Abstract
Recombinant protein translation in Escherichia coli may be limited by stable (i.e. low free energy) secondary structures in the mRNA translation initiation region. To circumvent this issue, we have set-up a computer tool called ‘ExEnSo’ (Expression Enhancer Software) that generates a random library of 8192 sequences, calculates the free energy of secondary structures of each sequence in the −70/+96 region (base 1 is the translation initiation codon), and then selects the sequence having the highest free energy. The software uses this ‘optimized’ sequence to create a 5′ primer that can be used in PCR experiments to amplify the coding sequence of interest prior to sub-cloning into a prokaryotic expression vector. In this article, we report how ExEnSo was set-up and the results obtained with nine coding sequences with low expression levels in E. coli. The free energy of the −70/+96 region of all these coding sequences was increased compared to the non-optimized sequences. Moreover, the protein expression of eight out of nine of these coding sequences was increased in E. coli, indicating a good correlation between in silico and in vivo results. ExEnSo is available as a free online tool.
INTRODUCTION
The production of soluble proteins is a known bottleneck of Structural Genomics (SG) programs. For practical reasons, Escherichia coli is the most widely used expression system for producing the recombinant proteins demanded by these programs. However, many recombinant proteins are either insoluble or poorly expressed in E. coli.
Increasing the proportion of soluble proteins can be obtained by tuning expression conditions (1) or by refolding inclusion bodies (2), but unexpressed proteins are often considered as a dead end, although they may represent a significant part of SG programs. For instance, 57 out of 151 prokaryotic expression constructs (38%) of the European VIZIER project (http://www.vizier-europe.org/) do not provide enough recombinant protein for crystallization trials (our unpublished data).
In some cases, the reason for this lack of expression can be found. The sequence of interest may contain codons rarely used by E. coli. In that case, expressing the recombinant protein in strains over producing the corresponding tRNA(s), such as Rosetta (Novagen) or BL21 RIL and RP (Stratagene) can solve the problem (3).
The lack of expression can also result from reduced interaction efficiency between mRNA ribosome-binding site (RBS) and 16S rRNA, and between translation initiation codon ATG and fMet-tRNA(fMet). This low efficiency interaction has been associated with stable mRNA secondary structures in the translation initiation region (TIR) (4).
To address this issue, 5′ coding sequences have been added or deleted resulting in a dramatic enhancement of protein expression (5–7). The same goal has been reached by increasing the free energy of TIR secondary structure by means of silent mutations (5). This latter strategy relies on the individual analysis of each sequence to optimize, followed by site-directed mutagenesis. The technique does improve protein expression but is rather time consuming, and the modifications usually take place within the coding sequence which limits the number of bases that can be mutated if the mutation is to be kept silent.
These two limitations can be circumvented by using libraries of randomly mutated 5′ untranslated region (5′UTR), following the rational used in directed evolution (8). In agreement with this approach, we have observed that independent clones of a library made of four randomly mutated positions in the vicinity of the RBS exhibited different recombinant protein expression levels (unpublished results).
Although fruitful, physically constructing and screening random libraries is an expensive and time consuming process. To avoid this double expense, we sought methods allowing the generation as well as the screening of random libraries to be performed in silico rather than at the bench. We have set-up an in silico screening of computer-generated libraries on the following hypothesis: since stable secondary structures in regions encompassing TIR were correlated with low protein expression (4–7, 9), molecular clones having unstable TIR secondary structures (i.e. high free energy) could be expected to provide higher translation levels.
On the basis of this hypothesis, we devised a computer tool called ‘ExEnSo’ (Expression Enhancer Software) intended to design a forward PCR primer with a sequence encoding a TIR with the most unstable mRNA secondary structure present in an in silico generated random library of TIR-containing sequences. In this article, we report the set up of ExEnSo and the results obtained on a set of nine recombinant proteins poorly expressed in E.coli.
MATERIALS AND METHODS
Cloning and expression of recombinant proteins
Although ExEnSo can virtually be used with any vector, the experiments reported in this article used Gateway recombination cloning (Invitrogen) and pDEST14 expression vector only.
All the proteins used in this study (Supplementary Figure 1) were expressed as N-terminal His-tag fusion. To that end, their coding sequence was PCR amplified using ‘standard’ forward (GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATGCCA CCATGAAACATCACCATCACCATCAC-21 first bases of the coding sequence) and reverse (GGGGACCACTTTGTACAAGAAAGCTGGGTCTTATTA-21 last bases of the coding sequence) primers (10).
