Abstract
Cell-death-inducing DFF45-like effector A (CIDE-A) belongs to a family of proapoptotic proteins, the expression of which is highly restricted in human tissues and cells. Here, the core region of the human CIDE-A promoter was characterized. Surprisingly, two Sp1/Sp3-binding sites, rather than tissue-specific transcription factors, were found to be required for the promoter activity. Although the ubiquitously expressed Sp1 and Sp3 were crucial, they alone could not adequately regulate the specific expression of CIDE-A. We found that the expression of CIDE-A was further regulated by CpG methylation of the promoter region. By performing bisulfite sequencing, we observed dense CpG methylation of the promoter region in tissues and cells with low or no expression of CIDE-A but not in tissues with high level of CIDE-A expression. In vitro methylation of this region showed significantly reduced transcriptional activity. Treatment of CIDE-A-negative cells with 5-aza-2′-deoxycytidine demethylated the CpG sites; this opened the closed chromatin conformation and markedly enhanced the binding affinity of Sp1/Sp3 to the promoter in vivo, thereby restoring CIDE-A expression. These data indicated that CpG methylation plays a crucial role in establishing and maintaining tissue- and cell-specific transcription of the CIDE-A gene through the regulation of Sp1/Sp3 binding.
GenBank Accession Number: EF672106
INTRODUCTION
Apoptosis, a morphologically distinct form of programmed cell death, is an evolutionary highly conserved phenomenon that plays an important role in the regulation of cellular activities in eukaryotes (1). Recently, a novel family of cell-death-inducing DFF45 (DNA fragmentation factor-45)-like effector (CIDE) proteins has been identified. The CIDE family includes three members (CIDE-A, CIDE-B and CIDE-C) in humans, and their mice homologues have also been identified (2–4). CIDE-A was initially identified by virtue of its sequence similarity to the N-terminal region of the apoptotic DNA fragmentation factors DFF40/CAD and DFF45/ICAD. The overexpression of CIDE-A in various cell lines induces caspase-independent cell death associated with the fragmentation of DNA (3). In addition to cell-death induction, recent studies have indicated a correlation between CIDE-A and obesity. CIDE-A-null mice are resistant to diet-induced obesity and diabetes and display higher metabolic rate and lipolysis in the brown adipose tissue than their wild-type littermates (5). In obese women undergoing weight reduction through a low-calorie diet, the CIDE-A gene is the highest upregulated gene in the subcutaneous adipose tissue among 8000 investigated genes (6). Furthermore, CIDE-A is considered to play a human-specific role in lipolysis regulation and metabolic complications of obesity, which is at least partly mediated by cross-talk between CIDE-A and tumor necrosis factor-α (TNF-α) (7). Thus, CIDE-A is also considered as a strong candidate gene for obesity and could serve as a novel target for therapeutic intervention of obesity and Type II diabetes (7–9).
According to the previous reports, the expression of CIDEs is strongly tissue and cell specific. In addition to high levels of expression in the adipose tissue, CIDE-A transcripts of various lengths are present in different human tissues such as the heart, muscle and thymus (3,7). The mice CIDE-A is expressed abundantly in the brown adipose tissue and could be transcriptionally activated by proliferator-activated receptor-α (PPARα) and PPARγ (10). Other CIDE genes also have multiple transcripts with different levels of tissue- and cell-specific expression. For example, the human CIDE-B gene is significantly expressed in the liver and spleen tissues (3), while the expression of the human CIDE-C gene was detected in the small intestine, heart, colon and stomach (4). Studies on the human CIDE-B gene showed that the cell-specific expression of two transcripts of CIDE-B was driven by two different promoters through epigenetic and genetic mechanisms (11). The expression of the adipocyte-specific gene FSP27, a member of the mice CIDE family, was regulated by CCAAT/enhancer-binding protein (C/EBP) and other C/EBP-like transcription factors (2). Thus, the transcription of the CIDE genes appears to be regulated in a strictly tissue- and cell-specific manner. However, the molecular mechanisms that regulate the tissue- and cell-specific transcription of the human CIDE-A gene have not been elucidated.
In the present study, we investigated the transcriptional regulation of the human CIDE-A gene in human tissues and several cancer cells and found that the ubiquitously expressed transcription factors Sp1 and Sp3 were crucial for CIDE-A promoter activity. In addition, we found that DNA methylation of the CIDE-A promoter played an important role in regulating tissue- and cell-specific expression of this gene by interfering with Sp1/Sp3 binding.
MATERIALS AND METHODS
Samples of human tissues and cell lines
Human adipose and liver tissue samples were obtained from Zhongshan Hospital (Shanghai, China). Informed consent was obtained from the patients. The total RNA and genomic DNA of human heart, spleen, kidney, lymph node, lung and stomach tissues were purchased from Shenzhen Innogent Bioscience Inc. All cell lines used in this study were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and were maintained in our laboratory. BEL-7404 cells (human hepatocellular carcinoma), U2OS cells (human osteosarcoma), HEK-293T cells (human embryonic kidney) and Hela cells (human cervical carcinoma) were grown in Dulbecco's modified Eagle's medium (DMEM, Gibco BRL, Grand Island, NY, USA) supplemented with 10% (v/v) fetal bovine serum (FBS, Gibco BRL). Saos-2 cells (human osteosarcoma) were grown in McCoy's 5A medium (Gibco BRL) supplemented with 15% FBS. All media were supplemented with 100 units/ml penicillin and 100 μg/ml streptomycin. All the cells were cultured at 37°C in 5% CO2.
RNA analysis
Total RNA was extracted from cultured cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Total RNA (5 μg) was reverse transcribed using oligo(dT)18 primer and SuperScript II First-Strand Synthesis System (Invitrogen) in a reaction volume of 20 μl according to the manufacturer's instructions. PCR was performed using two pairs of primers. The first primer pair was used for amplifying a 305-bp CIDE-A gene fragment (CIDE-A-F, 5′-AAGAGGTCGGGAATAGCGAGAGTC-3′; CIDE-A-R, 5′-CTGCATCCCTATCCACACGTGAACC-3′). The second primer pair was used for amplifying a 532-bp GAPDH gene fragment as an internal control (GAPDH-F, 5′-AGGTCGGAGTCAACGGATTTG-3′; GAPDH-R, 5′-GTGATGGCATGGACTGTGGT-3′). The amplification conditions for CIDE-A were as follows: initial denaturation at 94°C for 5 min; 30 cycles of 1 min at 94°C, 1 min at 58°C and 30 s at 72°C; and a final extension step for 10 min at 72°C. For GAPDH, the following PCR conditions were applied: 1 cycle at 94°C for 5 min; 25 cycles of 1 min at 94°C, 1 min at 55°C and 1 min at 72°C; and a final extension step for 10 min at 72°C. The PCR products were electrophoresed on a 1.5% agarose gel.
CpG island prediction
The presence of a putative CpG island in proximity to the CIDE-A gene was analysed using the CpG Island Searcher program (http://www.uscnorris.com/cpgislands/cpg.cgi) (12) with the default setting (%GC >55%, ObsCpG/ExpCpG >0.65, length >500 bp).
