Abstract
Mechanisms of gene repression by transforming growth factor-beta (TGF-beta) are not well understood. TGF-beta represses transcription of pulmonary surfactant protein-B gene in lung epithelial cells. Repression is mediated by SMAD3 through interactions with NKX2.1 and FOXA1, two key transcription factors that are positive regulators of SpB transcription. In this study, we found that SMAD3 interacts through its MAD domains, MH1 and MH2 with NKX2.1 and FOXA1 proteins. The sites of interaction on NKX2.1 are located within the NH2 and COOH domains, known to be involved in transactivation function. In comparison, weaker interaction of FOXA1 winged helix, and the NH2-terminal domains was documented with SMAD3. Both in vitro studies and in vivo ChIP assays show that interaction of SMAD3 MH1 and MH2 domains with NKX2.1 and FOXA1 results in reduced binding of NKX2.1 and FOXA1 to their cognate DNA-binding sites, and diminished promoter occupancy within the SpB promoter. Thus, these studies reveal for the first time a mechanism of TGF-beta-induced SpB gene repression that involves interactions between specific SMAD3 domains and the corresponding functional sites on NKX2.1 and FOXA1 transcription factors.
INTRODUCTION
Transforming growth factors-beta (TGF-beta) are a large family of secreted polypeptides with established roles in nearly every aspect of cellular physiology, pathology and tumorigenesis. In the lung, as in other organs, TGF-beta regulates tissue morphogenesis and cellular differentiation through regulation of target gene expression (1). Although regulation of gene expression by TGF-beta is complex, from a simplistic perspective, the binding of TGF-beta ligand to its receptors, TβRII and TβRI initiates a cascade of phosphorylation events that eventually result in nuclear translocation of SMAD2 and SMAD3 subsequent to their association with SMAD4. To date, much has been elucidated regarding the mechanisms of TGF-beta-induced gene activation. However, TGF-beta also inhibits gene transcription and comparatively much less is known about the latter mechanism.
Surfactant protein-B, SPB is a protein constituent of pulmonary surfactant, a surface-active material that stabilizes lung alveoli and is therefore critical to air breathing. As a key component of pulmonary surfactant, SPB is absolutely required for postnatal survival. In humans, inherited SPB deficiency is a lethal condition that requires lung transplantation. Targeted inactivation of SpB in mouse also causes perinatal lethality (2). Even reduced SPB perturbs lung function in both humans and transgenic mice (3,4). Increased bioactive TGF-beta is associated with a number of pathological lung conditions, including interstitial lung fibrosis (5,6) and bronchopulmonary dysplasia, BPD (7). TGF-beta was found to inhibit the expression of SpB, but the precise mechanism remained unknown (8).
Expression of SpB is dependent on the activity of a number of transcription factors. The role of the homeodomain transcription factor NKX2.1, (otherwise known as TTF-1 or TEBP) is central, illustrated by the observation that in Nkx2.1(–/–) embryonic lungs SpB gene activity is entirely lacking (9). Many signaling cues during lung development or in response to injury are mediated through physical and functional interactions of NKX2.1 with other nuclear factors. These interactions modulate the activity of NKX2.1 on its target gene promoters. For example, NKX2.1 interacts with the nuclear factors, TAZ (a transcriptional co-activator with PDZ-binding domain) and NFI (nuclear Factor I) to activate transcription from the lung-specific SpC gene (10,11). TAZ interacts with the amino-terminal domain of NKX2.1 increasing its transcription activation properties. The 5′ flanking region of the SpB gene also includes binding sites for members of the forkhead/winged helix transcription factors, FOXA1/FOXA2. In transient transfection studies, the latter binding sites were found to be critical for SpB promoter activity (12). Compound genetic disruption of Foxa1/Foxa2 loci eliminates epithelial cell differentiation and consequently results in the absence of SpB mRNA (13).