The standard forward primer is a 91 bases sequence made of (from 5′ to 3′): (i) a stretch of four Gs [(this four Gs clamp is required to make the minimal 25 base pairs attB1 sequence an efficient substrate for the recombination enzyme BP clonase (Invitrogen, Gateway user manual)]; (ii) the 5′ recombination cloning site attB1; (iii) 17 bases containing the RBS; (iv) the translation initiation codon; (v) a lysine codon [this A-rich codon improves protein expression by destabilizing mRNA secondary structures (11)] followed by a 6His tag encoding sequence, and (vi) the first 21 bases of the coding sequence of the protein of interest.
The standard reverse primer is made of (from 5′ to 3′): (i) a four Gs clamp; (ii) the 3′ recombination cloning site attB2; (iii) two stop codons (TAA) and (iv) the last 21 bases of the coding sequence of the protein of interest.
PCR products were sub-cloned in two steps by recombination cloning following the manufacturer's instructions (Gateway, Invitrogen), except that reaction volumes were 5 µl instead of 20 µl. In the first step (BP reaction), PCR products were sub-cloned into shuttle vector pDONR201, and recombinants were selected by transforming DH10B cells with the whole BP reaction volume, and then plating on kanamycin plates. Two kanamycin-resistant colonies were randomly picked-up and PCR screened using attL1 and attL2 primers. The sequence of one PCR-positive clone was verified by DNA sequencing using the same primers pair and used for sub-cloning (LR reaction) into pDEST14 expression vector. Recombinants were selected by transforming DH10B cells with the whole LR reaction volume and then plating on ampicillin plates. Two ampicillin-resistant colonies were randomly picked-up and PCR screened using attB1 and attB2 primers. Protein expression screening was as described (1,12).
Mutable bases and free energy calculation
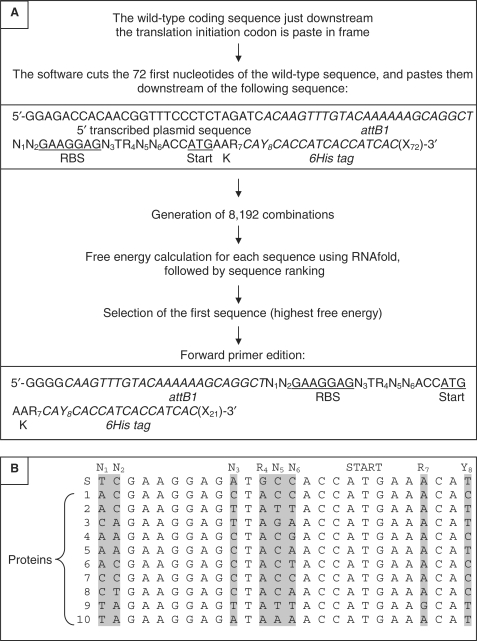
The mRNA region −70/+96 (base + 1 is the adenine of the translation initiation codon) was used in all the mutagenesis and free energy calculation experiments performed in this study. This 166 bases sequence can be divided in three parts (Figure 1A):
A non-mutable region. Bases –70 to –18 cannot be mutated because they are respectively provided by the plasmid (–70/–43) and by the 5′ recombination site attB1 (–42/–18).
A mutable region (−17/+9) made of (i) 17 bases containing the RBS (−17/−1), (ii) the translation initiation codon (ATG, +1/+3), (iii) the lysine codon (+4/+6) and (iv) the first codon of the 6His tag (+7/+9).
A non-mutable region made of the last five codons of the 6His tag (+10/+24) followed by the first 72 first bases of the coding sequence of the protein of interest (+25/+96). This sequence size was selected to comply with the recommendation of Cebe and Geiser (5).
Figure 1.
(A) Summary of the sequence of events performed by ExEnSo. (B) Sequence alignment of the mutable region (−17/+9) of the 10 sequences optimized by ExEnSo. S, sequence of the standard forward primer. 1 to 10, coding sequences used in this study (for details, see Supplementary Figure 1). On top of the alignment are indicated the mutable bases (N1 to Y8) and the translation initiation codon (ATG). Grey boxes contain the mutated bases.