Plasmid constructs
The 5′ region (−1556 to −13, relative to the translation start site of the human CIDE-A gene) of human CIDE-A was amplified by PCR using high-fidelity LA Taq polymerase (Takara, Dalian, China) from human genomic DNA isolated from HEK-293T cells and inserted into the pGL3-Basic vector (Promega, Madison, WI, USA). Nested deletions of the CIDE-A promoter region were carried out using the plasmid p(−1556/−13) as the template by PCR amplifications, and the sequences of the products were confirmed by direct sequencing. Primers for nested deletions are: -1556-F (5′-CCCTCGAGCTACAGGTGCTCGCCGTCAT-3′); -1112-F (5′-CCCTCGAGACCACGGTGACCCTTTCTGA-3′); -892-F (5′-CCCTCGAGCCGAGTCCCACCCCTCAACA-3′); -640-F (5′-CCCTCGAGTCACTGTCCCCAAATGCTCA-3′); -438-F (5′-CCCTCGAGGAGGCAAGTGGCGCTCACAT-3′); -250-F (5′-CCCTCGAGGGGCTTCCAAGGTCCTCGGT-3′); -196-F (5′-CCCTCGAGGCTGCGGGAGCCAGGACGAC-3′); -137-F (5′-CCCTCGAGGGAGAGCTCGTGCCAAAACG-3′); -13-R (5′-CCAAGCTTTAGCGGGCCTGGAGGTCTGT-3′); -137-R (5′-CCAAGCTTACCGCCGAGGATCGCGACTT-3′)
Site-directed mutagenesis of two Sp1/Sp3 response elements (RE1, −76/−70; RE2, −70/−64) in plasmid p(−137/−13) was performed by PCR (GGGCGGG in Sp1/Sp3 RE was changed to GGACAAG), and the mutagenesis was confirmed by direct sequencing. Primers for site-directed mutations are: Spm1-R (5′-CGGCCCCGCCCTTGTCCGCCTGCCAGCCCCGCGAA-3′); Spm1-F (5′-TTCGCGGGGCTGGCAGGCGGACAAGGGCGGGGCCG-3′); Spm2-R (5′-CGGCCTTGTCCCCGCCCGCCTGCCAGCCCCGCGAA-3′); Spm2-F (5′-TTCGCGGGGCTGGCAGGCGGGCGGGGACAAGGCCG-3′); Spm3-R (5′-CGGCCTTGTCCTTGTCCGCCTGCCAGCCCCGCGAA-3′); Spm3-F (5′-TTCGCGGGGCTGGCAGGCGGACAAGGACAAGGCCG-3′)
Transfections and luciferase assays
For transient transfections, Hela and HEK-293T cells were plated onto 24-well plates 1 day before transfection at a density of 5 × 104 and 2 × 105 cells/well, respectively. Cells were transfected with 0.4 μg of various luciferase constructs by using LipofectAMINE™ reagent (Gibco BRL) according to the manufacturer's instructions. The pRL-TK plasmid (Promega; 40 ng/sample) containing the Renilla luciferase gene driven by the herpes simplex virus thymidine kinase promoter was cotransfected with the constructs, and the luciferase activity was normalized. The preparation of cell lysates and measurements of luciferase activity were performed using the Dual Reporter assay system (Promega) and Lumat LB 9507 luminometer (EG & G Berthold, Berlin, Germany) according to the manufacturer's instructions. All transfections were performed three times in triplicate.
Electrophoretic mobility shift assays
Nuclear extracts were prepared from Hela cells as described by Gazzoli and Kolodner (13). The wild-type probe of the CIDE-A promoter (from −91 to −57) was generated by annealing two complementary oligonucleotides (SpWT: 5′-GCGGGGCTGGCAGGCGGGCGGGGGCGGGGCCGCCG-3′; 5′-GTGCGGCGGCCCCGCCCCCGCCCGCCTGCCAGCCC-3′, the core sequence of the Sp1/Sp3-binding site is underlined) and filling the 3′ recessive ends by repair synthesis with dATP, dTTP, dGTP, [α-32P]dCTP and the Klenow fragment of DNA polymerase I (Takara). The mutated probe of the CIDE-A promoter was generated by the same procedure, except that several oligonucleotides with mutations (SpM1: 5′-GCGGGGCTGGCAGGCGGACAAGGGCGGGGCCGCCG-3′, 5′-GTGCGGCGGCCCCGCCCTTGTCCGCCTGCCAGCCC-3′; SpM2: 5′-GCGGGGCTGGCAGGCGGGCGGGGACAAGGCCGCCG-3′, 5′-GTGCGGCGGCCTTGTCCCCGCCCGCCTGCCAGCCC-3′; SpM3: 5′-GCGGGGCTGGCAGGCGGACAAGGACAAGGCCGCCG-3′, 5′-GTGCGGCGGCCTTGTCCTTGTCCGCCTGCCAGCCC-3′, mutations are shown in bold) in the core sequence of the Sp1/Sp3-binding site were used. Nuclear extracts containing 10 μg of the protein were preincubated in 20 μl of binding buffer [10 mM Tris–HCl (pH 7.5), 50 mM NaCl, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, 50 μg/ml poly(dI-dC)· poly(dI-dC), and 4% glycerol] with or without unlabeled competitor (100-fold molar excess). For supershift assays, 1 μg of antibodies or rabbit IgG (Sigma) was added to the preincubation mixture. After 30-min preincubation on ice, the DNA probe (20 000 cpm) labeled with [α-32P] dCTP was added, and the samples were incubated at room temperature for 30 min. The reaction mixtures were resolved on 4% polyacrylamide gels. The gels were dried and subjected to PhosphorImager analysis using a Storm 860 system and ImageQuant TL software (Amersham Biosciences, Sunnyvale, CA, USA). Antibodies used in supershift assays were rabbit polyclonal antibodies against Sp1 (sc-59X) and Sp3 (sc-644) from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Sodium bisulfite genomic sequencing
Sodium bisulfite modification of genomic DNA was performed as described previously (14). Briefly, 2 μg of genomic DNA, which had been digested with EcoRI and BglII, was denatured in 0.3 M NaOH for 15 min at 37°C and then treated with 3 M sodium bisulfite (Sigma, St Louis, MO, USA) and 10 mM hydroquinone (Sigma) for 16 h at 50°C. The DNA was then desalted using the Wizard DNA Clean-Up System (Promega), desulphonated in 0.3 M NaOH, and precipitated. The treated DNA was amplified by PCR in a 50-μl reaction volume using the bisulfite-specific primers (BS-F, 5′-TTGGTAAGTGATTAAAAGAGATT-3′ and BS-R, 5′-CCTAATTATTTCTCCTACATAATC-3′). The amplification conditions for the 325-bp DNA fragment were as follows: 94°C for 5 min; 35 cycles of 1 min at 94°C, 1 min at 50°C, 30 s at 72°C; and a final extension step for 10 min at 72°C. The purified PCR products were cloned into the pMD18-T vector (Takara), and eight clones were picked out and sequenced for each sample.
Nuclease accessibility assay
The nuclei of cells were prepared as described previously (15) and digested with the restriction enzyme AluI (New England Biolabs, Beverly, MA, USA). DNA was then extracted from the digested nuclei with the proteinase K/phenol procedure (16). DNA from the AluI-digested nuclei was amplified by PCR using primers P1 and P3 and electrophoresed on a 2% agarose gel. The absence or presence of a 238-bp fragment indicated whether AluI is accessible to the chromatin in the CIDE-A promoter region. To assess the input of the chromatin, another primer, P2, was also included in the PCR reaction mixture. The amount of AluI-digested chromatin was represented by the ratio 238:113 bp. The primers used were as follows: P1, 5′-GGGCTTCCAAGGTCCTCGGT-3′; P2, 5′- ACCGCCGAGGATCGCGACTT-3′; P3, 5′- TAGCGGGCCTGGAGGTCTGT-3′.
In vitro methylation of the critical CIDE-A promoter region
The CIDE-A reporter construct p(−250/−13) and the pGL3-Basic vector were methylated by incubation with SssI methylase (New England BioLabs) for 3 h at 37°C in the presence of 160 μM S-adenosylmethionine. The methylation status was verified by digestion with HhaI, a methylation sensitive enzyme.
