Abstract
The in-depth structure/function analysis of large protein complexes, such as RNA polymerases (RNAPs), requires an experimental platform capable of assembling variants of such enzymes in large numbers in a reproducible manner under defined in vitro conditions. Here we describe a streamlined and integrated protocol for assembling recombinant archaeal RNAPs in a high-throughput 96-well format. All aspects of the procedure including construction of redesigned expression plasmids, development of automated protein extraction/in vitro assembly methods and activity assays were specifically adapted for implementation on robotic platforms. The optimized strategy allows the parallel assembly and activity assay of 96 recombinant RNAPs (including wild-type and mutant variants) with little or no human intervention within 24 h. We demonstrate the high-throughput potential of this system by evaluating the side-chain requirements of a single amino acid position of the RNAP Bridge Helix using saturation mutagenesis.
INTRODUCTION
Many of the enzymes involved in molecular information processing are large and complex molecular machines that are assembled from a variety of subunits. Recent technical advances in X-ray crystallography have started to yield insights into the structure of protein complexes, such as bacterial and eukaryotic RNA polymerases (RNAPs; 1–6). RNAPs are key components of the cellular transcriptional machineries. The availability of high-resolution structures of such enzymes has inspired numerous structural models attempting to explain key RNAP functions in mechanistic terms. Some of these models are based on features of individual structures [e.g. separation of the transcript from the DNA template strand (7)], whereas other mechanisms have been proposed by comparing the conformations of protein domains in different crystals or by co-crystallizing polymerases in the presence of various substrates or cofactors (e.g. 8,9). It is important to recognize that X-ray structures can only provide individual ‘snap-shots’ of molecular events likely to involve considerable conformational changes and/or short-lived intermediates. Predictions based on such static models must therefore be tested extensively by targeted mutagenesis of key domains (and individual residues therein) to assess their functional contributions (10). We believe that archaeal transcription systems, and specifically archaeal RNAPs, provide an ideal testing ground for the development of high-throughput experimental strategies to obtain comprehensive insights into their structure/function space. Archaea contain a single RNAP and a reduced set of basal transcription factors that closely resemble the eukaryotic core RNAPII transcriptional machinery (11). Technical obstacles preventing the in vitro assembly of catalytically active eukaryotic RNAPs were a key factor that originally motivated us to develop a method for the assembly of archaeal RNAPs from recombinant subunits (12). The fully recombinant system resulting from these efforts reproduces faithfully all known aspects of archaeal in vitro transcription systems, including promoter-directed transcription and activator-stimulated transcription (12–15).
The availability of such recombinant in vitro transcription systems sets the scene for the introduction of any desired mutation into any position within an archaeal RNAP by incorporating a suitably mutated subunit into the in vitro assembly reactions. Here we describe an integrated experimental strategy that takes full advantage of these features of the recombinant archaeal RNAP system and allows the formulation of a high-throughput approach by combining efficient mutagenesis with a highly automated protein purification/in vitro assembly process. We scrutinized every aspect of the previously developed in vitro assembly method to convert a long and complex manual method into a streamlined and simplified automated procedure that is capable of producing and assaying hundreds of individually assembled enzyme variants per week.
MATERIALS AND METHODS
Re-engineered bacterial expression vectors for targeted mutagenesis
The original expression vectors for the RNA polymerase subunits A′ and B′′ from the euryarchaeal hyperthermophile Methanocaldococcus jannaschii (mjA′ and mjB′′, respectively) were generated by subcloning the full-length, PCR-generated open reading frames into pET21a (12). For this study, the pET21a-mjA′ vector was modified by replacing the N-terminal portion of the reading frame (NdeI-AleI) with a synthetic gene fragment (GenScript) encoding the identical amino acid sequence, but containing codons optimized for Escherichia coli expression and with unique restriction enzyme targets flanking the Zipper and Lid encoding sequences. The C-terminal portion (BstBI-BamHI) of pET21a-mjA′ was also replaced with a synthetic gene fragment (GenScript) containing optimized codons and unique restriction enzyme targets flanking the Bridge Helix. A similar modification (replacement of the BseYI-EcoRI fragment with a synthetic gene construct) was carried out on pET21a-mjB′′ to allow mutagenesis of the Fork Loop domains. Further details/full sequences/samples of the modified constructs are available from the corresponding author on request.
