Abstract
The rezipping force of two complementary DNA strands under tension has been measured in the presence of Escherichia coli single-stranded-binding proteins under salt conditions ranging from 10– to 400 mM NaCl. The effectiveness of the binding protein in preventing rezipping is strongly dependent on salt concentration and compared with the salt dependence in the absence of the protein. At concentrations less than 50 mM NaCl, the protein prevents complete rezipping of λ-phage on the 2-s timescale of the experiment, when the ssDNA is under tensions as low as 3.5 ± 1 pN. For salt concentrations greater than 200 mM NaCl, the protein inhibits rezipping but cannot block rezipping when the tension is reduced below 6 ± 1.8 pN. This change in effectiveness as a function of salt concentration may correspond to salt-dependent changes in binding modes that were previously observed in bulk assays.
INTRODUCTION
The opening and maintenance of single-stranded DNA (ssDNA) within a double-stranded DNA (dsDNA) molecule is necessary for biological processes such as transcription, translation and replication. Nearly all organisms produce ssDNA-binding proteins, which show a high binding affinity for ssDNA. These proteins are necessary for replication, recombination and repair, and it is expected that their binding preference for ssDNA allows them to maintain open regions of ssDNA within a dsDNA molecule.
Escherichia coli single-stranded binding (SSB) protein is a well-known and documented ssDNA-binding protein. In this article, we present measurements of the effectiveness of E. coli SSB proteins in maintaining partially open DNA as a function of salt concentration. Salt is expected to play an important role in DNA stabilization for several reasons including salt-dependent changes in the free energy difference between ssDNA and dsDNA in the absence of SSB protein (1–3), as well as salt-dependent changes in the binding modes and binding affinities of the SSB protein (4–8). Though the binding affinities and binding modes have been extensively studied, the actual salt-dependent impact of the binding on the stability of the ssDNA within a dsDNA molecule has not yet been measured, prior to this work.
Many bulk studies have been carried out to measure the binding of SSB protein to ssDNA and salt concentrations has been shown to play a significant role in binding behavior (4–8). At room temperature, SSB protein exhibits two primary binding modes (5,6). In low salt buffers below 50 mM NaCl, the binding is highly cooperative, with a site size of 35 base pairs (bp). In higher salt buffers above 200 mM NaCl, the binding becomes less cooperative and the site size increases to 56 bp with a further increase to 65 bp at even higher salt concentrations. At intermediate salt concentrations, a mixture of both binding modes is present. The function of these binding modes in vivo is unclear, but it has been suggested that they are used selectively in different cell processes (9).
Salt concentrations not only determine which binding mode is dominant, but can also affect the binding affinity of a particular mode. While the binding affinity of SSB protein is independent of salt at concentrations below 150 mM (8), the binding affinity drops drastically with increasing salt concentrations from 200–400 mM (7). This drop in binding affinity is not due to any change in the binding mode, as the binding mode is constant in this salt range, but rather is due to electrostatic interactions between the protein and the DNA.
Even in the absence of SSB protein, the stability of dsDNA is salt dependent, partly due to changed screening charges between the phosphate backbones. Experiments in which dsDNA melting temperatures are measured as a function of salt show that the stability of dsDNA increases with salt concentration (10). Thus, based on the salt dependence of the DNA and the SSB protein binding, it is expected that the stability of partially open DNA in the presence of SSB protein will be salt dependent; however, previous studies have not allowed one to determine the extent of salt-dependent stabilization of ssDNA by the SSB protein.
The effectiveness of the protein in maintaining open regions of ssDNA can be directly studied through single DNA molecule unzipping and rezipping. In unzipping the dsDNA molecule, large regions of ssDNA are created to which the protein can bind. By decreasing the force and allowing rezipping to take place, the effectiveness of the proteins in preventing reannealing can be measured. Previous work has shown that in a PBS buffer, SSB protein inhibits rezipping, but is not capable of maintaining ssDNA under tensions of less than 5.5 ± 1 pN (11). Based on SSB protein's highly salt-dependent binding behavior, it is likely that the rezipping behavior measured in PBS, with a salt concentration of 137 mM NaCl, is not universal to all salt concentrations.