5-aza-2′-Deoxycytidine and trichostatin A treatment
Cells were treated with 1 and 10 μM 5-aza-2′-deoxycytidine (5-aza-dC, Sigma) for 72 h or with 300 nM trichostatin A (TSA) (Sigma) for 24 h, as described previously (16,17). Briefly, the cells were plated onto 6-well plates at a density of 2 × 104 at 24 h before treatment. The cells were then treated with 5-aza-dC or TSA. The same volume of dissolving stock was used as a negative control. Due to the chemical instability of 5-aza-dC, fresh inhibitor was added after every 24 h for a total of 72-h incubation.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was performed using the ChIP-IT kit (Active Motif, Carlsbad, CA, USA) according to the manufacturer's protocol. Briefly, cells were treated with 1 μM 5-aza-dC for 3 days, along with a negative control. Formaldehyde was added to the cells at a final concentration of 1% and incubated for 10 min at room temperature. The cells were pelleted, resuspended in SDS lysis buffer (1% SDS/10 mM EDTA/50 mM Tris–HCl, pH 8.1), and incubated on ice. The lysate samples were sonicated, and debris was removed by centrifugation. After obtaining the chromatin solution, salmon sperm DNA–protein G agarose slurry was added and incubated for 2 h at 4°C. Beads were pelleted by centrifugation, and the supernatants were incubated overnight at 4°C with 2 μg of specific antibodies [anti-Sp1 (sc-59X) and anti-Sp3 (sc-644), Santa Cruz Biotechnology; anti-acetyl-Histone H3 K9 and anti-dimethyl-Histone H3 K9, Upstate Biotechnology, Inc.]. The resultant DNA complexes were collected using a salmon sperm DNA–protein G agarose slurry. After reverse cross-linking of protein–DNA with 5 M NaCl followed by RNase A treatment, DNA was purified using mini-columns provided with the kit. The purified DNA was amplified by the promoter-specific primers (ChIp-F, 5′-GGGCTTCCAAGGTCCTCGGT-3′ and ChIp-R, 5′-CACAGCCTCTTAAAGTGCGG-3′), and PCR was performed under the following conditions: 1 cycle at 94°C for 5 min; 35 cycles of 1 min at 94°C, 1 min at 58°C, 30 s at 72°C; and a final extension step for 10 min at 72°C. The PCR products were analysed by electrophoresis on a 2% agarose gel.
RNA interference and western blot analysis
RNA interference was achieved by using siRNA pools of Sp1 and Sp3 (Genepharm Research Shanghai, China). A functional non-targeting siRNA was used as a control for potential non-specific effects caused by siRNA transfection. HEK-293T cells were plated in 6-well dishes and grown to ∼60% confluence. Cells were then transfected using Lipofectamine 2000 according to the manufacturer's protocol (100 nM of each siRNA pool).
Forty-eight hours after transfection, HEK-293T cells were harvested for detection of Sp1 and Sp3 proteins. Total cell samples were harvested by applying 2×SDS Laemmli sample buffer directly on to cell monolayers after three washes with PBS. Electrophoresis, transfer of proteins on to nitrocellulose membranes and blocking of membranes were performed as described previously (18). Antibodies specifically against Sp1 (anti-Sp1, sc-59X), Sp3 (anti-Sp3, sc-644) and tubulin (Ab-4; NeoMarkers, Freemont, CA, USA) were incubated with the membranes for 1 h at room temperature. Secondary antibody [goat anti-rabbit and donkey anti-goat IgG conjugated to horseradish peroxidase (Santa Cruz)] was used at a dilution of 1:2000, and incubated with membranes for 1 h at room temperature. Proteins revealed by western blotting were visualized by chemiluminescence.
RESULTS
Tissue- and cell-specific expression of CIDE-A and the structure of the CIDE-A locus
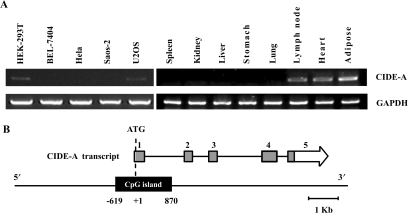
To confirm that the transcription of the CIDE-A gene is regulated in a cell- and tissue-specific manner, the expression of the CIDE-A gene in five cancer cell lines and eight human tissues was measured by reverse transcription-PCR using the GAPDH gene as an internal control (Figure 1A). A high level of expression of the CIDE-A gene was observed in adipose, lymph node and heart tissues, while no expression was detected in liver, kidney, lung, spleen and stomach tissues. Furthermore, the expression level in HEK-293T and U20S cells was very low and in Hela, BEL-7404 and Saos-2 cells was undetectable.
Figure 1.
Cell- and tissue-specific expression and genomic structure of the human CIDE-A gene. (A) RT-PCR analysis of CIDE-A expression in five human cell lines and eight human tissues as indicated. Five micrograms of total RNA from cells and tissues was analysed by RT-PCR using primers specific for CIDE-A CDS. A 305-bp fragment of CIDE-A CDS was amplified. A 532-bp fragment of the GAPDH gene was used as a positive control for RNA integrity. (B) Structure of the human CIDE-A locus. Boxed arrows represent published cDNAs encoding known proteins. The open, grey and black boxes represent the untranslated regions, ORFs and predicted CpG island, respectively.
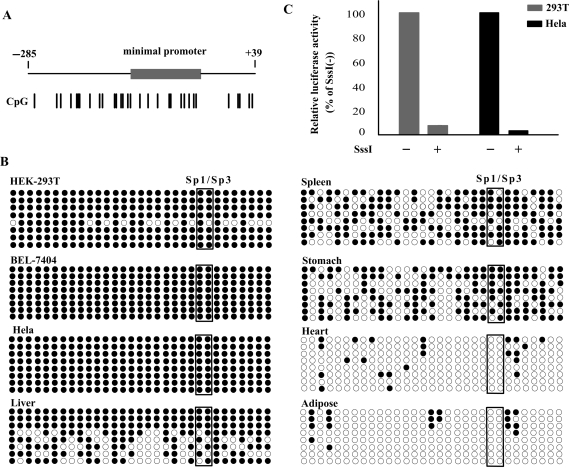
The genomic structure of CIDE-A was analysed and is shown in Figure 1B. The gene corresponding to the reported cDNA that encode CIDE-A (GenBank accession number NM_001279) is mapped to chromosome 18p11. The CIDE-A transcript contains 5 exons (exons 1, 2, 3, 4 and 5). The CpG Island Searcher program revealed a CpG island (from −619 to 870) spanning the 5′ upstream area and exon 1 of the CIDE-A gene. To simplify the nomenclature, we have attributed position number +1 to the translation start site of the CIDE-A gene.
Identification of the critical promoter region of CIDE-A
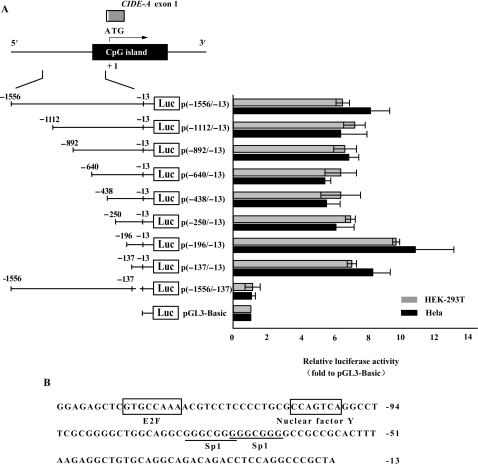
To investigate the transcriptional regulation of the CIDE-A gene, we identified the promoter region of this gene. We first constructed a luciferase reporter plasmid p(−1556/−13) that contained a 1544-bp genomic DNA fragment spanning the 5′ upstream region of CIDE-A. This plasmid was used for transient transfection of Hela and HEK-293T cells. HEK-293T cells expressed very low level of CIDE-A, whereas Hela cells did not (Figure 1A). The cloned fragment exhibited obvious promoter activity in both cell lines, indicating that an active promoter sequence was isolated (Figure 2A). Next, we developed a series of deletion constructs to identify the minimal promoter required for activation. Deletion in the 3′ region from −13 to −137 completely abolished the activity of the reporter gene and enabled to locate the minimal promoter within the 125-bp region between positions −137 and −13 (Figure 2A). The nucleotide sequence of the minimal promoter region is shown in Figure 2B and deposited in GenBank under the accession number EF672106. This sequence is located in the CpG island. We searched for transcription factor binding sites using the MatInspector program and found consensus sequences for the binding of Sp1/Sp3, E2F and Nuclear factor Y. Although Hela cells did not express CIDE-A, the different constructs exhibited obvious promoter activity, regardless of the CIDE-A expression status, suggesting additional mechanisms of CIDE-A gene regulation in vivo.