Robotic platforms
All robotic manipulations described here were carried out on either of the two Theonyx Liquid Performers (Aviso Trade GmbH), equipped with an 8-tip pipetting arm, separate gripper arm, barcode reader, vacuum manifold, microplate shaker, thermocycler, microplate reader, 96-well microdialyzer and E-Page facility (Invitrogen) and an integrated microplate centrifuge. Detailed descriptions of all robotic procedures, platform layouts and specialized hardware modifications are available from the corresponding author upon request.
Bacterial growth and harvest
Single E. coli ‘Acella’ (EdgeBio) colonies containing wildtype or mutant pET expression constructs were inoculated into 1.5 ml of autoinduction growth medium (16; Novagen ‘Overnight Express’) containing 100 μg/ml ampicillin in a 24-deepwell plate. The plates were covered with a ‘BugStopper’ venting capmat (Whatman), barcoded and incubated in a shaking incubator for 18 h at 37°C at 250 r.p.m. The plates were then transferred to the robotic platform (1–4 plates per run). After reading the barcode, each plate was transferred to the robotic shaker for 10 s (to ensure an even cell suspension) before 10 μl of each culture were transferred to a 96-well plate for automatically measuring the cell density at 600 nm (A600) after a 1/10 dilution step. This step documents cultures that either did not grow or failed to reach the expected cell density.
Recombinant protein extraction
All procedures described subsequently were carried out on the robotic platforms at room temperature. Nine hundred microlitres of each bacterial expression culture were transferred from the 24-well growth plates to a defined position in a 2.2 ml 96-deepwell block and then lysed directly, without prior centrifugation, by the addition of a mixture of 100 μl FastBreak reagent (Promega) and 2 μl Lysonase (Novagen). The lysates were mixed extensively by repeated up- and down-pipetting and occasional vortexing on the robotic microplate shaker at 5 min intervals for a total of 30 min at room temperature. Inclusion bodies were then separated from the lysate using the robotic centrifuge (10 min at 4700 r.p.m; ∼2300 g), washed once with 950 μl TG100 buffer (25 mM Tris base; 192 mM glycine; pH 8.9) by vigorous robotic pipetting and plate shaking. To ensure complete automation of the entire procedure we developed a custom-built microplate decanter that automatically decants the supernatants without disturbing the inclusion body pellets (R.O.J.W., unpublished data). After a further centrifugation/automatic decanting step the washed inclusion bodies were solubilized by the addition of 500 μl of TG100 buffer containing near-saturating urea (8.3 M) by robotic pipetting and plate shaking. We chose the TG buffer system to provide a vast excess of free amino groups which would compete for the carbamylation reactions that might occur in the urea-containing in vitro assembly reactions (see subsequently). The protein concentrations of the resulting solubilized inclusion body preparations were measured by mixing 10 μl with 190 μl Bicinchonic Acid (BCA) protein assay reagent (Sigma) in a clear microplate and measuring the absorption at 562 nm after incubation at 37°C for 2 h. Different concentrations of bovine serum albumin (BSA) dissolved in TG100/8.3 M urea were set up at the same time to establish a standard curve. Out of several other assays tested, we found that the BCA assay performed best in the presence of the high urea concentrations used and gave an essentially linear read-out in the 0.1–10 mg/ml range. We previously used β-mercaptoethanol (15 mM) in all of our subunit purification buffers (12), but this compound interferes with the BCA assay and was therefore omitted without any apparent loss of activity of the recombinant subunits after in vitro assembly (R.O.J.W., unpublished observations). Based on comparisons with the BSA standards and on the staining of the purified subunits on gels we typically obtained 250 μg of purified recombinant subunits from 900 μl expression culture with a standard deviation in the concentrations of individual subunit preparations of less than ±10%.
Automatic assembly of recombinant RNAPs in 96-well format
A commercially available 96-well microdialysis device (Spectrum Laboratories) was installed on top of a Hi300N magnetic stirrer (Hanna Instruments) positioned on the robotic platform. The dialyzer position is fully accessible to the pipetting and gripper arms of the robots. The perspex lid was customized for handling by the gripper (details available from the corresponding author). A peristaltic pump (Watson Marlow Sci400) was calibrated to deliver a flow-rate of 1 ml/min and switched on and off under robotic control. In vitro assembly reactions were set up in a total volume of 100 μl per dialysis well and a dialysis membrane with a 3 K cut-off point was used for all assemblies (Spectrum Laboratories; a narrow pore membrane is required to prevent the smallest subunits from leaking into the buffer chamber under denaturing conditions). The concentrations of the various RNAP subunits and subunit ratios were empirically determined to yield recombinant RNAPs displaying the same specific activities in non-specific and factor-dependent transcription assays as previously described for the wild-type enzyme (12). For the example shown here (mjA′ G825-X mutagenesis set), 90 μl of a ‘Master Mix’ containing 3.2 μg A′′, 4.2 μg B′, 6 μg B′′, 7.2 μg D, 8.7 μg L, 6.5 μg H, 5 μg N and 3 μg P were mixed with 10 μl of the robotically purified mjA′ mutant subunit (∼12 μg). The stoichiometric ratios of the subunits required for optimal in vitro assembly most likely reflect differences in the relative refolding efficiencies. Under these conditions the subunits present in the Master Mix (-A′) are rate-limiting in the assembly reactions, thus ensuring that the added mutant mjA′ subunits are present in >2 molar excess. Any minor variation in the concentrations of various mjA′ samples is thus compensated for and does not affect the amount of assembled polymerase (the presence of excess mjA′ has no detectable negative effect on the efficiency of the assembly reaction).