We present a study of the salt dependence of the SSB protein function in vitro by measuring the rezipping of DNA in the presence of SSB proteins. Results show that the effectiveness of SSB protein in preventing rezipping is extremely dependent on salt. This dependence might have been due to salt–DNA interactions, the salt dependence of single SSB protein-binding modes or the salt dependence of the variation between binding modes. We show that the salt-dependent stability of DNA in buffers with low SSB protein concentration exhibits no transitions and eliminates salt–DNA interactions as a cause of the salt-dependent stability of ssDNA in the presence of SSB proteins. Thus, we demonstrate that the salt dependence at higher protein concentrations is due to changes in binding behavior of the proteins, and note that this dependence correlates to the different binding modes detected in bulk assays.
MATERIALS AND METHODS
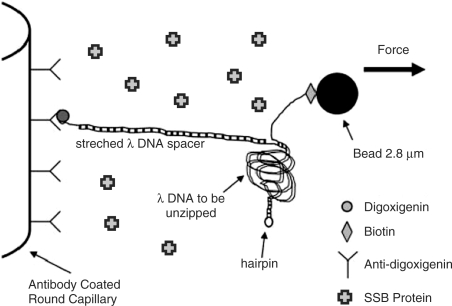
The DNA construct used for unzipping measurements has been described previously (12) and is shown schematically in Figure 1. Briefly, it consists of a linker λ-DNA (New England Biolabs), which is hybridized and ligated to one end of the λ-DNA strand that is to be opened. The second strand of the λ-DNA to be opened is hybridized and ligated to a biotinylated oligonucleotide. The other end is closed with a hairpin loop to prevent complete separation of the construct in an unzipping event. The linker λ-DNA is tagged with a digoxigenin-labeled oligonucleotide. It is attached to a glass capillary coated with an antidigoxigenin antibody. The λ-DNA strand to be opened is bound to a 2.8-μm streptavidin-coated magnetic bead (Dynabeads) via the biotinylated oligonucleotide. The λ-DNA and beads are stored at 4°C after preparation and incubated with E. coli SSB protein (Epicentre), at room temperature for 15 min prior to the experiment. The buffer of the DNA-bead mixture is 10 mM Tris, pH 7.5, with salt concentrations ranging from 10–400 mM NaCl. The protein buffer contains 50% glycerol, 0.1% Triton-X, 50 mM Tris, pH 7.5, 0.1 mM EDTA, 0.1 M NaCl and 1 mM DDT; 1 μl of the protein buffer is added to 69 μl of the DNA-bead mixture in the experiment.
Figure 1.
Schematic of the DNA binding to the inner glass capillary in the presence of SSB proteins and the magnetic bead such that pulling the bead away from the surface will cause the dsDNA shown on the right side of the diagram to be separated into two single DNA strands. Note that the figure is not to scale, considering that λ-DNA contains 48 502 bp.
Mechanical unzipping of dsDNA is carried out by a magnetic tweezers apparatus (13). A stack of magnets exerts a force, F, on the magnetic beads by F = m∇B, where m is the magnetization of the bead and B is the magnetic field. The force on the beads is controlled by the distance of the magnet from the beads, with a force range of 1–30 pN. Spread in magnetization of the beads leads to a standard deviation of ∼30% in the force measurements. The force measurements are taken by incrementally increasing or decreasing the position of the magnet by 100 μm every 2 s. The instantaneous position of the bead is measured immediately following the magnet movement and again 2 s later.