Within the mutable region (−17/+9), mutations were allowed to occur at the following eight positions (Figure 1A): N1 (−17) and N2 (−16) are two bases following attB1. N3 (−8) is the first base 3′ to the RBS (N can be A, C, G, or T). Following N3, a T (−7) was found in all the expression plasmids we have analysed [pIVEX (Roche), pET (Novagen), pDEST17 (Invitrogen)], and in the complementary sequence of the 16S rRNA. R4 (A or G, −6) is an A in this series of vectors, and a G in our standard forward primer for pDEST14 hence the use of R instead of N for that position. In contrast, N5 (−5) and N6 (−4) were not conserved and could therefore undergo full range mutagenesis. R7 (A or G, +6) and Y8 (C or T, +9) are two silent mutations located at the beginning of the translated sequence, respectively in the lysine (K = AAA or AAG) and in the first histidine (CAC or CAT) codon of the 6His tag.
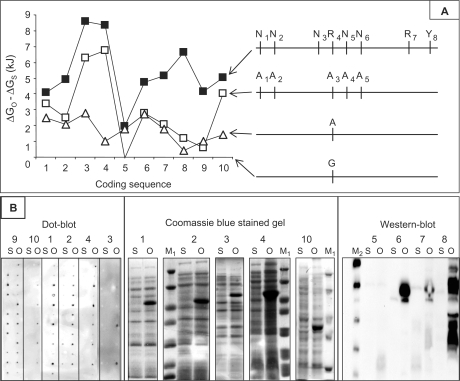
In the mono adenosine (1A) mutagenesis experiment (Figure 2A), R4 was mutated in A. R4 was chosen for that experiment because it allows for a simple binary choice (A or G).
Figure 2.
(A) Left, graph representing the calculated free energy of region −70/+96. The Y-axis (ΔGO − ΔGS) is the difference between the free energy of the mutated sequence (ΔGO) and the free energy of the standard sequence (ΔGS). Raw ΔGO and ΔGS are in Supplementary Figure 1. The X-axis is made of the 10 coding sequences (the numbering from 1 to 10 is that used in Supplementary Figure 1). The negative control (a protein highly expressed in E. coli although bearing a standard −70/+96 sequence) is number 9. Open triangle, mono-adenine (1A) substituted sequence; open square, penta-adenine (5A) substituted sequence; black filled square, sequence optimized by ExEnSo. Right, the different mutations of the mutable region (−17/+9) are indicated in front of the corresponding curve [the baseline (X-axis) is the standard sequence (R4 = G)]. (B) In vivo protein expression of coding sequences 1–10 (protein numbering on top of gels, western- and dot-blots is the same as in A) was performed following a fractional factorial approach made of 12 expression conditions (12), and then analyzed by dot-blot using anti-His antibodies (left panel). The results provided by the 12 conditions are individually displayed from top to bottom. The condition providing the best optimized to standard expression ratio was used for analysis by SDS-PAGE followed by Coomassie blue staining (1–4, 10, middle panel). Proteins with low expression levels were analysed by western blotting and anti-His antibodies (5–8, right panel). S, standard sequence; O, sequence optimized by ExEnSo. M1, molecular weight marker (Proteins 2 and 4: 116, 66.2, 45, 35, 25, 18.4, 14.4 kDa. Protein 10: 116, 66.2, 45, 35, 25). M2, His-tagged molecular weight marker (40, 30, 20, 15, 10 kDa).
In the poly adenosine (5A) mutagenesis experiment (Figure 2A), N1, N2, R4, N5 and N6 were mutated in A. Note that only five of the eight mutable bases could undergo a N to A substitution because N3 and R7 were already an A in the standard sequence (Figure 1B), and Y8 could not be an A for the afore mentioned reason.
In all cases, the free energy of −70/+96 region secondary structures was calculated using RNAfold 1.6.4 (http://www.tbi.univie.ac.at/~ivo/RNA/) (13).
Set-up and use of ExEnSo
ExEnSo generates a set of partially random 166 bases RNA sequences spanning the region −70/+96, selects the sequence having the highest free energy and then provides a forward primer with an optimized sequence (Figure 1A).
In practice, the software inserts the first 72 bases of the wild-type coding sequence of the protein of interest downstream of the first 94 bases of the sequence spanning the region −70/+96 of the mRNA.
Considering the number of mutable bases (as mentioned earlier) and the degeneracy code used, each random library is made of a total of 8192 sequences (N1 × N2 × N3 × R4 × N5 × N6 × R7 × Y8, with N = 4 and R = Y = 2).