High-throughput transcription assays
The following procedures were carried out automatically without human intervention. Non-specific transcription assays (measuring the incorporation of α-32P-rUTP into acid insoluble transcripts) were set up as 50 μl reactions in a 96-well thin wall plate using reaction conditions previously described (12,14). After incubation for 45 min at 70°C (using the integrated thermocycler with heated lid) the plate was transferred to a temperature-regulated block maintained at 1°C. The precipitation of the radiolabelled transcripts was initiated by the addition of 150 μl 15% ice-cold trichloroacetic acid (TCA), followed by incubation for 30 min at 1°C. The precipitation reactions were then transferred by pipetting onto a 96- GF/F glass fiber filter plate (Whatman) positioned on the robotic vacuum platform. The soluble portion of the reactions was filtered to waste. The precipitates immobilized on the filter surface were washed eight times with 200 μl ice-cold 10% TCA to remove traces of unincorporated nucleotides, twice with 200 μl ice-cold 95% ethanol and then dried by a final application of vacuum. The incorporated α-32P-rUTP levels were measured after addition of 50 μl of scintillant (Optifluor-O) on a microplate counter (TopCount NXT, Packard) and were found to be identical to the results obtained with the previously described manual technique (12,14).
RESULTS
Expression vector redesign
Structural studies of RNAPs have revealed that only a relatively small proportion of amino acid residues are likely to play any direct functional role in transcription mechanisms. These residues are typically arranged within structurally distinct domains [such as Fork Loops, Bridge Helix, Lid, Zipper, Rudder (2,3)] that contact substrates (DNA, RNA, nucleotides) directly and/or interact with other protein domains within the active site (1–4,7,8). Residues located within such domains are thus prime targets for site-directed mutagenesis projects. Moreover, these functional domains tend to be either inherently unstructured [e.g. Fork and Trigger Loops (2–4)] or are part of flexible α-helical structures [e.g. Bridge Helix (1,2) and Trigger Loop (8)] so that single amino acid substitutions are unlikely to interfere with subunit assembly or overall stability of the enzyme.
The RNAP from the archaeon M. jannaschii (mjRNAP) contains 12 different subunits that are structurally and functionally highly similar to their counterparts in eukaryotic RNAPII (11,12). A high-throughput functional analysis of a particular functional domain, such as the Bridge Helix located in the archaeal A′ subunit, requires an efficient method for creating the desired mutants, followed by a robust method for purifying the mutated subunits in a highly reproducible manner. The resulting purified mutant protein preparations then need to be combined with a ‘master mix’ of all other essential subunits and assembled into separate recombinant RNAPs. Finally, the enzymes containing the mutated subunits need to be assayed and compared to the wild-type polymerase to assess the phenotypic consequence of the introduced mutations.