RESULTS
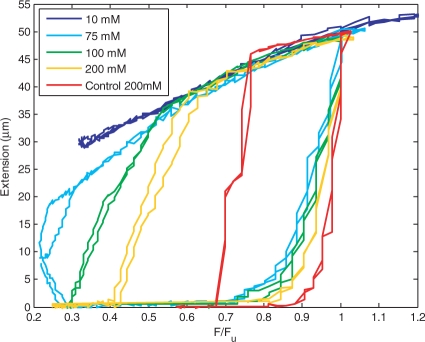
A single DNA molecule is unzipped and subsequently allowed to rezip in the presence of SSB protein. Typical extension versus force curves of a single molecule for 10, 75, 100 and 200 mM NaCl buffers in the presence of 1.3 μM SSB protein are shown in Figure 2. For comparison, a typical unzipping and rezipping curve in the absence of SSB protein at 200 mM NaCl is shown as well. The force, F, is normalized by the unzipping force measured for the individual molecule, Fu, to remove variation due to the spread in bead magnetization. While Fu varies from ∼10–20 pN for different salt concentrations, due to changes in dsDNA stability, it does not vary significantly with protein concentration, in agreement with previous work (11). The constant applied force is increased or decreased in steps every 2 s and the position of the magnetic bead is measured. As F is initially increased, no unzipping occurs, and the bead extension remains zero. When F increases to 0.7–0.8 Fu, the extension of the bead begins to increase, indicating unzipping of the DNA molecule. At F = Fu, the molecule is completely unzipped. Further increase in F has little effect on the bead extension. Next, F is incrementally decreased. The bead extension initially decreases due to the elasticity of ssDNA, but as F is decreased more, rezipping begins. In the 75, 100 and 200 mM NaCl buffers, the bead extension decreases rapidly for F < 0.6Fu, indicative of rezipping. For 100 and 200 mM NaCl, complete rezipping occurs and Fz can be measured. It is difficult to distinguish protein binding from rezipping, as both shorten the region of ssDNA under force (4,14). Protein binding has been shown to shorten ssDNA by 35% in the absence of tension (14), whereas its extension as a function of tension is unknown. For this reason, we only measure complete rezipping events, where the extension of the ssDNA under force is zero, implying that the region of the ssDNA has also gone to zero and that the molecule has completely rezipped. For 75 mM NaCl, it is clear that rezipping does take place, however, since complete rezipping of the molecule does not occur on the timescale of the experiment, a value for the force at which complete rezipping occurs, Fz, cannot be measured. In the case of 10 mM NaCl, however, the bead extension does not show a rapid decrease, even below F = 0.3Fu, suggesting that rezipping may not occur at all on the timescale of the experiment.
Figure 2.
Typical results for unzipping and rezipping a single DNA molecule in 10, 75, 100 and 200 mM NaCl in the presence of 1.3 μM SSB protein. For comparison, a typical unzipping and rezipping curve in the absence of SSB protein at 200 mM NaCl is shown as well. Unzipping and rezipping is repeated twice to demonstrate reproducibility.
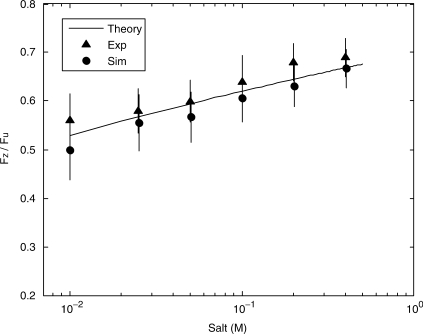
The stability of naked DNA is known to be salt dependent (1–3) and thus it is necessary to separate the effects of DNA salt dependence and protein salt dependence in this experiment. In the absence of protein, Fz is measured as a function of salt, shown in Figure 3, where the Fz is normalized by Fu, to remove bead variation.
Figure 3.
Fz/Fu from both experiments (triangles) in the absence of protein and simulation (circles) as a function of salt. The solid line is the ratio expected from theory based on the salt dependence of ssDNA persistence length and the salt dependence of enthalpy.
The ratio of Fz/Fu can be predicted theoretically based on the salt dependence of ssDNA and dsDNA in the absence of proteins. The energy difference, ΔG, between an open base pair and a closed base pair is determined by the free energy of ssDNA under an applied force, gu(F), and the enthalpic difference, ΔH, and entropic difference, ΔS, between dsDNA and ssDNA:
| 1 |
At forces ranging from 1 to 100 pN, the free energy term, gu(F), can be determined by modeling ssDNA as a freely jointed chain (15),
| 2 |
where L is the ssDNA nucleotide length, 5.6Å (16), and a is the persistence length. The force required for unzipping can be determined as the force at which ΔG = 0. The salt dependence of the melting temperature, Tm(X), of dsDNA has been shown experimentally to follow a logarithmic form,
| 3 |
where X is the salt concentration in moles/liter (10). Several equations exist for predicting Tm(X), including equations which predict that A-T and G-C sequences differ in their salt dependence (1). Analysis of these various equations have shown little variation in the salt dependence of the final predicted Fz/Fu ratio over the range of salt investigated. We assume ΔS to be independent of X (17), and find ΔH(X) by the relation,
| 4 |
Diffusion measurements of ssDNA have yielded the persistence length of ssDNA as a function of salt (2), and can be fit by the equation
| 5 |
where a is given in Angstroms.