Figure 2.
Identification of the minimal promoter region of the CIDE-A gene. (A) Identification of the critical CIDE-A promoter region using luciferase assays. The CIDE-A transcripts (open and grey boxes) and CpG island (black box) are indicated. Hela cells (black bars) and HEK-293T cells (grey bars) were transfected with the indicated constructs and assayed for luciferase activity 24 h after transfection. Numbers above the constructs indicate the positions relative to the translation start site of CIDE-A. The activity of each promoter construct is shown as the luciferase activity relative to that of the pGL3-Basic vector (a promoterless vector). Values were normalized for transfection efficiency by cotransfection with the Renilla expression plasmid and are shown as means ± SD for three independent experiments. (B) Nucleotide sequence of the CIDE-A minimal promoter region. The E2F and nuclear factor Y response elements are shown in boxes. The consensus sequences for two potential Sp1/Sp3-binding sites are underlined.
Sp1/Sp3-binding motifs are important for CIDE-A promoter activity
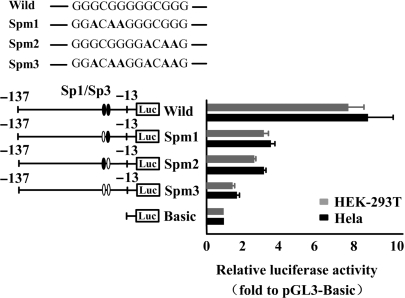
As shown in Figure 2B, two putative binding sites for Sp1/Sp3 were found in the minimal promoter region of CIDE-A. To determine whether the Sp1/Sp3-binding elements are important in CIDE-A promoter activity, we generated several constructs of p(−137/−13) that contained mutations in the Sp1/Sp3-binding site by site-directed mutagenesis. The reporter constructs containing mutated sequences were then transfected into Hela and HEK-293T cells and assayed for promoter activities. The mutation induced in either of the Sp1/Sp3 sites decreased the promoter activity level to ∼50% of that of the wild type, whereas the activity decreased to a basal level when both the Sp1/Sp3-binding sites were mutated (Figure 3). These results clearly indicate that both the Sp1/Sp3-binding sites are essential for the activity of the CIDE-A minimal promoter.
Figure 3.
Sp1/Sp3-binding motifs are important for CIDE-A promoter activity. The effect of Sp1/Sp3-binding site on the regulation of CIDE-A promoter activity is shown. Values of luciferase activities due to the wild-type construct p(−137/−13) and the mutated constructs (Spm1, Spm2 and Spm3) in Hela cells (black bars) and HEK-293T cells (grey bars) are shown as means ± SD for three independent experiments, each of which was performed using triplicate samples. Mutations are shown in bold above the histogram.
Determination of the specific binding of Sp1 and Sp3 to the critical CIDE-A promoter region
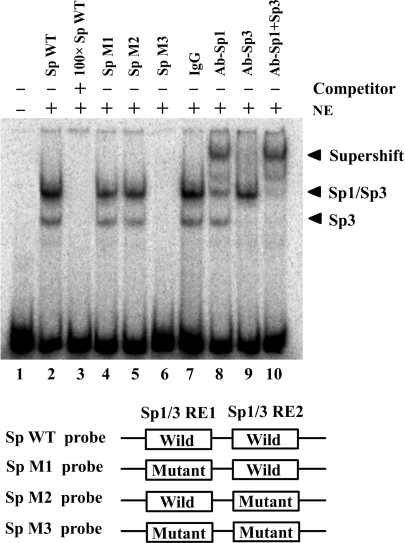
To directly demonstrate that Sp1 and Sp3 bind to the putative binding site within the critical CIDE-A promoter region, EMSA was performed using α-32P-labeled oligonucleotides spanning the two putative Sp1/Sp3-binding sites with nuclear extracts generated from Hela cells. Shifted complexes were observed when the labeled wild-type probe was used (Figure 4, lane 2); these complexes were eliminated by the addition of 100-fold molar excess of unlabeled wild-type probe (Figure 4, lane 3). EMSA was also performed on mutant probes to understand the contribution of each of the Sp1/Sp3-binding sites to the observed binding patterns. Any mutation in the two Sp1/Sp3-binding sites (SpM1 or SpM2 probe) slightly decreased the level of complex formation, while these bands were not observed when using the SpM3 probe with mutations in both the Sp1/Sp3-binding sites (Figure 4, lanes 4–6). To further confirm the specific binding of Sp1/Sp3 to the CIDE-A minimal promoter, supershift assay was performed using anti-Sp1 and anti-Sp3 antibodies. The slowest migrating protein–DNA complex was supershifted by anti-Sp1 antibodies (Figure 4, lane 8), whereas the rapidly migrating bands were supershifted by anti-Sp3 antibodies (Figure 4, lane 9). However, no supershift was observed when control IgG was used instead of specific antibodies (Figure 4, lane 7). The EMSA results demonstrated that Sp1 and Sp3 bound specifically to the two Sp1/Sp3-binding sites in the CIDE-A minimal promoter.
Figure 4.
Specific binding of Sp1 and Sp3 to the critical CIDE-A promoter region. The binding of Sp1 and Sp3 to the CIDE-A core promoter was determined by electrophoretic mobility shift analysis (EMSA). By using 32P-labeled 35-bp double-stranded oligonucleotides containing wild or mutated Sp1/Sp3-binding sites as probes, EMSAs were performed with the same amount of nuclear extracts (NE) from Hela cells, and the products were separated on a 4% polyacrylamide gel (lanes 2–10). Lane 1, free probe; lane 2, 32P-labeled wild-type Sp1/Sp3 consensus oligonucleotides were mixed with nuclear proteins; lane 3, the same reaction was performed as that in lane 2, except for the presence of a 100-fold excess of unlabeled wild-type Sp1/Sp3 consensus oligonucleotides as a competitor; lanes 4–6, binding assays of 32P-labeled mutant-type Sp1/Sp3 consensus oligonucleotides mixed with nuclear proteins; lanes 7–10, 1 μg each of IgG, anti-Sp1, anti-Sp3 or both anti-Sp1 and anti-Sp3 antibodies were added to the binding reaction mixtures with 32P-labeled wild-type probe.
Methylation status of CpG sites in the CIDE-A promoter
Sp1 and Sp3 are ubiquitously expressed transcription factors; therefore, they might not be responsible for the tissue- and cell-specific expression of the CIDE-A gene. The CIDE-A promoter is located in a CpG island (Figure 5A), and the Sp1/Sp3-binding sites in the CIDE-A gene promoter contain a CpG dinucleotide, a potential target for DNA methylation in mammals; this suggests that DNA methylation may play a role in the regulation of tissue- and cell-specific expression of the CIDE-A gene. We then examined the methylation status of 31 CpG sites in the promoter region by sodium bisulfite sequencing (Figure 5B). The results from the sequences of eight clones showed that in CIDE-A expressed tissues the overall percentage of methylated CpG sites was very low, only ∼7% in adipose and ∼9% in heart, indicating the CpG sites in CIDE-A promoter region were unmethylated in adipose and heart tissues; In contrast, in tissues that did not express CIDE-A (liver, spleen and stomach), the percentage of methylated CpG sites was higher, about 73%, 63% and 57%, respectively, indicating the hypermethylated CpG sites in CIDE-A absent expressed tissues. In the cells in which CIDE-A was silenced (Hela and BEL-7404), a high degree of methylation was observed at each CpG site, including the Sp1/Sp3-binding sites. However, in HEK-293T cells, that with a lower expression of CIDE-A, the methylation patterns were more mosaic, in eight clones sequenced we detected one clone with lower percentage of methylation (∼55%) and other clones were fully methylated. These results revealed that the methylation status of the CpG dinucleotide reflected the tissue and cell specificity of the CIDE-A gene expression. The effect of in vitro methylation on the promoter activity of the CIDE-A gene was then investigated through transient transfection of CpG-methylated reporter plasmids into HEK-293T and Hela cells. We modified the promoter constructs with SssI methylase and studied the activity of the methylated promoter. The luciferase activities of CpG-methylated reporter plasmids were calculated relative to those of unmethylated plasmids (unmethylated controls). The luciferase activity of SssI-methylated pGL3-(−250/−13) decreased to ∼5% and 4% of that of the unmethylated controls in HEK-293T and Hela cells, respectively (Figure 5C), suggesting a direct association between CpG methylation and CIDE-A promoter activity.