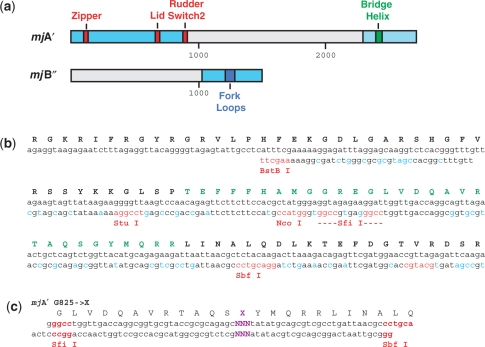
We started the design of the high-throughput assembly strategy by redesigning the previously used bacterial expression vectors encoding the mjA′ and mjB′′ subunits (12). These two subunits include many of the identified functional motifs surrounding the active site (mjA′: Zipper, Lid, Rudder, Switch-2, Bridge Helix; mjB′′: Fork Loop 1, Fork Loop 2; Figure 1a). The redesigned sequences were codon-optimized for optimal translation efficiency in E. coli, and also contain unique 6- and 8-bp target sites for restriction enzymes that flank the nucleotide sequences encoding the various functional motifs (Figure 1b). The introduced restriction enzyme sites are silent, i.e. encode the ‘wild-type’ primary amino acid sequence of the encoded proteins. Cutting these modified expression vectors with two restriction enzymes flanking a particular functional domain releases a short piece of double-stranded DNA and allows this sequence to be replaced by a double-stranded synthetic oligonucleotide encoding the desired codon replacement(s) (Figure 1c). Using this method, we have up to now successfully created systematic substitution mutants in several RNAP motifs, including Fork Loop 1 and 2, Lid, Zipper and the Bridge Helix (L.T., D.T., H.C. and R.O.J.W., unpublished results). Apart from the ease of creating a variety of mutations, one of the main advantages of this method of creating mutants is that it allows the saturation mutagenesis of a short region (typically 30–80 nt, depending on the length of the chosen motif) without affecting any of the other regions outside the targeted area. This mutagenesis strategy therefore offers many advantages over frequently used ‘shotgun’ mutagenesis methods, such as error-prone PCR (17), by limiting the mutagenic changes to distinct regions of an open reading frame.
Figure 1.
Redesign of the bacterial expression vectors. (a) Schematic diagram of the redesigned coding regions of the mjA′ and mjB′′ bacterial expression vectors. The full-length open reading frames are shown with the codon-optimized sequences in light blue. The position of the sequences encoding key functional RNAP domains are shown in color (Zipper, Lid, Rudder, Switch-2 in red, Bridge Helix in dark green and Fork Loops in blue). The numbers refer to nucleotide positions. (b) Original and redesigned sequence encoding the Bridge Helix domain and surroundings (positions 2281-2550 in the mjA′ open reading frame). The original nucleotide sequence (as found in the M. jannaschii genome; 23) is shown on the top line with the redesigned sequence (starting at the BstBI site) beneath. The silently introduced restriction sites flanking various portions of the Bridge Helix-encoding sequence are in red, whereas other sequence alterations that improve codon usage in E. coli are shown in light blue. The amino acid sequence encoded is shown in single-letter code in black and the Bridge Helix primary sequence is highlighted in green. (c) Example of mutagenic oligonucleotides for creating targeted substitutions in Bridge Helix residues mjA′-G825. The two strands are designed to hybridize to each other to create a double-stranded ‘cassette’ bearing single-stranded extensions (shown in red) suitable for ligating into cloning vectors cleaved with suitable restriction enzymes. The cassettes shown have been used to replace the wild-type Bridge Helix sequence between the SfiI and SbfI sites (as shown in panel B) with a sequence containing a single randomized codon (shown in purple; 18).
High-throughput expression and purification of RNAP subunit variants
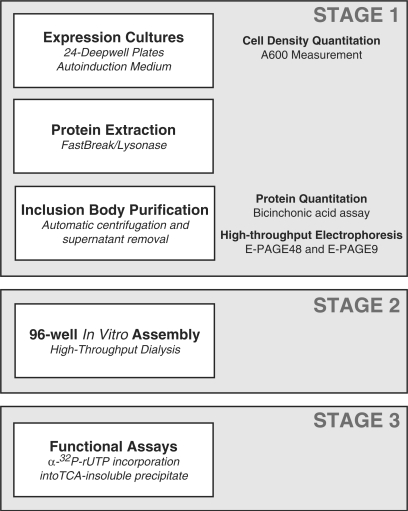
The previous section described an approach capable of creating a broad range of mutations restricted to ‘functional’ residues. The resulting bacterial expression vectors encode recombinant RNAP subunits (mjA′ or mjB′′) that contain a variety of predetermined mutations in one or more residues. This is, however, only the first step of a more complex procedure since the mutant subunits need to be assembled in the presence of all other essential subunits into an intact recombinant RNAP before the functional consequences of a particular mutation can be assessed. These robotically implemented procedures take place in parallel (up to 96 independent purification/assemblies per run) to maximize consistency and to create optimal conditions for functional comparisons. In addition, the technical solutions outlined below take full advantage of efforts to miniaturize protein expression/purification methods and the extensive use of automation/robotic capabilities. A flowchart that summarizes the entire sequence of expression/purification procedures is shown in Figure 2.
Figure 2.