Monte Carlo simulations using nearest-neighbor energy parameters (10) have been carried out to examine the expected salt dependence on the ratio Fz/Fu. Figure 3 shows the simulated ratio when the applied force is incrementally increased by 0.5 pN every 3 × 105 iterations. The difference in Fu and Fz is due to the heterogeneity of the DNA sequence. For example, G-C base pairs require a high force for unzipping while A-T base pairs require a low force for rezipping. In this case, reasonable agreement with experiment and simulation can be made when the Fu is approximated by the force at which ΔG = 0, where ΔH and ΔS are the averages of the five most strongly bound base-pair neighbors. Fz is approximated in the same manner for the five most weakly bound base-pair neighbors. The predicted ratio Fz/Fu as a function of salt is shown in Figure 3, and shows good agreement with both experimental and simulation results, demonstrating that the salt dependence of DNA stability is very gradual and shows no significant transitions. Thus, we will assume that salt-dependent transitions of DNA stability in the presence of SSB protein are due to binding changes in the protein.
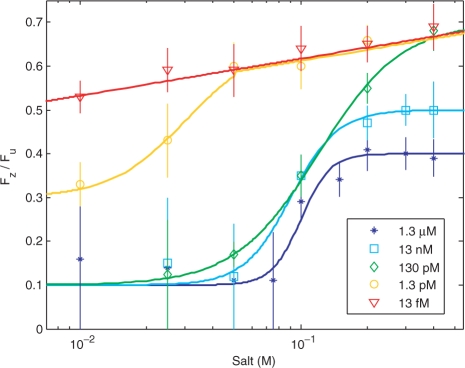
The ratio of Fz/Fu at multiple protein concentrations ranging from 13 fM to 1.3 μM as a function of salt is shown in Figure 4. Each data point represents the measurement for a single molecule, and the error is determined by the variation of magnetization in the beads. For high protein concentrations at salt concentrations less than 50 mM NaCl, Fz/Fu cannot be measured due to the fact that rezipping does not occur on the timescale of the experiment; this is indicated by large error bars at these points.
Figure 4.
The ratio Fz/Fu as a function of salt concentration for protein concentrations ranging from 13 fM to 1.3 μM. The solid lines are a guide to the eye.
The curve for 13 fM protein is similar to the curve shown in Figure 3, where measurements in the absence of protein were compared with simulation and theory for unzipping and rezipping in the absence of protein. This similarity suggests that at these very low concentrations the protein has very little impact on the stability of the DNA. For 1.3 pM, the protein has little effect on the stability for salt concentrations above 50 mM; however, at low salt concentrations the protein increases the stability of the ssDNA. At 130 pM, the protein has little effect at the highest salt concentration of 400 mM; however, stability in the presence of the protein increases with decreasing salt concentration from 200 to 10 mM. At protein concentrations of 13 nM and 1.3 μM, the increase in stabilization as a function of salt concentration appears to saturate at high salt concentrations, while at low salt concentration the stabilization is so great that Fz can no longer be measured. The rezipping behavior seen at protein concentrations in which the protein response is saturated indicates two rezipping modes. For salt concentrations less than 50 mM, a rezipping force cannot be measured, while above 100 mM, the protein cannot maintain ssDNA at tensions less than 0.3Fu. Intermediate salt concentrations show a gradual transition between these two modes, as seen in Figure 4.