Figure 5.
Methylation status of CpG sites in the CIDE-A promoter. (A) Schematic diagram of the CIDE-A gene 5′ region. Numbers indicate the positions relative to the translation start site of CIDE-A. The CIDE-A minimal promoter is indicated by the grey box. The position of the 31 CpG sites detected by bisulfite sequencing is indicated by a short vertical line. (B) Bisulfite sequencing of the CIDE-A promoter region. The genomic DNA from BEL-7404, Hela and HEK-293T cells and from the liver, spleen, stomach, heart and adipose tissues was treated with bisulfite and amplified by PCR. The PCR products were then subcloned and sequenced. For each cell line and tissue, the methylation status of 31 CpGs in the promoter region is shown for eight clones. White and black circles indicate unmethylated and methylated CpGs, respectively. Circles enclosed by boxes represent the CpGs in the Sp1/Sp3-binding sites. (C) Effect of in vitro methylation on the activity of the CIDE-A promoter. The CIDE-A promoter-luciferase plasmid p(−250/−13) was methylated in vitro by SssI methylase. Methylated and unmethylated p(−250/−13) were then transiently transfected into Hela and HEK-293T cells and assayed for luciferase activity. Results are shown as percentages of luciferase activity, with the activity due to unmethylated p(−250/−13) considered as 100%. The average ± SD values from three independent experiments performed in triplicates are shown.
Induction of CIDE-A gene expression by 5-aza-dC
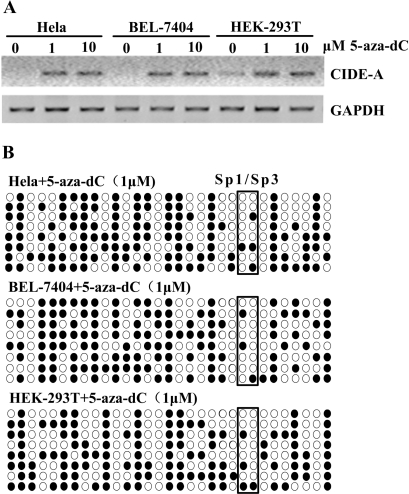
To confirm the role of DNA methylation in the CIDE-A minimal promoter in silencing gene expression, two CIDE-A-negative cell lines, i.e. Hela and BEL-7404, and one cell line with low-level expression of CIDE-A, i.e. HEK-293T, were treated with the DNA methyltransferase inhibitor 5-aza-dC (1 and 10 μM). As shown in Figure 6A, CIDE-A expression was induced in Hela and BEL-7404 cells after 72 h of exposure to the reagent, while the expression in HEK-293T cells was also increased significantly. The expression of CIDE-A was induced at a similar level following treatment with 5-aza-dC at 1 and 10 μM concentrations. The methylation status in Hela, BEL-7404 and HEK-293T cells after 5-aza-dC treatment was then determined by sodium bisulfite sequencing. Methylation was significantly decreased in the CIDE-A critical promoter region in Hela, BEL-7404 and HEK-293T cells after 5-aza-dC treatment (Figure 6B). The non-expressing cell lines Hela and BEL-7404 were also treated with 300 nM TSA, a histone deacetylase inhibitor. No significant induction of CIDE-A expression was detected by reverse transcription-PCR (data not shown), suggesting that histone deacetylase inhibition by TSA was not sufficient to reactivate the CIDE-A gene that was silenced following hypermethylation of the promoter region.
Figure 6.
Induction of the CIDE-A gene expression by 5-aza-dC. (A) HEK-293T, Hela and BEL-7404 cells were treated with 1 or 10 μM 5-aza-dC. Seventy-two hours after 5-aza-dC treatment, total RNA was isolated from treated or untreated cells and subjected to RT-PCR to amplify CIDE-A. The GAPDH gene was used as a positive control for RNA integrity. (B) Bisulfite sequencing to determine the methylation status of the critical CIDE-A promoter region after 5-aza-dC treatment. Circles enclosed by boxes represent the CpGs in the Sp1/Sp3-binding sites.
5-aza-dC treatment changes the chromatin conformation around the CIDE-A promoter
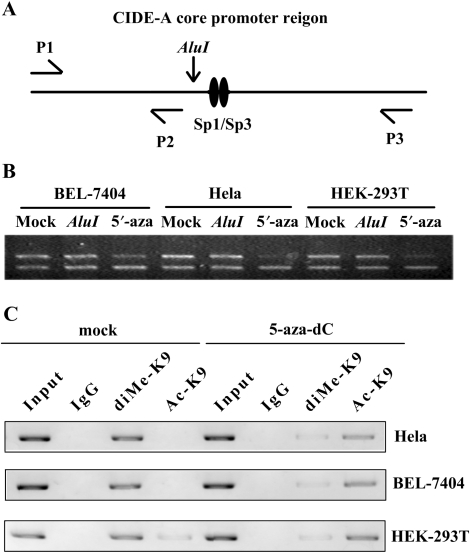
Chromatin conformation near the CIDE-A promoter was studied by the nuclease accessibility assay and ChIP assay. The nuclei from Hela, BEL-7404 and HEK-293T cells were digested with the restriction enzyme AluI. DNA extracted from the nuclei was amplified by PCR and analysed by electrophoresis. The ratio 238:113 bp represents the amount of undigested chromatin at the AluI site (−135/−130) in the CIDE-A promoter (Figure 7A). This ratio remained unchanged in these cells when compared with DNA from undigested nuclei, indicating that the AluI enzyme was not accessible to the AluI sequence in the chromatin and that the chromatin conformation was closed in these cells. After treatment with 1 μM 5-aza-dC, the ratio slightly decreased in BEL-7404 cells, while the decrease in the ratio was considerably more significant in Hela or HEK-293T cells (Figure 7B); this indicates that the chromatin around the CIDE-A promoter adopted a more open conformation after 5-aza-dC treatment.
Figure 7.
5-aza-dC treatment changes the chromatin conformation around the CIDE-A promoter. (A) Procedure of nuclease accessibility assay. Nuclei were digested with the restriction enzyme AluI. DNA was extracted from the nuclei and amplified by PCR using a set of three primers (P1, P2 and P3). The decrease in the ratio 238:113 bp after AluI digestion indicates that the chromatin is accessible to AluI. (B) Chromatin accessibility changes in the critical CIDE-A promoter region in Hela, HEK-293T and BEL-7404 cells following treatment with 5-aza-dC. The ratio decreased significantly after treating with 1 M 5-aza-dC for 3 days. (C) In vivo H3K9 di-methylation and acetylation levels in the CIDE-A core promoter before and after 3 days’ 1 M 5-aza-dC treatment were analysed by Chromatin immunoprecipitation (ChIP) assay using antibodies that recognize Ac-H3K9 and di-methyl H3K9. The normal rabbit IgG was used as a negative control and Input indicates 5% input DNA, a positive amplification control.