Previously, we showed that TGF-beta repression of SpB transcription in lung epithelial cells occurs through TβRI signaling and mediated by SMAD3 (14). No evidence for direct binding of SMAD3 to the SpB promoter was found, and a DNA-binding mutant of SMAD3 effectively repressed SpB, eliminating direct DNA-binding requirement. Mutations within a 70 bp domain of SpB promoter that includes binding sites for NKX2.1 and FOXA proteins eliminated SMAD3-dependent repression of transcription. Further studies suggested interactions between SMAD3 and both NKX2.1 and FOXA1 proteins. We therefore proposed that SMAD3 interactions with NKX2.1 and FOXA1 underlie the molecular basis for TGF-beta-induced repression of SpB transcription. In the present study, we have elucidated the precise nature and the sites of interaction between SMAD3 and the two transcription factors NKX2.1 and FOXA1 using a number of experimental approaches. The sum of the results show that interaction of SMAD3 occurs through the MH1 and MH2 domains of SMAD3 and lead to inhibition of NKX2.1 and FOXA1 binding to their cognate sites within the SpB promoter.
MATERIALS AND METHODS
Plasmid construction
Complete coding region of rat Foxa1, rat Foxa2 and human Nkx2.1 were PCR amplified from cDNA clones (a gift from Dr Costa, University of Illinois at Chicago) verified by sequencing and cloned into EcoRI and BamHI sites (for Foxa1 and Foxa2) or EcoRI and HindIII sites (for Nkx2.1), in translational frame with VP16 coding region of pVp16 vector (Clontech, CA, USA), and designated Vp16-Foxa1, Vp16-Foxa2 and Vp16-Nkx2.1 respectively. Vp16-Foxa1N, Vp16-Foxa1DB and Vp16-Foxa1C were constructed by cloning the specific Foxa1 fragments that encodes amino acids 1–137, amino acids 138–317 and amino acids 318–466, into EcoRI and BamHI sites of pVp16. Similar strategy was used for making GAL4 fusion constructs containing specific domain fragments of Nkx2.1. Gal4-Nkx2.1N, Gal4-Nkx2.1HD and Gal4-Nkx2.1C were constructed by cloning the specific Nkx2.1 fragments that encodes amino acids 1–141, amino acids 142–253 and amino acids 254–371, into EcoRI and BamHI, BamHI and MluI, MluI and HindIII sites of pM (Clontech, CA, USA), respectively.
Gal4-Smad3, Vp16-Smad3 and glutathione-S-transferase, GST-Smad3 were provided by Dr R. Derynck (UCSF, CA, USA). Construction of Gal4-MH1, Gal4-MH2 and Gal4-LNKR have been described (15).
Cell culture and transient transfection assays
The human pulmonary epithelial cell lines A549 and H441 were maintained in F-12K Nutrient Mixture (Amersham Biosciences, NJ, USA) containing 10% fetal bovine serum and 1% penicillin–streptomycin. All plasmids used in transfection studies were purified on QIAGEN columns (Qiagen, CA, USA). Transient transfection of A549 cells was performed with SuperFect as described by the manufacturer (Qiagen, CA). After transfection, cells were lyzed and the extracts were collected as described (Promega, WI, USA). Supernatants of the cell extracts were used for assay of β-galactosidase and luciferase as described by the manufacturer (Promega, WI). MLE15 cells were cultured as described (16) and used for extracting nuclear protein for EMSA.
Mammalian two-hybrid assay
Expression constructs for GAL4-fusion protein (1.125 µg) and VP16-fusion protein (5.625 µg) were cotransfected with the GAL4-luciferase reporter constructs Gal4-Lux (3.375 µg, pFR-Luc, Stratagene) and pSV-β-gal (Promega, WI) into A549 cells. The interaction between GAL4-fusion proteins and VP16-fusion protein as indicated in each experiment was measured as a function of transactivation of the heterologous GAL4 promoter as quantified by luciferase production. Vectors including pM and pVp16 (Clontech, CA), were used in transfection experiments as control for GAL4-fusion protein and VP16-fusion protein.
Oligonucleotides
Synthetic oligonucleotides were annealed and diluted as described (12) and used directly in EMSA as cold competitor. For use as probe in EMSA reactions, the annealed oligonucleotides were purified by gel electrophoresis on 3% low melting agarose (Promega, WI), excised and then eluted using QIAEXII (Qiagen, CA). Two picomoles of the purified, annealed oligonucleotides were end-labeled with T4 polynucleotide kinase and γ-33P-ATP as described (14). The labeled probes were purified from unincorporated γ-33P-ATP using G-25 Sephadex column (Roche Applied Science, IN, USA). The DNA sequence of the oligonucleotides are as follows. For NKX2.1 binding: 5′-CAC CTG GAG GGC TCT TCA GAG C-3′ (12). For FOXA1 binding: 5′-CAA AGA CAA ACA CTG AGG TCG-3′ (12).