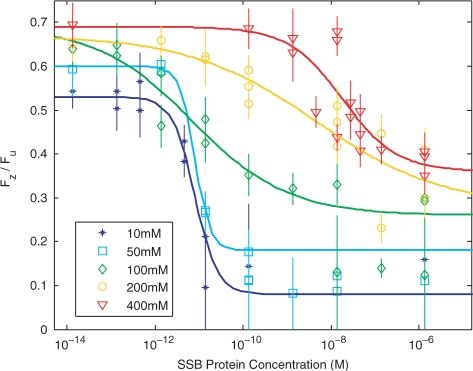
In Figure 4, the increase in stability as a function of salt concentration was shown for different protein concentrations. However, if there is a transition between a less collective and more collective binding mode, then a plot of Fz/Fu at a given salt concentration should be well described by a dose response curve, if the protein binding changes collectivity at that given salt concentration. Figure 5 shows such curves for salt concentrations ranging from 10 to 400 mM. For all of the salt concentrations shown, Fz/Fu shows a protein-concentration-dependent transition that saturates at both high and low protein concentrations. Each data point represents the measurement for a single molecule, and the error is determined by the variation of magnetization in the beads. In the regime where the protein concentration is saturated, the measurements are fairly reproducible from molecule to molecule, with an average standard deviation between 2 and 5 individual molecule measurements of 2–6% of the actual measurement value. Greater variation is seen in the region where protein concentration is not saturated, where the average standard deviation is 15–20% of the actual measurement value. This is most likely due to local variations in protein concentration. At very low protein concentrations, the protein has no effect on the stability of the DNA and the salt dependence of Fz/Fu simply represents the inherent change in the stability of the DNA as a function of salt. At all salt concentrations, there is a sharp change in stability as a function of protein concentration, where the change is largest at salt concentrations below 100 mM, where the protein can keep the DNA partially open for several seconds at tensions as low as 3.5 ± 1 pN. At higher salt concentrations, tensions of at least 6 ± 1.8 pN is required to maintain the partially open DNA, even at the highest protein concentrations.
Figure 5.
The ratio of Fz/Fu as a function of protein concentration for 10–400 mM NaCl. The solid lines represent best-fit curves using the dose–response equation, calculated in the case where the data points which could not be measured (indicated by large error bars) were excluded.
Recent binding assays (18) have shown that at salt concentrations less than 50 mM NaCl, SSB proteins bind almost exclusively in the 35-bp mode, while at concentrations greater than 200 mM NaCl, SSB proteins bind nearly exclusively in the 65-bp mode. This correlation between the salt-dependent binding mode transition and the rezipping transition suggests that the stabilization of ssDNA by the SSB protein is determined by its binding mode. The 35-bp-binding mode is highly cooperative and the protein forms a tight filament on the ssDNA (19–21), and in this mode, little to no rezipping as observed. The 65-bp-binding mode shows little cooperativity and leaves gaps of unbound ssDNA (19,22) between bound proteins. In this case, the unbound ssDNA can bind and possibly rezip around the SSB proteins or physically remove them, resulting in complete rezipping.
The data shown in Figure 5 is fit by a dose–response equation
| 6 |
where X is the protein concentration, Fz(0) is the rezipping force in the absence of proteins, and Fz(∞) is the rezipping force at saturation. C is the EC50, or the concentration of protein required for the rezipping force to be halfway between Fz(∞) and Fz(0), and b is a ‘slope factor’ determined by the cooperativity of the binding and is the mathematical equivalent of the Hill coefficient (23). By fitting the measured rezipping force to this equation, we can extract b and C. Best-fits to the data are shown in Figure 4, where data points at which Fz could not be measured have been excluded.
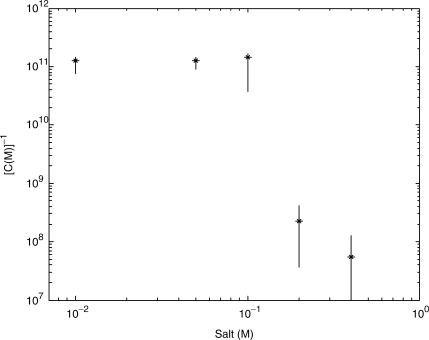
If one assumes that the EC50 point for rezipping correlates to the EC50 point for binding, then the binding affinity of the protein can be approximated by C−1. Previous studies of binding affinity report that the binding affinity is independent of salt for concentrations below 150 mM (8), while binding affinity, K, decreases significantly with increasing salt above 150 mM, such that −d [log(K)/log(X)] = 6−7 (7). The approximate binding affinity extracted from this technique, shown in Figure 6, qualitatively agrees with previous findings. Below 100 mM NaCl, C−1 is constant, while above 100 mM NaCl, C−1 decreases with salt such that −d(log(C−1)/log(X) = 5.6 ± 3. Thus the assumption that Fz/Fu is directly related to the binding of the protein to DNA is reasonably well justified.
Figure 6.
The inverse of EC50 as a function of salt concentration, which can be approximated as the binding affinity if one assumes that the EC50 for rezipping response will be the same as EC50 for binding.