Once the library of mutants has been generated, the software individually calculates the free energy of the 8192 mRNA structures using RNAfold, and then ranks the 8192 combinations from most (highest free energy) to least (lowest free energy) unstable. The sequence ranked first is the optimized sequence.
To create the 5′ primer, the software deletes the first 28 (–70 to –43) and last 51 (+46 to +96) bases of the first sequence (i.e. encoding the most unstable structure), and then adds four Gs at the 5′ end of the deleted sequence. The resulting 91 bases sequence is identical to the standard forward primer, except that it contains substitutive mutations. This sequence can be used as 5′ primer in PCR experiments to amplify the coding sequence prior to sub-cloning into pDEST14. In Figure 1B, the mutable region (−17/+9) of the sequences generated and selected by ExEnSo (optimized sequences) for the 10 coding sequences used in the present study have been aligned.
RESULTS AND DISCUSSION
In SG programs, we produce recombinant proteins mainly in E. coli using the prokaryotic expression vector pDEST14 (Invitrogen). This transcriptional vector is devoid of translational elements such as RBS or translation initiation codon, which must be provided by the inserted coding sequence. To that end, we use a couple of ‘standard’ forward and reverse primers for PCR amplifying the coding sequence to express prior to sub-cloning into pDEST14.
In the present study, this couple of primers was used to amplify nine sequences encoding proteins with low expression level in E. coli. A negative control (i.e. a coding sequences with a non-optimized TIR but highly expressed in E. coli) was also included in the test (Supplementary Figure 1).
Influence of the adenine content on the free energy of the translation initiation region
According to previous studies, increasing TIR adenine (A) content could improve protein expression by reducing the stability (increasing free energy) of mRNA structures in that region (11,14). To check whether this would apply to our set of proteins, a guanine (G) located between the RBS and the translation initiation codon in the standard forward primer [mutable position R4 (-6), see Materials and Methods section] was mutated in A. The efficiency of this substitution was evaluated by calculating the free energy of the mRNA region −70/+96 containing R4, as described in Materials and Methods section. The results are reported in Figure 2A. This G to A mutation resulted in all cases in a free energy increase of sequence −70/+96 from 0.4 kJ (coding sequence 8) to 2.8 kJ (coding sequence 3), suggesting that a single G to A switch effectively destabilized the region −70/+96 presumably by preventing the formation of a previously existing GC pair.
On the basis of this result, we speculated that substituting with A more bases in the vicinity of mutable position R4 could further increase this positive effect. In addition to this first G to A mutation, four other N to A mutations were introduced as described in Materials and Methods section. The free energy of mRNA sequence −70/+96 was again calculated for the 10 coding sequences, and compared to the results obtained in the previous single G to A mutation experiment. Compared to the 1A-substituted sequence, the 5A-substituted sequence provided three kinds of results (Figure 2A): (i) surprisingly, coding sequence 5 showed a return to the baseline (i.e. the 1A to 5A switch cancelled the previous G to 1A effect); (ii) for four coding sequences (2, 6, 7, 9), increasing from 1 to 5 the adenine content of sequence −70/+96 was more or less neutral; (iii) the remaining five coding sequences (1, 3, 4, 8, 10) definitely benefited from this ‘adenine scanning’, although to a limited extent for coding sequences 1 and 8. In conclusion, the free energy of region −70/+96 of five mRNAs out of 10 was increased to different extents by increasing its adenine content from 1 to 5 via substitutive mutations.
Unfortunately, the number of N to A substitutions is limited because it can generate non silent mutations as far as coding sequences are concerned and because As are already present in the non mutated sequences. Since we (our unpublished data) and others (8) have experienced that random mutagenesis of 5′UTR and TIR can modulate protein expression, we reasoned that random mutagenesis of those regions could provide a mean to further extend the ability of a mutagenesis approach to increase the instability of region −70/+96, while circumventing the limitations inherent to A scanning. To that end, we devised ExEnSo a computer tool for generating and screening random libraries of the −70/+96 region (see Materials and Methods section and Figure 1A for a detailed description of ExEnSo).
Random versus directed mutagenesis
To evaluate whether the virtual random mutagenesis and screening performed by ExEnSo provided a benefit over 1A or 5A mutagenesis, the free energy of region −70/+96 was calculated using the same set of 10 coding sequences, and compared to the results obtained in 1A and 5A directed mutagenesis experiments (Figure 2A).