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared from MLE15 cells using a mini-extraction procedure (12). Five micrograms of the nuclear extract were incubated with 33P-end-labeled oligonucleotide probe with or without cold competitor in 12.5 mM Tris–HCl (pH 7.5), 62.5 mM NaCl, 0.62 mM DTT, 10% glycerol, 0.05% NP-40, 0.05 µg/µl Poly(dI-dC) in a total volume of 20 µl at 4°C for 15 min. For experiments with antibody or GST fusion proteins, 4 µl antibody or 4.8 µg GST fusion protein were mixed with the nuclear extracts in reaction mixture and incubated at 4°C for 15 min before addition of the 33P-end-labeled probe. Bound and free probes were separated by gel electrophoresis on a 4.5% non-denaturing polyacrylamide gel. The NKX2.1 antibody was purchased from Lab Vision Corporation (CA, USA). The FOXA1 antibody was purchased from (CeMines, Co, USA).
Glutathione-S-transferase (GST) fusion protein purification
Cultures of Escherichia coli were transformed with plasmids expressing either GST or GST-SMAD3 fusion proteins. Recombinant proteins were isolated by ultra-sonication, followed by purification using Bulk GST purification module as described by the manufacturer (Amersham Biosciences, NJ, USA).
Co-immunoprecipitation and western blotting
Co-immunoprecipitation was performed as previously described (14). Briefly, proteins were extracted from H441 cells transfected with expression of the appropriate construct and incubated with antibody against NKX2.1, FOXA1 or FLAG (SMAD3-fusion). The Protein G beads (Roche Applied Science, IN, USA) were washed and added to the reaction mixture. After incubation for 1 h at 4°C on a rotator, the beads were washed thoroughly and resuspended in 2× reducing loading buffer (Active Motif, CA, USA) for western blot analysis. As negative control, a parallel reaction was performed without the inclusion of the antibody. Antibodies used for immunoprecipitation were mouse anti-FLAG (Sigma, MI), mouse anti-NKX2.1 (Lab Vision Corporation, CA, USA), and mouse anti-FOXA1 (Seven Hills Bioreagents, OH, USA) antibodies. Western blots were performed with either rabbit anti-NKX2.1 (Seven Hills Bioreagents, OH), rabbit anti-FOXA1 (CE Mines, CO, USA), or rabbit anti-SMAD3 (Abcam, MA, USA) antibodies.
Chromatin immunoprecipitation assay (ChIP)
ChIP assays were performed using a ChIP-IT™ kit (Active Motif, Inc., Carlsbad, CA, USA) following the manufacturer′s instructions. Unless otherwise stated, all reagents, buffers and supplies were included in the kit. Briefly, MLE15 cells were cross-linked with 1% formaldehyde for 10 min at room temperature. After washing and treatment with glycine Stop-Fix solution, the cells were resuspended in lysis buffer and incubated for 30 min on ice. The cells were homogenized and nuclei were resuspended in shearing buffer and subjected to optimize enzymatic shearing conditions to yield 100–400 bp DNA fragments. The chromatin was pre-cleared with protein G beads and incubated (overnight at 4°C) with 3 µg of negative control mouse IgG (provided in the ChIP-IT™ kit) and anti-NKX2.1 polyclonal antibody (Seven Hills Bioreagents, Cincinnati, OH). Protein G beads were then added to the antibody/chromatin incubation mixtures and incubated for 1.5 h at 4°C. After extensive washings, the immunoprecipitated DNA was removed from the beads in an elution buffer. To reverse cross-links and remove RNA, 5 M NaCl and RNase were added to the samples and incubated for 4 h at 65°C. The samples were then treated with proteinase K for 2 h at 42°C, and the DNA was purified using DNA purification mini-columns. The purified DNA was subjected to PCR amplification (1 cycle of 94°C for 3 min, 35 cycles of 94°C for 20 s, 58°C for 30 s and 72°C for 30 s). For assessment of NKX2.1 occupancy of the SpB promoter, the primers used in PCR were: ATT TGA CGG TGA ACA AAG TCA GGC T (forward for NKX2.1 and FOXA1-binding sites); GAC CTC AGT GTT TGC CTG TGT CT (reverse for NKX2.1); and TCT TAT AGT AGG GGA GAG GAC CTG (reverse for FOXA1) (17).