Flow diagram of the automated subunit purification and assembly process. See text for further details.
The need for processing a large number of expression constructs resulting from the mutagenesis strategy described above preclude the previously described methods of growth of bacterial cultures on a ‘preparative’ scale (typically 0.5–2.0 l), followed by sonication and chromatographic purification of the recombinant subunits (12). Instead, by growing the constructs in 24-deepwell plates containing autoinduction medium (16) we were routinely able to obtain ∼250 μg of recombinant wild-type or mutant mjA′ and mjB′′ subunits from 900 μl overnight cultures at a satisfactory level of purity using robotic procedures. Briefly, after transfer to a 96-deepwell culture plate, 900 μl of each bacterial expression culture were lysed directly (i.e. without centrifugation) by the addition of a detergent/lysozyme/nuclease mixture. Since the mjA′ and mjB′′ subunits form insoluble inclusion bodies they can be purified from the lysate by robotic centrifugation and automatic supernatant decanting. The retained inclusion bodies are recovered by solubilizing them directly in a low ionic-strength buffer containing saturated urea. At this stage, the mjA′ or mjB′′ subunit preparations are >95% pure and are ready for assembly into recombinant RNAPs. The protein concentrations of the purified subunits are automatically determined on the robot using a colorimetric assay and, as an optional step the proteins can be characterized further by high-throughput electrophoresis (Supplementary Figure S1). The simplicity of the automated procedure ensures an exceptionally high degree of consistence in the quality and quantity of the recombinant subunits prepared in this manner, thus contributing greatly to the ability to compare the activities of the recombinant RNAPs produced at the next stage.
High-throughput in vitro assembly of recombinant RNAP variants
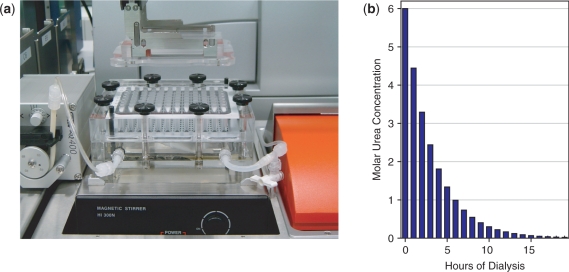
Active archaeal RNAP can be reconstituted by mixing the various subunits in appropriate ratios under denaturing conditions (6 M urea), followed by the time-controlled removal of urea by dialysis against buffers containing decreasing urea concentrations (12,15). The originally described methods were time-consuming (because the buffers were changed manually according to a strictly defined timetable) and low-throughput (parallel assembly of only a few recombinant RNAP preparations per experiment). In addition, variations in the assembly conditions (e.g. due to differences in protein/buffer preparations) contributed to poor quality control in such procedures. We therefore introduced two significant technical changes aimed at increasing throughput and reproducibility. First, we changed the dialysis set-up by using a microdialyzer device on the robotic platform that allows the in vitro assembly of up to 96 recombinant RNAPs in parallel (Figure 3a). The dialysis wells on the microdialyzer are arranged in the standardized 96-well format that facilitates the robotic transfer of liquids in and out of the dialysis block. Each well typically holds 100 μl of dialyzate that is separated from a lower communal buffer chamber by a dialysis membrane. The communal buffer chamber contains inlet and outlet ports that allow dialysis buffer from an external supply to be pumped through the chamber (Figure 3a). This is an important consideration in light of the other modification we made in the procedure: instead of manual changes of dialysis buffer to lower the urea concentration during the assembly reaction we decided that the reproducibility of the procedure could be improved substantially if the buffer changes were carried out automatically (similar to an independently developed method; 18). We therefore modified the renaturation protocol by initially filling the chamber with ∼200 ml of buffer containing 6 M urea. The RNAP subunit mixtures are then robotically pipetted into the wells and dialyzed against the 6 M urea buffer. After 1 h a pump is switched on under robotic control to pump urea-free buffer into the chamber at a low rate (typically 1 ml/minute). The urea-free buffer is immediately mixed with the 6 M urea buffer in the chamber by a magnetic stirrer and excess liquid is allowed to drain through the outlet port. This simple set-up therefore leads to a slow and continuous dilution of the urea concentration in the dialysis buffer in the lower chamber, resulting in a corresponding drop of the urea concentration in the RNAP subunit mixtures located across the dialysis membrane. Over a period of around 16–17 h the urea concentration in the dialysis chamber approaches zero and the assembly reaction is complete (Figure 3b). The assembled recombinant RNAPs are robotically pipetted out of the dialysis wells and transferred to 96-well microplates for immediate use in various functional assays. The robotic systems carrying out these procedures thus constitute a genuine ‘RNA polymerase factory’ capable of purifying and assembling up to 96 recombinant archaeal multi-subunit complexes with little or no human intervention.