The dose–response equation above is valid in the case of a single binding mode. As SSB protein exhibits multiple binding modes in equilibrium at intermediate salts, such an equation cannot provide a completely accurate picture of the response. However, as an approximation, it still provides useful information in comparing the stabilization of partially open DNA as measured by Fz/Fu to previous assays (7,8,21) that simply measured the binding of the protein to ssDNA in the absence of a matching strand that competes with the protein for binding to the single strand.
Escherichia coli SSB proteins have been shown to bind to ssDNA by wrapping the ssDNA around the protein (9,19,24). It is possible that applying a force to the ssDNA may inhibit this wrapping process. However, whether in a cell or in vitro, something must be done initially to create and maintain ssDNA within a dsDNA molecule, and this requires a change in the free energy of the ssDNA which may be accomplished through applied force, torque, etc. Since the salt-dependent change in the ssDNA stability measured in this work correlates with the previously observed salt-dependent change in binding modes, it seems unlikely that any inhibition of stability due to the tension on the strand has significantly affected the salt-dependent transition between binding modes.
CONCLUSIONS
We have measured the salt dependence on the effectiveness of E. coli SSB proteins in maintaining ssDNA and have shown that salt-dependent changes in the protein significantly alter the stability of the protein–ssDNA complex. In low salt buffers, the protein effectively blocks rezipping at tensions as low as 3.5 ± 1 pN, while at high salts, the protein cannot maintain ssDNA under tensions less than 6 ± 1.8 pN. We note that this change in the effectiveness of the SSB in maintaining the stability of partially open DNA correlates with the previously measured salt-dependent transition between the two primary binding modes of SSB proteins; therefore, these results suggest that the change in stability is due to the previously observed change in binding modes implying that there are significant functional differences between the two binding modes.
ACKNOWLEDGEMENTS
We acknowledge useful conversations with Nancy Kleckner, Guido Guidotti and Charles Limouse. This material is based upon work supported under a National Science Foundation Graduate Research Fellowship and research was funded by grants: Office of Naval Research and Defense Advanced Research Project Agency [N00014-01-1-0782]; Materials Research Science and Engineering Center: National Science Foundation Division of Materials Research [0213805]; Army Research Office [W911NF-04-1-0170]. Funding to pay the Open Access publication charges for the article was provided by Harvard University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Owczarzy R, You R, Moreira BG, Manthey JA, Huang L, Behlke MA, Walder JA. Effects of sodium ions on DNA duplex oligomers: improved predictions of melting temperatures. Biochemistry. 2004;43:3537–3554. doi: 10.1021/bi034621r. [DOI] [PubMed] [Google Scholar]
- 2.Tinland B, Pluen A, Sturm J, Weill G. Persistence length of single-stranded DNA. Macromolecules. 1997;30:5763–5765. [Google Scholar]
- 3.Wenner JR, Williams MC, Rouzina I, Bloomfield VA. Salt dependence of the elasticity and overstretching transition of single DNA molecules. Biophys. J. 2002;82:3160–3169. doi: 10.1016/S0006-3495(02)75658-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chase JW, Williams KR. Single-stranded DNA binding proteins required for DNA replication. Annu. Rev. Biochem. 1986;55:103–136. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- 5.Lohman TM, Overman LB. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J. Biol. Chem. 1985;260:3594–3603. [PubMed] [Google Scholar]
- 6.Bujalowski W, Lohman TM. Escherichia coli single-strand binding protein forms multiple, distinct complexes with single-stranded DNA. Biochemistry. 1986;25:7799–7802. doi: 10.1021/bi00372a003. [DOI] [PubMed] [Google Scholar]
- 7.Overman LB, Bujalowski W, Lohman TM. Equilibrium binding of Escherichia coli single-strand binding protein to single-stranded nucleic acids in the (SSB)65 binding mode. Cation and anion effects and polynucleotide specificity. Biochemistry. 1988;27:456–471. doi: 10.1021/bi00401a067. [DOI] [PubMed] [Google Scholar]