RESULTS
SMAD3 interacts with FOXA1& NKX2.1 in a mammalian two-hybrid assay
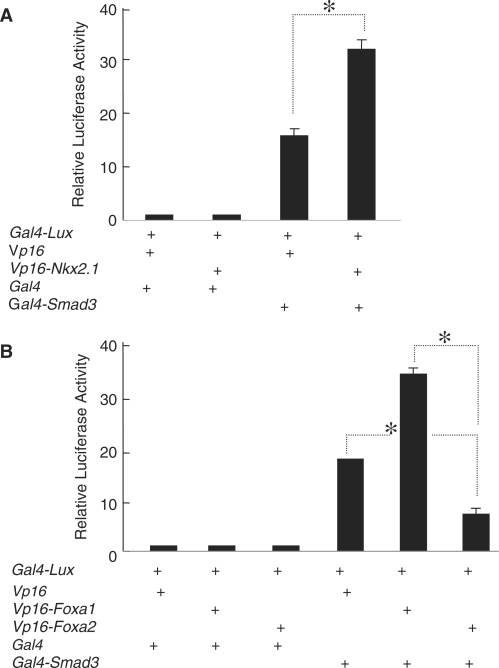
A mammalian two-hybrid assay in the lung carcinoma, A549 cell line was used to determine physical interactions between SMAD3 and NKX2.1 or FOXA1. Accordingly, a construct expressing GAL4-fused-SMAD3 was transiently cotransfected with another construct that expresses a VP16-fused-NKX2.1 protein and the interactions were monitored by luciferase production from a Gal4-luciferase reporter construct (Figure 1, Panel A). In a separate set of experiments, we used VP16-fused-FOXA1 to determine its interactions with SMAD3 (Figure 1, Panel B). In both sets of studies SMAD3 interacted physically with either NKX2.1 or FOXA1 to bring about stimulation of the Gal4-luciferase reporter construct. FOXA1 and FOXA2 are nearly 93% identical in their primary structure (18). Using a VP16-FOXA2 fusion protein in a mammalian two-hybrid assay showed that FOXA2 also interacts with SMAD3 in a similar manner as FOXA1 (Figure 1, Panel B). The above results clearly indicated physical and robust interactions between FOXA1, NKX2.1 and the TGF-beta-activated SMAD3.
Figure 1.
SMAD3 interacts with NKX2.1 (Panel A) and both FOXA1 and FOXA2 (Panel B) in lung epithelial cells. Mammalian two-hybrid assays were performed in A549 cells using Vp16-Nkx2.1, Vp16-Foxa1 or Vp16-Foxa2 expression plasmids according to the protocol described in Materials and Methods section. Luciferase readings from the controls consisting of, empty Vp16 and Gal4 plasmids alone, or in combination with Vp16-Nkx2.1, Vp16-Foxa1 or Vp16-Foxa2 were adjusted to unity and used to normalize all experimental results. Representative results from three independent experiments are shown. Asterisks denote P < 0.05.
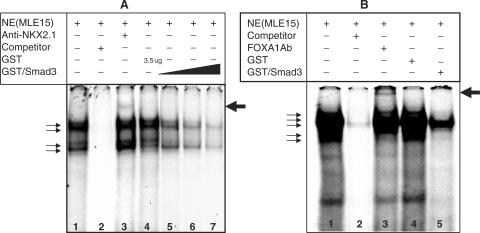
To verify the two-hybrid assay data, we further examined the impact of interactions of bacterially expressed and purified glutathione-S-transferase, GST-fused-SMAD3 on the binding of NKX2.1 and FOXA1 to their cognate-binding sites by EMSA. Using MLE15 nuclear extracts, we examined the binding of nuclear proteins to a 22 bp oligonucleotide representing the cognate-binding site for NKX2.1 (Materials and Methods section). This analysis showed the formation of at least four nucleoprotein complexes (Figure 2, Panel A, Lane 1). Incubation of the nuclear extract with an unlabeled oligonucleotide that has identical DNA sequence to that of the probe competed effectively for the formation of the four complexes (Figure 2, Panel A, Lane 2), whereas inclusion of a monoclonal anti-NKX2.1 antibody in the reaction mixture supershifted a complex as designated by the arrow (Lane 3). Inclusion of a bacterially expressed and purified GST-fused-SMAD3 protein competed, in a dose-dependent manner for the formation of all four complexes, but particularly the larger complexes (lanes 5–7). Importantly in the control reaction, 3.5 µg of GST protein alone had no impact on the formation of any of the four complexes (Lane 4). These results clearly show that GST-SMAD3 protein interferes, in a dose-dependent manner, with the binding of NKX2.1 to its target sequences under in vitro settings (Figure 2).
