroxide spin label: a fluorophore in disguise. Angew. Chem., Int. Ed. 2007;46:2655–2658. doi: 10.1002/anie.200603993. [DOI] [PubMed] [Google Scholar]
- 55.Dawson RMC, Elliot DC, Elliot WH, Jones KM. Data for biochemical reaserch. New York: Oxford University Press; 1986. [Google Scholar]
- 56.Melhuish WH. Quantum efficiencies of fluorescence of organic substances – effect of solvent and concentration of fluorescent solute. J. Phys. Chem. 1961;65:229–235. [Google Scholar]
- 57.Lakowicz JR. Principles of Fluorescence Spectroscopy. New York: Kluwer Academic/Plenum Publishers; 1999. [Google Scholar]
- 58.Tirado MM, Garcia de la Torre J. Rotational-dynamics of rigid, symmetric top macromolecules – application to circular-cylinders. J. Chem. Phys. 1980;73:1986–1993. [Google Scholar]
- 59.Nordén B. Formalistic framework of linear dichroism spectroscopy. III. Simple formulas for dichroism analysis – orientation of solutes in stretched polymer matrices. J. Chem. Phys. 1980;72:5032–5038. [Google Scholar]
- 60.Duhamel J, Kanyo J, Dinter-Gottlieb G, Lu P. Fluorescence emission of ethidium bromide intercalated in defined DNA duplexes: evaluation of hydrodynamics components. Biochemistry. 1996;35:16687–16697. doi: 10.1021/bi9610919. [DOI] [PubMed] [Google Scholar]
- 61.Eimer W, Williamson JR, Boxer SG, Pecora R. Characterization of the overall and internal dynamics of short oligonucleotides by depolarized dynamic light-scattering and NMR relaxation measurements. Biochemistry. 1990;29:799–811. doi: 10.1021/bi00455a030. [DOI] [PubMed] [Google Scholar]
- 62.Nuutero S, Fujimoto BS, Flynn PF, Reid BR, Ribeiro NS, Schurr JM. The amplitude of local angular motion of purines in DNA in solution. Biopolymers. 1994;34:463–480. doi: 10.1002/bip.360340404. [DOI] [PubMed] [Google Scholar]
- 63.Strickler SJ, Berg RA. Relationship between absorption intensity and fluorescence lifetime of molecules. J. Chem. Phys. 1962;37:814–822. [Google Scholar]
- 64.Bloomfield VA, Crothers DM, Tinoco I. Nucleic Acids; Structures, Properties, and Functions. Sausalito, CA: University Science Books; 1999. [Google Scholar]