Figure 3.
Robotic microdialysis set-up for assembly of recombinant RNAPs. (a) The photograph shows the placement of the 96-well microdialysis device on the robotic platform. A magnetic stirrer is sunk into the platform with full manual access to the stir speed control dial. The lid of the microdialysis device is held by the gripper arm of the robot during the loading and removal of the assembly reactions. On the left hand side, the peristaltic pump controlling the flow/dilution rate of the dialysis buffer can be seen. (b) Calculated urea concentration of the dilution of 200 ml 6M urea-containing buffer with urea-free buffer at a flow of 1 ml/min. After 17 h, the urea concentration in the dialysis buffer has effectively fallen to zero. The change of urea concentration (dc) over time (dt) can be expressed as a differential function (dc/dt) that is equal to the dilution (-D) of the initial concentration (c0) of urea according to the following equation ct = c0 × e −Dxt [D, Dilution (i.e. flowrate/volume); ct, urea concentration at time t; e, Euler's constant].
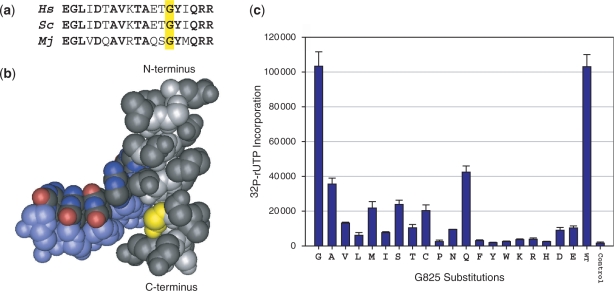
Case study: saturation mutagenesis of mjA′-G825
We demonstrate the effectiveness, reproducibility and high-throughput potential of the RNA polymerase factory strategy by showing examples of results obtained by saturation mutagenesis of a single residue of the Bridge Helix of archaeal RNAP. Briefly, the Bridge Helix is a highly conserved structure (Figure 4a) spanning the cleft near the active site in bacterial, archaeal and eukaryotic multi-subunit RNAPs (1–4). It is thought to play an important (though still controversial) role during the RNAP transcription cycle by defining the position of the template strand and of the DNA/RNA hybrid in the catalytic site (Figure 4b). Inspection of the yeast RNAPII X-ray structure of an elongation complex (7; PDB #1SFO) suggests that an evolutionary invariant residue (scRPB1-G835 in yeast RNAPII, orthologous to mjA′-G825 in M. jannaschi; Figure 4a) may have been specifically selected for the small size of its side chain to allow an unimpeded passage of the DNA template strand into the active site. The available structural data predicts that any other amino acid substitutions in this particular location would result in decreased catalytic activity due to steric clashes between the larger side chains and the phosphodiester backbone of the DNA template strand. To test this hypothesis we created a set of mjA′ mutants containing all 19 possible amino acid substitutions (the ‘mjA′ G825-X’ set) by substituting the SfiI-SbfI fragment of the redesigned mjA′ expression vector with a double-stranded synthetic oligonucleotide containing a randomized codon (19) in the position encoding G825 (Figure 1c). A complete set of amino acid substitutions in G825 was selected from ∼60 randomly picked clones and by cloning double-stranded oligonucleotides designed to contain any missing substitutions. Plasmids encoding the mjA′ G825-X subunit variants were grown in expression strains and processed by the RNA polymerase factory. The results obtained confirm that the size of the side chain does indeed appear to play a key role for the residue at position 825 (Figure 4c). The activities of recombinant RNAPs containing the other 19 possible residues in this position reach at the most 40% of the wild-type catalytic activity and many of the more bulky amino acid substitutions (F, Y, W, K, R, H) result in <5% activity in non-specific transcription assays. The wide dynamic range of reproducible activity measurements demonstrated by the mjA′ G825-X mutant set show that the RNA polymerase factory is a robust experimental tool capable of providing functional data at an unprecedented speed and accuracy.
Figure 4.