Figure 2.
EMSA analysis of SMAD3 interactions with NKX2.1 (Panel A) and FOXA1 (Panel B). A representative EMSA showing the results of an experiment with nuclear extracts from MLE15 cells to analyze nucleoproteins bound to an oligonucleotide of the binding sites for NKX2.1 or FOXA1 within the SpB promoter. Please note supershifted complex with either NKX2.1 or FOXA1 antibodies in Lane 3, Panel A and Lane 3 Panel B, respectively (arrows). In panel A, increasing amounts of GST-SMAD3 were used to examine the effect on the binding affinity of NKX2.1.
Similar analyses for FOXA1 using an oligonucleotide representing the cognate FOXA1-binding site on the SpB promoter identified multiple nucleoprotein complexes (Figure 2, Panel B, Lane 1). The formation of the majority of these complexes was inhibited upon inclusion of an unlabeled oligonucleotide (Lane 2). Inclusion of a polyclonal anti-FOXA1 antibody resulted in the formation of a supershifted complex (Lane 3, Arrow). As in Figure 2, Panel A, a recombinant GST protein alone did not compete for the formation of any of the complexes, whereas inclusion of a recombinant GST-SMAD3 fusion protein significantly reduced the formation of the complexes (Lanes 4 and 5, respectively). Dose-dependent inhibition was not conducted for GST-SMAD3 with FOXA1. These data demonstrate that interaction between SMAD3 and FOXA1 reduce the binding of the latter transcription factor to its cognate-binding site in vitro.
SMAD3 interacts with specific domains of NKX2.1 and FOXA1
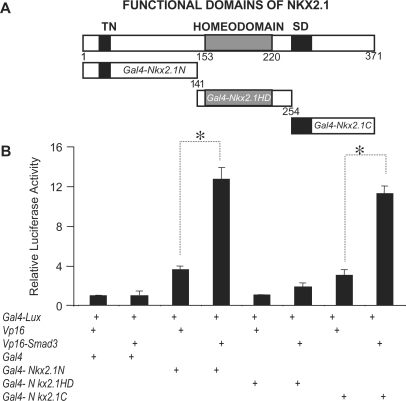
To identify the specific domains on the NKX2.1 molecule that mediate the NKX2.1–SMAD3 interactions, we constructed Gal4-fusion plasmids containing one of the three functional domains on the NKX2.1 protein; the NH2-terminus, the homeodomain and the COOH-terminus (Figure 3, Panel A). Each of the latter domains is thought to render distinct roles in transcriptional activation function of NKX2.1. The use of the mammalian two-hybrid assay in A549 cells showed little, if any physical interactions between SMAD3 and the DNA binding, homeodomain region of NKX2.1 (Figure 3, Panel B). In contrast, as evidenced by the level of luciferase production, both the NH2- and the COOH-terminal ends of NKX2.1 interacted strongly with SMAD3 indicating the two regions flanking the NKX2.1 homeodomain to be the sites of interaction involved in inhibition of DNA-binding function.
Figure 3.
Physical interactions between SMAD3 and functional domains of NKX2.1. GAL4 or VP16 fused NKX2.1 and its various functional domains were used in mammalian two-hybrid assays to determine the site of interactions with SMAD3. Vp16 or Gal4 plasmids were used either alone or in combination with other expression vectors as control. Luciferase reading from combination of empty Vp16 and Gal4 plasmids were adjusted to unity and used for normalization of all experimental values. Representative results from three independent experiments are shown. Asterisks denote P < 0.05.