Compared to 1A mutagenesis, the free energy of region −70/+96 of nine coding sequences out of 10 was increased. Only that of coding sequence five remained unchanged.
Compared to 5A mutagenesis, the free energy of region −70/+96 of all 10 coding sequences was increased.
When the results obtained with the 10 coding sequences were considered collectively, an average increase of 1.75 (± 0.79) kJ, 2.95 (± 2.25) kJ, and 5.4 (± 2) kJ were respectively obtained per coding sequence by 1A, 5A, and random mutagenesis with respect to the standard sequence (R4 = G).
Since all N to A substitutions had been performed in the 5A mutagenesis experiment, ExEnSo could not further destabilize region −70/+96 by substituting the remaining three mutable bases by A (see Materials and Methods section).
Nevertheless, a close examination of the optimized sequences indicates that ExEnSo makes some use of A for optimizing standard sequences (Figure 1B). When the eight mutable positions are considered in the 10 optimized sequences, A, C, T and G were respectively used 34, 26, 16 and 3 times by ExEnSo to optimize the standard sequence. For instance, mutable base R4 is a G in the standard sequence and an A in all the optimized sequences. Similarly, the A at mutable position R7 of the standard sequence is conserved in 9 out of the 10 optimized sequences. Thus, a bias towards the preferential use of A seems to exist in ExEnSo. However, the opposite situation also exists: N3 is an A in the standard sequence, but this A is conserved in only 1 out of the 10 optimized sequences. Taken together, these observations suggest that the combinatorial rational used by ExEnSo relies only partially on A scanning. Conversely, G seems to be particularly unfavourable for increasing the energy of region −70/+96 of this set of coding sequences for an unknown reason.
In vivo protein expression
In the next step, the ability of the optimized sequences provided by ExEnSo to effectively improve protein expression was assessed. Using the same set of 10 coding sequences, the level of protein expression obtained with the standard sequences was compared to the level obtained with the optimized sequences (Figure 2B). Protein expression was significantly increased in 8 cases out of 10 (proteins 1–4, 6–8, 10), thereby underlining a good correlation between in silico (10 out of 10) and in vivo (8 out of 10) results. Although this correlation suggests that the latter results from the former via improved translation initiation (4–7, 9), it does not exclude other explanations such as increased mRNA lifespan [e.g. by elimination of RNase E/III cleavage sites (15)] or altered action of small non-coding RNAs (16) or of ‘riboswitches’ (15). Upper and lower limits exist to this correlation.
Firstly, increasing the free energy of region −70/+96 of negative control coding sequence 9 (ΔGO − ΔGS = 4.17 kJ) did not result in an increased protein expression, suggesting that increasing the expression of an already over-expressed protein is more limited by other parameters of protein expression (availability of tRNA, transcript lifespan, etc.) than by the stability of mRNA secondary structures in the −70/+96 region.
Secondly, a situation opposite to that of protein 9 also exists. Expression of protein 5 did not increase when the optimized sequence was used, although protein 5 combined a positive ΔGO − ΔGS and no protein expression when the standard sequence was used. Interestingly, protein 5 had the lowest ΔGO − ΔGS of this set of protein (1.98 kJ), and was also refractory to 5A directed mutagenesis (Figure 2A). This result allows speculating that a ΔGO − ΔGS threshold might exist below which no protein expression improvement can be obtained. On the other hand, factors such as mRNA (15) or protein stability could also account for this negative result.
Whatever may be the reason(s), these two negative results seem to be marginal, at least within the set of proteins used in this study.
One-fourth of proteins with improved expression (two proteins out of eight, i.e. protein 2 and protein 4) were expressed soluble (data not shown). From 1 l culture, enough protein encoded by optimized sequences 2 and 4 could be purified by affinity chromatography on Ni column (Supplementary Figure 2) to be used in crystallization trials. The remaining optimized proteins are potentially good candidates for refolding screening of inclusion bodies (2).
CONCLUSION
Considering the cost price of the process ranging from primer ordering to protein expression trials, we believe that systematically ordering PCR primers via ExEnSo could save a significant amount of both working time and funding of SG programs.
ExEnSo is available as a free online tool at the following URL: http://exenso.afmb.univ-mrs.fr.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Dr Xavier De Lamballerie, Dr Jacques Rohayem and Dr Eric Snijder for kindly providing the viral cDNAs. This work was funded by the European VIZIER project (contract number, LSHG-CT-2004-511960; web site, http://www.vizier-europe.org/). Funding to pay the open Access publication charges for their article was provided by the European VIZIER project.