Saturation mutagenesis of mjA′-G825. (a) The sequence alignment shows part of the central and C-terminal region of the Bridge Helix [Hs, Homo sapiens RPB1 (positions 845–863); Sc, Saccharomyces cerevisae RPB1 (positions 822–840); Mj, M. jannaschii A′ (positions 812–830)]. Bold residues are identical in all three species and the residues orthologous to mjA′-G825 are marked by an yellow box. (b) The position of the glycine residue in yeast RNAPII corresponding to mjA′-G825 is shown in yellow. The Bridge Helix (including the residues aligned in a) is in gray and the backbone of the DNA template strand is shown in light blue. Note the close spatial proximity between the G825 side chain and the phosphodiester backbone of the DNA [data from PDB #1SFO (5) visualized with Cn3D; www.ncbi.nlm.nih.gov/Structure/CN3D/cn3d.shtml]. (c) Transcription activity assays of RNAP variants containing all 19 possible substitutions of mjA′-G825 carried by the RNA polymerase factory. The two columns on the right represent the positive (WT; wild-type) and negative (RNAP containing a mjA′-T821 termination codon substitution) control reactions. Error bars shown represent standard deviations (n = 4) based on independently expressed, assembled and assayed mutants carried out in parallel.
DISCUSSION
The ability to routinely produce and assay up to 96 recombinant RNAPs within 24 h in a highly automated manner has many advantages. The most obvious advantage is that this system is capable of testing sample numbers that exceed the capability of most human laboratory workers, especially if carried out on a continuous basis over longer periods. The increased throughput translates into an ability to screen a larger number of mutants (e.g. a library of random mutants spread over a large segment of a subunit) for particular functional properties using a variety of automated assays. Alternatively, the ability to process a substantial number of samples can be used to measure the activity of a smaller number of mutants with a high degree of accuracy by increasing the amount of information available for statistical analysis. In the examples shown here, it is clear that a comparison of various mutant and wild-type activities can be achieved with a high degree of accuracy due to the optimized and robust design of the procedure.
A slightly less obvious (but in our view equally important) key feature of the RNA polymerase factory is that the robotic work flow does not merely speed up a particular subset of steps within a long protocol, but actually encompasses the entire procedure. We have even gone as far as eliminating a rather trivial human intervention (decanting of supernatants from deep-well plate after centrifugation) by constructing a custom-built device specifically for the purpose of removing a possible source of human error (R.O.J.W., unpublished data). Using the procedure as currently implemented it is therefore possible to start with a bacterial expression strain (containing a plasmid with an easily verifiable DNA sequence) and to end up with a fully assembled RNAP and an activity result within 24 h. All the intermediate steps are carried out on robotic platforms, thus essentially eliminating the possibility of error and/or sample mix-ups. In the worst case scenario, any unexpected or questionable result can be easily checked by repeating the process and, ultimately by checking the DNA sequence of the expression clone(s) used. The high degree of reproducibility and comparability observed and documented is also due to the fact that variants located within a single subunit (such as the Bridge Helix mutants shown in Figure 4) are assembled with a constant cocktail of all the other subunits, which ensures a consistency not only within a single experiment, but also between independent robotic runs.
In summary, the robotic platform for the automated purification of mutant RNAP subunit variants, combined with reproducible methods for assembling them into recombinant RNAPs, provides a powerful new tool for carrying out a detailed analysis of various functional domains. The time-consuming nature of studying the effects of mutations in the context of multi-protein complexes means that conventional mutagenesis studies are typically restricted to deleting whole domains, ‘alanine scanning’ (20), or the replacement of a small number of key residues with a few selected substitutions. The robotic RNA polymerase factory approach eliminates all the bottlenecks typically encountered in the manual procedure and thus increases experimental throughput by at least one order of magnitude. This enhanced capacity has encouraged us to initiate high-throughput mutagenesis studies based on hundreds of site-specific and random mutations in various functional domains of archaeal RNAPs. In preliminary work, we have already obtained several unexpected results that would probably have gone undetected with the conventional manual approaches (R.O.J.W. et al., unpublished observations). This shows that this experimental strategy, although initially driven by an assumption-free systematic mutagenesis process, has a high degree of ‘hypothesis-generating’ potential, similar to other current structural genomics endeavors (21).