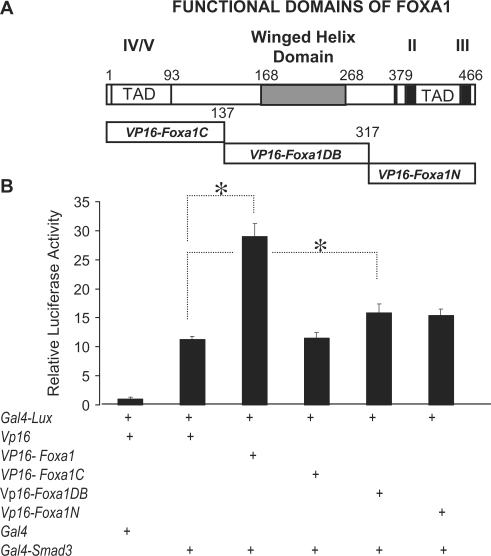
Similarly, we determined the functional domains of FOXA1 molecule that are the sites of physical interactions with SMAD3 (Figure 4, Panel A). In order to eliminate spurious artifacts stemming from specific fusion constructs, we generated the SMAD3 constructs as GAL4, rather than VP16-fusion proteins. The results with the full-length FOXA1 (Vp16-Foxa1) were identical, whether Vp16-Smad3 or Gal4-Smad3 fusion constructs were used, thereby validating the results of the mammalian two-hybrid assay. Figure 4, Panel B also shows evidence for physical interaction between SMAD3 and the winged helix DNA-binding domain of FOXA1. In addition, there was measurable interaction between SMAD3 and the transactivation domain of FOXA1 localized to the NH2 terminus. However, this interaction only approached, but did not reach statistical significance (P = 0.071). SMAD3 interaction with the latter two functional domains of FOXA1 was weak when compared to the full-length protein, suggesting that overlapping domains may be required for optimized interactions (Figure 4).
Figure 4.
Physical interactions between SMAD3 and functional domains of FOXA1. GAL4 or VP16 fused FOXA1 and its functional domains were used in mammalian two-hybrid assays to determine the site of interactions with SMAD3. Empty Vp16 or Gal4 plasmids were used either alone or in combination with other expression plasmids as controls. Luciferase reading from combination of empty Vp16 and Gal4 plasmids were adjusted to unity and used for normalization of all experimental values. Representative results from three independent experiments are shown. Asterisks denote P < 0.05.
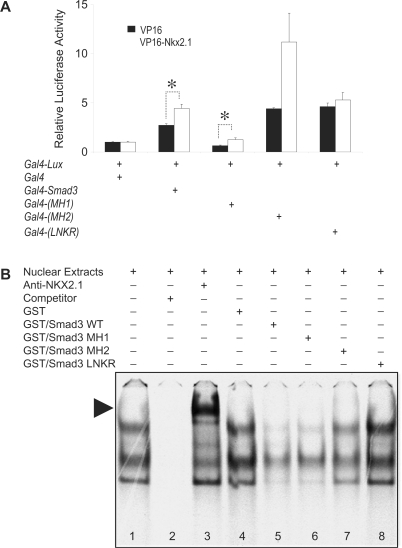
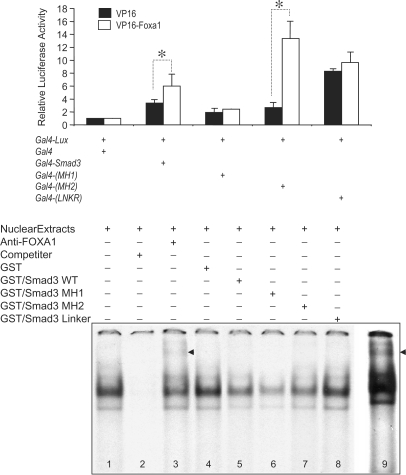
SMAD3 domains involved in NKX2.1 and FOXA1 interactions
Three specific functional domains, which cover the entire SMAD3 molecule were generated as Gal4-fusion constructs. These are the domains in the NH2-terminus (amino acids 1–130), also known as the MH1 domain, the linker domain represented by amino acids 130–230 (LNKR) and the COOH-terminus or MH2 domain (amino acids 230–424). These constructs were then used in a mammalian A549 cell two-hybrid assay with either GAL4-fused-NKX2.1 (Figure 5, Panel A) or GAL4-fused-FOXA1 expression constructs (Figure 6 Panel A). The results showed significant interactions between full-length NKX2.1 and the MH1 domain of SMAD3 (Figure 5). The interaction between NKX2.1 and the SMAD3 MH2 domain did not reach statistical significance (P = 0.083). Little if any interaction was observed with the SMAD3 LNKR domain (P = 0.25). Furthermore, EMSA with GST-fused domains of SMAD3 verified the two-hybrid assay findings (Figure 5, Panel B). Consistent with the two-hybrid results, robust interaction was found between NKX2.1 and the SMAD3 MH1 domain. Weak interaction of NKX2.1 with the MH2 domain was also documented. However, no interaction was observed between LNKR domain of SMAD3 and NKX2.1 (Figure 5, Panel B, Lane 8). In contrast to the interactions with NKX2.1, significant interaction with FOXA1 occurred mainly with the SMAD3 MH2 domain (Figure 6). The two-hybrid assay showed only weak interactions between FOXA1 and either SMAD3 MH1 or LNKR peptides. However, GST-fused-MH1 competed effectively against FOXA1 binding to its target DNA, either indicating the inherent differences between the two techniques or suggesting more complex interactions between the two proteins (Figure 6, Panel B).