Conflict of interest statement. None declared.
REFERENCES
- 1.Berrow NS, Bussow K, Coutard B, Diprose J, Ekberg M, Folkers GE, Levy N, Lieu V, Owens RJ, et al. Recombinant protein expression and solubility screening in Escherichia coli: a comparative study. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1218–1226. doi: 10.1107/S0907444906031337. [DOI] [PubMed] [Google Scholar]
- 2.Vincentelli R, Canaan S, Campanacci V, Valencia C, Maurin D, Frassinetti F, Scappucini-Calvo L, Bourne Y, Cambillau C, et al. High-throughput automated refolding screening of inclusion bodies. Protein Sci. 2004;13:2782–2792. doi: 10.1110/ps.04806004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dabrowski S, Kiaer Ahring B. Cloning, expression, and purification of the His6-tagged hyper-thermostable dUTPase from Pyrococcus woesei in Escherichia coli: application in PCR. Protein Expr. Purif. 2003;31:72–78. doi: 10.1016/s1046-5928(03)00108-6. [DOI] [PubMed] [Google Scholar]
- 4.de Smit MH, van Duin J. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc. Natl Acad. Sci. USA. 2003;87:7668–7672. doi: 10.1073/pnas.87.19.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cebe R, Geiser M. Rapid and easy thermodynamic optimization of the 5'-end of mRNA dramatically increases the level of wild type protein expression in Escherichia coli. Protein Expr. Purif. 2006;45:374–380. doi: 10.1016/j.pep.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Griswold KE, Mahmood NA, Iverson BL, Georgiou G. Effects of codon usage versus putative 5'-mRNA structure on the expression of Fusarium solani cutinase in the Escherichia coli cytoplasm. Protein Expr. Purif. 2003;27:134–142. doi: 10.1016/s1046-5928(02)00578-8. [DOI] [PubMed] [Google Scholar]
- 7.Wallis OC, Sami AJ, Wallis M. The effect of changes in nucleotide sequence coding for the N-terminus on expression levels of ovine growth hormone variants in Escherichia coli. Biochim. Biophys. Acta. 1995;26:360–368. doi: 10.1016/0167-4781(95)00035-f. [DOI] [PubMed] [Google Scholar]
- 8.Zhelyabovskaya OB, Berlin YA, Birikh KR. Artificial genetic selection for an efficient translation initiation site for expression of human RACK1 gene in Escherichia coli. Nucleic Acids Res. 2004;32:e52. doi: 10.1093/nar/gnh050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bucheler US, Werner D, Schirmer RH. Generating compatible translation initiation regions for heterologous gene expression in Escherichia coli by exhaustive periShine-Dalgarno mutagenesis. Human glutathione reductase cDNA as a model. Nucleic Acids Res. 1992;20:3127–3133. doi: 10.1093/nar/20.12.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matthes N, Mesters JR, Coutard B, Canard B, Snijder EJ, Moll R, Hilgenfeld R. The non-structural protein Nsp10 of mouse hepatitis virus binds zinc ions and nucleic acids. FEBS Lett. 2006;580:4143–4149. doi: 10.1016/j.febslet.2006.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komarova AV, Tchufistova LS, Dreyfus M, Boni IV. AU-rich sequences within 5' untranslated leaders enhance translation and stabilize mRNA in Escherichia coli. J. Bacteriol. 2005;187:1344–1349. doi: 10.1128/JB.187.4.1344-1349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benoit I, Coutard B, Oubelaid R, Asther M, Bignon C. Expression in Escherichia coli, refolding and crystallization of Aspergillus niger feruloyl esterase A using a serial factorial approach. Protein Expr. Purif. 2007;55:166–174. doi: 10.1016/j.pep.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brock JE, Paz RL, Cottle P, Janssen GR. Naturally occurring adenines within mRNA coding sequences affect ribosome binding and expression in Escherichia coli. J. Bacteriol. 2007;189:501–510. doi: 10.1128/JB.01356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaberdin VR, Bläsi U. Translation initiation and the fate of bacterial mRNAs. FEMS Microbiol. Rev. 2006;30:967–979. doi: 10.1111/j.1574-6976.2006.00043.x. [DOI] [PubMed] [Google Scholar]
- 16.Storz G, Opdyke JA, Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.