One of the major challenges that will need to be addressed in the foreseeable future is the development of new types of high-throughput assays capable of analyzing the functional properties of the large number of mutants that can be generated by the RNAP factory approach. For studying mutants in regions that have a direct influence on the catalytic activity of the polymerase (such as the Bridge Helix example described here), relatively simple assays capable of measuring overall nucleotide incorporation can be used to compare the activities of targeted substitutions directly with the wild-type enzyme (Figure 4 and R.O.J.W. et al., unpublished observations). It is, however, likely that the full characterization of the functional properties of mutants in other domains will depend on the development of additional high-throughput assays designed to test specific subfunctions of the enzymes (e.g. elongation rate). Most of the currently available manual in vitro transcription assays are gel- and autoradiography based (and thus not suitable for implementation on robotic platforms), but the development of new fluorescence-based methodologies (e.g.22) is likely to change this situation in the foreseeable future. The capacity for producing large volumes of functional data generated by such high-throughput robotic approaches will supplement available structural insights and may eventually need to be systematically stored in standardized electronic formats similar to the current literature, sequence and structural databases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
ACKNOWLEDGEMENTS
The Work described here was funded by grants to R.O.J.W. (BBSRC: BB/E000975/1 and BB/D5230001/1, MRC G0501703 and Wellcome Trust: 072563/Z/03/Z and 078043/Z/05/Z). We would like to thank M. Chatzigeorgiou for his help with the preparation of some of the G825 mutants. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 2.Cramer P, Bushnell DA, Fu J, Gnatt AL, Maier-Davis B, Thompson NE, Burgess RR, Edwards AM, David PR, Kornberg RD. Architecture of RNA polymerase II and implications for the transcription mechanism. Science. 2000;288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- 3.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 Å resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 4.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 5.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 6.Boeger H, Bushnell DA, Davis R, Griesenbeck J, Lorch Y, Strattan JS, Westover KD, Kornberg RD. Structural basis of eukaryotic gene transcription. FEBS Lett. 2005;579:899–903. doi: 10.1016/j.febslet.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 7.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science. 2004;303:1014–1016. doi: 10.1126/science.1090839. [DOI] [PubMed] [Google Scholar]
- 8.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 10.Trinh V, Langelier MF, Archambault J, Coulombe B. Structural perspective on mutations affecting the function of multisubunit RNA polymerases. Microbiol. Mol. Biol. Rev. 2006;70:12–36. doi: 10.1128/MMBR.70.1.12-36.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartlett MS. Determinants of transcription initiation by archaeal RNA polymerase. Curr. Opin. Microbiol. 2005;8:677–684. doi: 10.1016/j.mib.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Werner F, Weinzierl RO. A recombinant RNA polymerase II-like enzyme capable of promoter-specific transcription. Mol. Cell. 2002;10:635–646. doi: 10.1016/s1097-2765(02)00629-9. [DOI] [PubMed] [Google Scholar]
- 13.Ouhammouch M, Werner F, Weinzierl RO, Geiduschek EP. A fully recombinant system for activator-dependent archaeal transcription. J. Biol. Chem. 2004;279:51719–51721. doi: 10.1074/jbc.C400446200. [DOI] [PubMed] [Google Scholar]
- 14.Werner F, Weinzierl RO. Direct modulation of RNA polymerase core functions by basal transcription factors. Mol. Cell. Biol. 2005;25:8344–8355. doi: 10.1128/MCB.25.18.8344-8355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naji S, Grunberg S, Thomm M. The RPB7 ortholog E' is required for transcriptional activity of a reconstituted archaeal core enzyme at low temperatures and stimulates open complex formation. J. Biol. Chem. 2007;282:11047–11057. doi: 10.1074/jbc.M611674200. [DOI] [PubMed] [Google Scholar]
- 16.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 17.Ling MM, Robinson BH. Approaches to DNA mutagenesis: an overview. Anal. Biochem. 1997;254:157–178. doi: 10.1006/abio.1997.2428. [DOI] [PubMed] [Google Scholar]
- 18.Sorensen HP, Sperling-Petersen HU, Mortensen KK. Dialysis strategies for protein refolding: preparative streptavidin production. Protein Expr. Purif. 2003;31:149–154. doi: 10.1016/s1046-5928(03)00133-5. [DOI] [PubMed] [Google Scholar]
- 19.Steffens DL, Williams JGK. Efficient site-directed saturation mutagenesis using degenerate oligonucleotides. J. Biomol. Techniques. 2007;18:147–149. [PMC free article] [PubMed] [Google Scholar]
- 20.Cunningham BC, Wells JA. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Montelione GT, Rost B. Novel leverage of structural genomics. Nature Biotechnol. 2007;25:849–851. doi: 10.1038/nbt0807-849. [DOI] [PubMed] [Google Scholar]
- 22.Mestas SP, Sholders AJ, Peersen OB. A fluorescence polarization-based screening assay for nucleic acid polymerase elongation activity. Anal. Biochem. 2007;365:194–200. doi: 10.1016/j.ab.2007.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–73. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.