Figure 5.
Localization of SMAD3 functional domains, which interact with NKX2.1. Panel A, mammalian two-hybrid assays were used to test the ability of GAL4-fused MH1, GAL4-fused MH2 or GAL4-fused LNKR domains of SMAD3 to interact with a full-length VP16-fused NKX2.1. Empty Gal4 plasmid was used as control and the results were adjusted to unity. All experimental values were normalized against the latter (Materials and Methods section). Panel B, representative EMSA analysis of interactions between various domains of SMAD3 with NKX2.1 on the SpB promoter. Supershifted nucleoprotein complex with anti-NKX2.1 antibody is indicated with an arrow (Lane 3). Please note that inclusion of GST-LNKR in EMSA does not interfere with formation of nucleoprotein complexes (Lane 8). Asterisks denote P < 0.05.
Figure 6.
Localization of SMAD3 functional domains, which interact with FOXA1. Panel A, mammalian two-hybrid assay was used to test the ability of GAL4-fused MH1, GAL4-fused MH2 or GAL4-fised LNKR domains of SMAD3 to interact with a full-length VP16-fused FOXA1 protein. Results with the empty Gal4 plasmid were adjusted to unity and used to normalize all experimental values (Materials and Methods section). Panel B, representative EMSA analysis of interactions between various domains of SMAD3 with FOXA1 on the SpB promoter. Supershifted nucleoprotein complex with anti-FOXA1 antibody is indicated with an arrow (Lane 3, and a longer exposure in Lane 9). Please note that only GST-LNKR does not interfere with formation of nucleoprotein complexes (Lane 8). Asterisks denote P < 0.05.
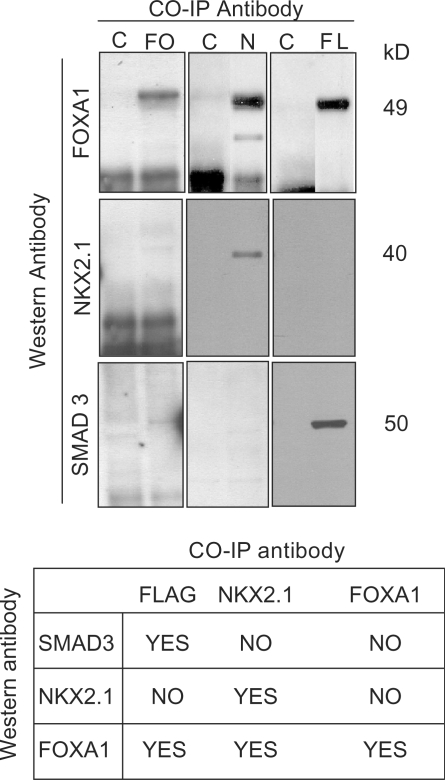
Co-immunoprecipitation of FOXA1, SMAD3 and NKX2.1
To further examine the interactions amongst NKX2.1, FOXA1 and SMAD3 proteins, we used co-immunoprecipation (co-IP). A Smad3-Flag expression construct was transfected into H441 cells, which exhibit endogenous Foxa1 and Nkx2.1 gene expression, and the cellular extracts were used for co-IP with mouse anti-FLAG, anti-NKX2.1 and anti-FOXA1 antibodies. We used three antibodies to minimize the possibility of false-negative results stemming from their intrinsic properties that may interfere with protein–protein interactions in co-IP studies. The immunoprecipitated complexes were resolved by gel electrophoresis and western blot analysis using rabbit-derived antibodies to the three proteins. The results are summarized in the table in Figure 7. Anti-FLAG, as expected, immunoprecipitated SMAD3 but also FOXA1, indicating measurable physical interactions by this antibody between SMAD3 and FOXA1, but not NKX2.1. Anti-NKX2.1 immunoprecipitated a complex that included only NKX2.1 and FOXA1, but not SMAD3. Finally, only FOXA1 was brought down by anti-FOXA1 antibodies, even though SMAD3–FOXA1 and NKX2.1–FOXA1 intracellular associations were established by anti-FLAG and anti-NKX2.1 co-IP results (Figure 7). Therefore, failure of anti-FOXA1 to co-immunoprecipitate NKX2.1 or SMAD3, a false-negative result, may be explained by an intrinsic property of this antibody (e.g. its epitope specificity may interfere with protein–protein interactions). Similarly, it is possible that absence of demonstrable NKX2.1–SMAD3 interactions by co-IP may simply be related to specific aspects of the antibodies used.