The epigenetic silencing of genes can result from the close connection between DNA methylation and histone code modifications in regions of transcriptional regulation. Dimethylated lysine 9 (di-methyl H3K9) and acetylated lysine 9 (Ac-H3K9) are two of the most widely studied histone modifications. Ac-H3K9 are typically associated with transcriptionally active promoters, whereas di-methyl H3K9 is associated with repressed promoters (19). We then utilized chromatin immunoprecipitation to determine the levels of the two histone H3 tail lysine modifications at CIDE-A gene promoters (Figure 7C). In Hela and BEL-7404 cells, where CIDE-A is none-expressed, high levels of di-methyl H3K9 were detected, whereas H3K9 acetylation was not detectable, consistent with their transcriptionally silent state. However, in HEK-293T cells, the H3K9 di-methylation levels of the promoter regions was similar with Hela and BEL-7404 cells, but having very low levels of detectable Ac-H3K9, which could explain the very low-level expression of CIDE-A. After treatment with 1 μM 5-aza-dC, H3K9 di-methylation was significantly diminished and H3K9 acetylation was obviously increased in all cases, which closely linked with gene reactivation. In light of our data above, we conclude that the activation of the CIDE-A locus was associated with chromatin remodeling after demethylation of the promoter region.
CpG methylation inhibits the binding of Sp1 and Sp3 to the CIDE-A promoter in vivo
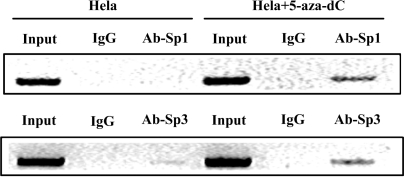
To elucidate the effects of methylation of the CIDE-A promoter on the binding of Sp1 and Sp3 to the promoter, ChIP assays were performed in CIDE-A-non-expressing Hela cells. No detectable Sp1 and very weak Sp3 binding were observed in Hela cells, whereas treatment with 1 μM 5-aza-dC for 72 h markedly enhanced the binding affinity of Sp1 and Sp3 to the CIDE-A promoter along with the induction of CIDE-A expression (Figure 8). These findings suggested that methylation of CpG sites in the CIDE-A promoter interfered with the binding of Sp1 and Sp3 in vivo.
Figure 8.
CpG methylation inhibits the binding of Sp1 and Sp3 to the CIDE-A promoter in vivo. ChIP was performed using the chromatin from Hela cells with or without 3 days’ 1 uM 5-aza-dC treatment. Protein–DNA complexes were subjected to immunoprecipitation using anti-Sp1 or anti-Sp3 antibodies. The normal rabbit IgG was used as a negative control. Protein-bound immunoprecipitated DNA was eluted and recovered for the PCR analysis of the CIDE-A promoter region by using a primer set specific for the CIDE-A promoter. Input represents the relative amounts of protein–DNA complex subjected to immunoprecipitation.
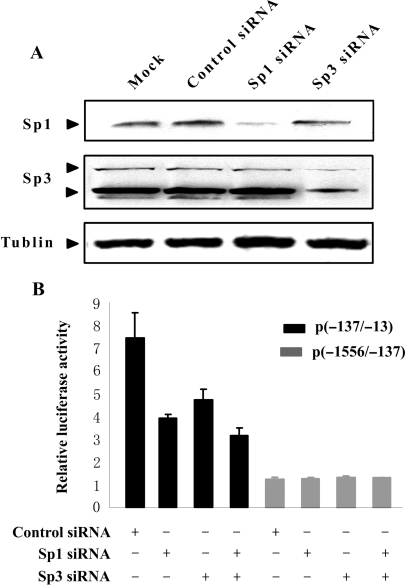
Knockdown of Sp1 and Sp3 expression by siRNA reduces CIDE-A promoter activity
To confirm the role of Sp1 and Sp3 in the regulation of CIDE-A promoter activity, we used RNA interference to knock down Sp1 and Sp3 expression in HEK-293T cells. Strongly reduced Sp1 and Sp3 protein levels were observed by Western blot analysis, whereas control siRNA had no effect on Sp1 or Sp3 protein levels (Figure 9A). HEK-293T cells were transfected with Sp1 and Sp3 siRNAs individually or simultaneously and then transfected again with the luciferase reporter plasmid containing the 125 bp CIDE-A core promoter. This resulted in a 38–58% reduction of luciferase activity in the presence of Sp1- or Sp3-specific siRNA compared to that with control siRNA. In contrast, no influence on transcriptional activity of control plasmid p(-1556/-137), which does not contain the Sp1/Sp3-binding motif, was observed (Figure 9B). These results indicate that Sp1 and Sp3 positively regulate the expression of CIDE-A by transactivating the promoter.
Figure 9.
Knockdown of Sp1 and Sp3 expression by siRNA reduces CIDE-A promoter activity. (A) Immunoblotting analysis of Sp1 and Sp3 protein levels in HEK-293T cells transfected with Sp1 and Sp3 siRNA. Control siRNA was used as negative control and Tublin as a loading control. (B) The effect of Sp1 or Sp3 siRNA on CIDE-A promoter activity detected by luciferase reporter assay. HEK-293T cells were transfected with Sp1 and Sp3 siRNAs individually or simultaneously and then transfected again with the luciferase reporter plasmid p(−137/−13) and p(−1556/−137). The activity of each promoter construct is shown as the luciferase activity relative to that of the pGL3-Basic vector. Values were normalized for transfection efficiency by cotransfection with the Renilla expression plasmid and are shown as means ± SD for three independent experiments.
DISCUSSION
Recent studies have shown that CIDE-A, which was originally identified as a proapoptotic protein, is strongly correlated with obesity. The expression pattern and functions of CIDE-A are well established; however, little is known about its transcriptional regulation. Since CIDE-A is considered as a novel target for therapeutic intervention of obesity and Type II diabetes, a better understanding of the mechanism responsible for the expression of this gene may provide new opportunities for therapy.
In the present study, we aimed to characterize the mechanism that regulated the tissue- and cell-specific expression of the CIDE-A gene. By using 5′ flanking region-luciferase fusion plasmids, we identified the core CIDE-A promoter as a 125-bp region located between the base pairs −137 to −13 that contained two Sp1/Sp3-binding sites. The activity of the CIDE-A promoter was greatly reduced when site-directed mutations were introduced in each of these binding sites; this finding demonstrated a crucial role of the Sp1/Sp3-binding sites in CIDE-A promoter activity. In addition, EMSAs also confirmed that Sp1 and Sp3 could specifically bind to the putative binding sites identified in the minimal region. In view of this finding, the fact that the CIDE-A gene was expressed in a tissue- and cell-specific manner was rather confusing because Sp1 and Sp3 are two ubiquitously expressed mammalian transcription factors that bind to DNA with similar specificity and affinity and participate in the regulation of the expression of genes involved in almost all cellular processes (20,21). Thus, it appears that Sp1 and Sp3 alone might not determine tissue- and cell-specific expression of the CIDE-A gene. Interestingly, the basal promoter activity of the core region was observed even in cells that did not endogenously express CIDE-A. Similar phenomenon was also observed in previous studies on CIDE-B, NF1 (neurofibromatosis type 1) and APC (adenomatous polyposis of the colon) promoters (11,22,23). Since the transiently transfected constructs were unlikely to adopt the native chromatin structure, this observation prompted us to investigate whether DNA methylation was involved in the mechanisms underlying the silencing of the CIDE-A gene in non-expressing cells and tissues.