- 8.Ruyechan WT, Wetmur JG. Studies on the noncooperative binding of the Escherichia coli DNA unwinding protein to single-stranded nucleic acids. Biochemistry. 1976;15:5057–5064. doi: 10.1021/bi00668a017. [DOI] [PubMed] [Google Scholar]
- 9.Kuznetsov SV, Kozlov AG, Lohman TM, Ansari A. Microsecond dynamics of protein-DNA interactions: direct observation of the wrapping/unwrapping kinetics of single-stranded DNA around the E. coli SSB tetramer. J. Mol. Biol. 2006;359:55–65. doi: 10.1016/j.jmb.2006.02.070. [DOI] [PubMed] [Google Scholar]
- 10.SantaLucia J, Allawi HT, Senviratne A. Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry. 1996;35:3555–3562. doi: 10.1021/bi951907q. [DOI] [PubMed] [Google Scholar]
- 11.Hatch K, Danilowicz C, Coljee V, Prentiss M. Direct measurements of the stabilization of single stranded DNA under tension by single stranded binding proteins. Phys. Rev. E. 2007;76 doi: 10.1103/PhysRevE.76.021916. 021916. [DOI] [PubMed] [Google Scholar]
- 12.Danilowicz C, Coljee VW, Bouzigues C, Lubensky DK, Nelson DR, Prentiss M. DNA unzipped under a constant force exhibits multiple metastable intermediates. Proc. Natl Acad. Sci. USA. 2003;100:1694–1699. doi: 10.1073/pnas.262789199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Assi F, Jenks R, Yang J, Love C, Prentiss M. Massively parallel adhesion and reactivity measurements using simple inexpensive magnetic tweezers. J. Appl. Phys. 2002;92:5584–5586. [Google Scholar]
- 14.Sigal N, Delius H, Kornberg T, Gefter ML, Alberts B. A DNA-unwinding protein isolated from Escherichia coli: its interaction with DNA and with DNA polymerases. Proc. Natl Acad. Sci. USA. 1972;69:3537–3541. doi: 10.1073/pnas.69.12.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith SB, Cui Y, Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 16.Dessinges MN, Maier B, Zhang Y, Peliti M, Bensimon D, Croquette V. Stretching single stranded DNA, a model polyelectrolyte. Phys. Rev. Lett. 2002;89:248102. doi: 10.1103/PhysRevLett.89.248102. [DOI] [PubMed] [Google Scholar]
- 17.Rouzina I, Bloomfield VA. Force-induced melting of the DNA double helix. 2. Effect of solution conditions. Biophys. J. 2001;80:894–900. doi: 10.1016/S0006-3495(01)76068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy R, Kozlov AG, Lohman TM, Ha T. Dynamic structural rearrangements between DNA binding modes of E. Coli SSB protein. J. Mol. Biol. 2007;369:1244–1257. doi: 10.1016/j.jmb.2007.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chrysogelos S, Griffith J. Escherichia coli single-strand binding protein organizes single-stranded DNA in nucleosome-like units. Proc. Natl Acad. Sci. USA. 1982;79:5803–5807. doi: 10.1073/pnas.79.19.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamon L, Pastre D, Dupaigne P, Le Breton C, Le Cam E, Pietrement O. High-resolution AFM imaging of single-stranded DNA-binding (SSB) protein – DNA complexes. Nucleic Acids Res. 2007;35:e58. doi: 10.1093/nar/gkm147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrari ME, Bujalowski W, Lohman TM. Co-operative binding of Escherichia coli SSB tetramers to single-stranded DNA in the (SSB)35 binding mode. J. Mol. Biol. 1994;236:106–123. doi: 10.1006/jmbi.1994.1122. [DOI] [PubMed] [Google Scholar]
- 22.Griffith JD, Harris LD, Register J., III Visualization of SSB-ssDNA complexes active in the assembly of stable RecADNA filaments. Cold Spring Harb. Symp. Quant. Biol. 1984;49:553–559. doi: 10.1101/sqb.1984.049.01.062. [DOI] [PubMed] [Google Scholar]
- 23.De Lean A, Muson PJ, Rodbard D. Simultaneous analysis of families of sigmoidal curves: application to bioassay, radioligand assay, and physiological dose-response curves. Am. J. Physiol. 1978;235:E97–E102. doi: 10.1152/ajpendo.1978.235.2.E97. [DOI] [PubMed] [Google Scholar]
- 24.Krauss G, Sindermann H, Schomburg U, Maass G. Escherichia coli single-strand deoxyribonucleic acid binding protein: stability, specificity and kinetics of complexes with oligonucleotides and deoxyribonucleic acid. Biochemistry. 1981;20:5346–5352. doi: 10.1021/bi00521a040. [DOI] [PubMed] [Google Scholar]