We investigated the DNA methylation profiles of a portion of the CpG island covering the Sp1/Sp3-binding sites. Sodium bisulfite sequencing analysis revealed that the CpG sites in the minimal promoter region were hypomethylated in CIDE-A expressed tissues (heart and adipose), but they were hypermethylated in CIDE-A absent or low expressed tissues (liver, spleen and stomach) and cell lines (Hela, BEL-7404 and HEK-293T cells). The involvement of methylation in the CIDE-A gene silencing was further supported by the finding that cells transfected by constructs densely methylated by SssI methylase in vitro showed a significant decrease in luciferase activities. Moreover, we found that after treatment with the methyltransferase inhibitor 5-aza-dC, the gene expression was restored in all the CIDE-A non-expressing cells. In addition, the nuclease accessibility assay demonstrated that the chromatin conformation around the Sp1/Sp3-binding sites in the CIDE-A non-expressing cells changed from ‘closed’ to ‘open’ after 5-aza-dC treatment. We further used ChIP to determine the levels of two critical histone H3 modifications (Ac-H3K9 and di-methyl H3K9) at CIDE-A gene core promoter. Consistent with the studies of other genes, we find that increased levels of di-methyl H3K9 at CIDE-A promoter correlates with a lack of gene expression, whereas an increased abundance of Ac-H3K9 correlates with increased gene expression. However, no significant changes were observed in CIDE-A expression levels, methylation status or chromatin conformation after the cells were treated with TSA, a histone deacetylase inhibitor. This observation is consistent with the previous reports on the regulation of hypermethylated genes such as hMLH1, CDKN2B (p15INK4B) and CDKN2A (p16INK4) (16). In these studies, 5-aza-dC treatment induced the expression of these hypermethylated genes and reversed histone modification, whereas TSA treatment did not induce these changes (16,24). Taken together, all these findings satisfy two well-known criteria for defining the role of DNA methylation in tissue- and cell-specific gene repression: (i) CIDE-A was densely methylated in non-expressing tissue and cells and (ii) induced demethylation resulted in the activation of CIDE-A.
The methylation of genomic DNA at CpG dinucleotides is a major epigenetic modification of the mammalian genome (25), and it functions in several aspects of gene expression such as X chromosome inactivation, genomic imprinting, immobilization of transposons and suppression of transcriptional noise (26–28). Methylation of CpG sites in the promoter region is also believed to ensure the silencing of tissue-specific genes (29). Several mechanisms have been proposed for methylation-induced gene repression. Among these mechanisms, the direct blocking of the binding of transcription factors to their CpG-containing binding sites is an important mechanism that contributes to transcriptional repression. In the case of c-Myb (30), c-Myc/Myn (31), E2F (32), CREB (cAMP-response element-binding protein) (33), AP2 (activator protein 2) (34) and NF-κB (nuclear factor κB) (35), methylated CpGs in their recognition site preclude the binding of the transcription factor and thereby directly inhibit gene expression.
Previous studies have shown that the Sp1-binding affinity to its responding element is not consequentially affected by DNA methylation, which appears to be promoter dependent. The Sp1-binding affinity to the luteinizing hormone receptor (LHR) or CLDN4 promoter was not affected by DNA methylation (36,37). In contrast, in the case of p21Cip1, 11 β-hydroxysteroid dehydrogenase type 2, GSTP1 (glutathione S-transferase p1) or IGFBP-3 (insulin-like growth factor-binding protein-3), the Sp1-binding affinity to the respective promoter region was found to be abolished by DNA methylation (38–41). Because of the vital roles of Sp1/Sp3 in maintaining CIDE-A basal promoter activity, we further investigated whether DNA methylation could impair the binding of Sp1/Sp3 to the CIDE-A promoter in non-expressing cells. In our study, the ChIP assay clearly showed that the binding of Sp1 and Sp3 to the CIDE-A promoter could be diminished by methylation of the Sp1/Sp3 motif, similar to that observed for p21Cip1, GSTP1 and IGFBP-3 promoters; furthermore, treatment of Hela cells with 1 μM 5-aza-dC for 3 days markedly enhanced the binding affinity of Sp1/Sp3 along with restoration of the expression of CIDE-A. We further confirmed the roles of Sp1 and Sp3 in regulation of CIDE-A promoter activity by using RNA interference. Consistent with our reporter assays demonstrating a crucial role for Sp1/Sp3 sites in CIDE-A promoter activity, Sp1 and Sp3 knockdown significantly reduced transcription levels controlled by the CIDE-A promoter. Taken together, our results clearly indicate that the methylation status of the CIDE-A promoter correlated well with CIDE-A gene expression in a subset of cell lines and tissues, and that DNA methylation was the key mechanism of regulating tissue- and cell-specific expression of CIDE-A through blocking of the binding of Sp1 and Sp3 to the CIDE-A promoter. Our findings also suggest that the switch between transcriptional silencing and expression of the tissue-specific gene could depend on varying epigenetic modification, which is driven by basal transcription factors such as Sp1/Sp3 and activation of the conserved promoter sites. It would be interesting to investigate the change in the methylation status of the CIDE-A promoter during adipose differentiation and to determine whether the regulation of higher-order chromatin structures by methyl-CpG-binding proteins such as MeCP1 (42), MeCP2 (43,44) and MBD2 (43,45) is involved in CIDE-A gene silencing in non-expressing cells and tissues. Such information would provide further insights into the role of CpG methylation in the regulation of tissue-specific expression of the CIDE-A gene.
In conclusion, our data revealed that DNA methylation of CpG sites in the CIDE-A promoter was the primary mechanism through which the transcription of CIDE-A was silenced or repressed in non-expressing cells and tissues. In addition, we showed that the Sp1/Sp3 consensus sequence—the critical regulatory elements—were methylated, and methylation of this motif prohibited the access of Sp1 and Sp3 transcriptional factors, resulting in the downregulation of the activity of the CIDE-A promoter.
ACKNOWLEDGEMENTS
We are grateful to other members of our group for technical assistance and helpful discussion. This work was supported by grants from the National High Technology Research and Development of China (863 Program; No.2006AA02Z190 and No.2002BA711A02), the National Natural Sciences Foundation of China (No.30571059). Funding to pay the Open Access publication charges for this article was provided by 863 Program (No.2006AA02Z190).
Conflict of interest statement. None declared.
REFERENCES
- 1.Wyllie AH, Kerr JF, Currie AR. Cell death: the significance of apoptosis. Int. Rev. Cytol. 1980;68:251–306. doi: 10.1016/s0074-7696(08)62312-8. [DOI] [PubMed] [Google Scholar]
- 2.Danesch U, Hoeck W, Ringold GM. Cloning and transcriptional regulation of a novel adipocyte-specific gene, FSP27. CAAT-enhancer-binding protein (C/EBP) and C/EBP-like proteins interact with sequences required for differentiation-dependent expression. J. Biol. Chem. 1992;267:7185–7193. [PubMed] [Google Scholar]
- 3.Inohara N, Koseki T, Chen S, Wu X, Nunez G. CIDE, a novel family of cell death activators with homology to the 45 kDa subunit of the DNA fragmentation factor. EMBO J. 1998;17:2526–2533. doi: 10.1093/emboj/17.9.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang L, Zhao M, Xu Z, Yokoyama KK, Li T. Molecular cloning and characterization of CIDE-3, a novel member of the cell-death-inducing DNA-fragmentation-factor (DFF45)-like effector family. Biochem. J. 2003;370:195–203. doi: 10.1042/BJ20020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Z, Yon Toh S, Chen Z, Guo K, Ng CP, Ponniah S, Lin SC, Hong W, Li P. Cidea-deficient mice have lean phenotype and are resistant to obesity. Nat. Genet. 2003;35:49–56. doi: 10.1038/ng1225. [DOI] [PubMed] [Google Scholar]
- 6.Dahlman I, Linder K, Arvidsson Nordstrom E, Andersson I, Liden J, Verdich C, Sorensen TI, Arner P. Changes in adipose tissue gene expression with energy-restricted diets in obese women. Am. J. Clin. Nutr. 2005;81:1275–1285. doi: 10.1093/ajcn/81.6.1275. [DOI] [PubMed] [Google Scholar]
- 7.Nordstrom EA, Ryden M, Backlund EC, Dahlman I, Kaaman M, Blomqvist L, Cannon B, Nedergaard J, Arner P. A human-specific role of cell death-inducing DFFA (DNA fragmentation factor-alpha)-like effector A (CIDEA) in adipocyte lipolysis and obesity. Diabetes. 2005;54:1726–1734. doi: 10.2337/diabetes.54.6.1726. [DOI] [PubMed] [Google Scholar]
- 8.Lin SC, Li P. CIDE-A, a novel link between brown adipose tissue and obesity. Trends Mol. Med. 2004;10:434–439. doi: 10.1016/j.molmed.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 9.Dahlman I, Kaaman M, Jiao H, Kere J, Laakso M, Arner P. The CIDEA gene V115F polymorphism is associated with obesity in Swedish subjects. Diabetes. 2005;54:3032–3034. doi: 10.2337/diabetes.54.10.3032. [DOI] [PubMed] [Google Scholar]
- 10.Viswakarma N, Yu S, Naik S, Kashireddy P, Matsumoto K, Sarkar J, Surapureddi S, Jia Y, Rao MS, et al. Transcriptional regulation of Cidea, mitochondrial cell death-inducing DNA fragmentation factor alpha-like effector A, in mouse liver by peroxisome proliferator-activated receptor alpha and gamma. J. Biol. Chem. 2007;282:18613–18624. doi: 10.1074/jbc.M701983200. [DOI] [PubMed] [Google Scholar]
- 11.Da L, Li D, Yokoyama KK, Li T, Zhao M. Dual promoters control the cell-specific expression of the human cell death-inducing DFF45-like effector B gene. Biochem. J. 2006;393:779–788. doi: 10.1042/BJ20051027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takai D, Jones PA. The CpG island searcher: a new WWW resource. In Silico Biol. 2003;3:235–240. [PubMed] [Google Scholar]
- 13.Gazzoli I, Kolodner RD. Regulation of the human MSH6 gene by the Sp1 transcription factor and alteration of promoter activity and expression by polymorphisms. Mol. Cell. Biol. 2003;23:7992–8007. doi: 10.1128/MCB.23.22.7992-8007.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osada H, Tatematsu Y, Masuda A, Saito T, Sugiyama M, Yanagisawa K, Takahashi T. Heterogeneous transforming growth factor (TGF)-beta unresponsiveness and loss of TGF-beta receptor type II expression caused by histone deacetylation in lung cancer cell lines. Cancer Res. 2001;61:8331–8339. [PubMed] [Google Scholar]
- 16.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 17.Deng G, Chen A, Hong J, Chae HS, Kim YS. Methylation of CpG in a small region of the hMLH1 promoter invariably correlates with the absence of gene expression. Cancer Res. 1999;59:2029–2033. [PubMed] [Google Scholar]
- 18.Thomas M, Kalita A, Labrecque S, Pim D, Banks L, Matlashewski G. Two polymorphic variants of wild-type p53 differ biochemically and biologically. Mol. Cell. Biol. 1999;19:1092–1100. doi: 10.1128/mcb.19.2.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lachner M, O'Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. J. Cell Sci. 2003;116:2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 20.Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 21.Philipsen S, Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999;27:2991–3000. doi: 10.1093/nar/27.15.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zou MX, Butcher DT, Sadikovic B, Groves TC, Yee SP, Rodenhiser DI. Characterization of functional elements in the neurofibromatosis (NF1) proximal promoter region. Oncogene. 2004;23:330–339. doi: 10.1038/sj.onc.1207053. [DOI] [PubMed] [Google Scholar]
- 23.Deng G, Song GA, Pong E, Sleisenger M, Kim YS. Promoter methylation inhibits APC gene expression by causing changes in chromatin conformation and interfering with the binding of transcription factor CCAAT-binding factor. Cancer Res. 2004;64:2692–2698. doi: 10.1158/0008-5472.can-03-3000. [DOI] [PubMed] [Google Scholar]
- 24.Fahrner JA, Eguchi S, Herman JG, Baylin SB. Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer Res. 2002;62:7213–7218. [PubMed] [Google Scholar]
- 25.Gruenbaum Y, Stein R, Cedar H, Razin A. Methylation of CpG sequences in eukaryotic DNA. FEBS Lett. 1981;124:67–71. doi: 10.1016/0014-5793(81)80055-5. [DOI] [PubMed] [Google Scholar]
- 26.Bird AP, Wolffe AP. Methylation-induced repression–belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 27.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 28.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 29.Eden S, Cedar H. Role of DNA methylation in the regulation of transcription. Curr. Opin. Genet. Dev. 1994;4:255–259. doi: 10.1016/s0959-437x(05)80052-8. [DOI] [PubMed] [Google Scholar]
- 30.Klempnauer KH. Methylation-sensitive DNA binding by v-myb and c-myb proteins. Oncogene. 1993;8:111–115. [PubMed] [Google Scholar]
- 31.Prendergast GC, Lawe D, Ziff EB. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991;65:395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- 32.Campanero MR, Armstrong MI, Flemington EK. CpG methylation as a mechanism for the regulation of E2F activity. Proc. Natl Acad. Sci. USA. 2000;97:6481–6486. doi: 10.1073/pnas.100340697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weih F, Nitsch D, Reik A, Schutz G, Becker PB. Analysis of CpG methylation and genomic footprinting at the tyrosine aminotransferase gene: DNA methylation alone is not sufficient to prevent protein binding in vivo. EMBO J. 1991;10:2559–2567. doi: 10.1002/j.1460-2075.1991.tb07796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Comb M, Goodman HM. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 1990;18:3975–3982. doi: 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirillov A, Kistler B, Mostoslavsky R, Cedar H, Wirth T, Bergman Y. A role for nuclear NF-kappaB in B-cell-specific demethylation of the Igkappa locus. Nat. Genet. 1996;13:435–441. doi: 10.1038/ng0895-435. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Fatima N, Dufau ML. Coordinated changes in DNA methylation and histone modifications regulate silencing/derepression of luteinizing hormone receptor gene transcription. Mol. Cell. Biol. 2005;25:7929–7939. doi: 10.1128/MCB.25.18.7929-7939.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Honda H, Pazin MJ, Ji H, Wernyj RP, Morin PJ. Crucial roles of Sp1 and epigenetic modifications in the regulation of the CLDN4 promoter in ovarian cancer cells. J. Biol. Chem. 2006;281:21433–21444. doi: 10.1074/jbc.M603767200. [DOI] [PubMed] [Google Scholar]
- 38.Zhu WG, Srinivasan K, Dai Z, Duan W, Druhan LJ, Ding H, Yee L, Villalona-Calero MA, Plass C, et al. Methylation of adjacent CpG sites affects Sp1/Sp3 binding and activity in the p21(Cip1) promoter. Mol. Cell. Biol. 2003;23:4056–4065. doi: 10.1128/MCB.23.12.4056-4065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alikhani-Koopaei R, Fouladkou F, Frey FJ, Frey BM. Epigenetic regulation of 11 beta-hydroxysteroid dehydrogenase type 2 expression. J. Clin. Invest. 2004;114:1146–1157. doi: 10.1172/JCI21647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stirzaker C, Song JZ, Davidson B, Clark SJ. Transcriptional gene silencing promotes DNA hypermethylation through a sequential change in chromatin modifications in cancer cells. Cancer Res. 2004;64:3871–3877. doi: 10.1158/0008-5472.CAN-03-3690. [DOI] [PubMed] [Google Scholar]
- 41.Chang YS, Wang L, Suh YA, Mao L, Karpen SJ, Khuri FR, Hong WK, Lee HY. Mechanisms underlying lack of insulin-like growth factor-binding protein-3 expression in non-small-cell lung cancer. Oncogene. 2004;23:6569–6580. doi: 10.1038/sj.onc.1207882. [DOI] [PubMed] [Google Scholar]
- 42.Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64:1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- 43.Meehan RR, Lewis JD, Bird AP. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992;20:5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 45.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]