Figure 7.
Co-immunoprecipitation of FOXA1, SMAD3 and NKX2.1. Co-immunoprecipitation with three antibodies, anti-FOXA1, anti-NKX2.1 and anti-FLAG (against Smad3) was performed from extracts of lung carcinoma cells transfected with a Smad3-Flag expression construct. The co-IP complexes were analyzed by western blotting using rabbit-derived antibodies to the three target proteins. C = Control (no co-IP antibody), FO = anti-FOXA1 antibody, N = anti-NKX2.1 antibody and FL = anti-FLAG (SMAD3) antibody. The table is a summary of the findings.
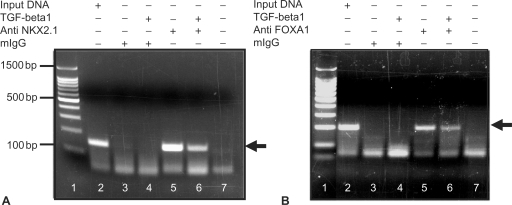
TGF-beta treatment of MLE15 cells reduces the binding of NKX2.1 and FOXA1 to their target genomic sites in vivo
To examine the in vivo relevance of the studies described above, we performed chromatin immunoprecipitation studies (ChIPs) using nuclear extracts from MLE15 cells. Chromatin fractions were prepared from MLE15 cells treated with and without 10 ng/ml of human recombinant TGF-beta and immunoprecipitated using an anti-NKX2.1 monoclonal antibody and a polyclonal anti-FOXA1 antibody. Subsequent to isolation of DNA from the immunoprecipitate, specific oligonucleotide primers were used to PCR amplify a region of the SpB promoter located between –222 and –74 nt, for the NKX2.1 reaction and –222 and –24 nt, for the FOXA1 reaction. This region includes the binding sites for both NKX2.1 and FOXA1. The experiments were repeated three times with identical results. For NKX2.1 reaction (Figure 8, Panel A), comparison between the intensities of the PCR bands obtained in chromatin prepared from TGF-beta-treated (Figure 8, Panel A, Lane 6) versus untreated (Lane 5) samples supports the concept that TGF-beta treatment diminishes the binding of NKX2.1 to its target sequences in vivo. Similarly, comparison between PCR products in Lanes 5 and 6 (Figure 8, Panel B) clearly shows that treatment of MLE15 cells with TGF-beta ligand diminishes the binding of FOXA1 to its target sequences on the SpB promoter in vivo.
Figure 8.
Chromatin immunoprecipitation (ChIP) analysis of SpB promoter occupancy for NKX2.1 (Panel A) and FOXA1 (Panel B) in response to TGF-beta in MLE15 cells. TGF-beta-treated MLE15 nuclear extracts were immunoprecipitated with either anti-NKX2.1 or anti-FOXA1 antibodies and specific regions of the SpB promoter (Please see Materials and Methods section) were analyzed by PCR. Arrows indicate the PCR products. Lanes 1 are molecular weight markers. Representative results from two independent experiments are shown.
DISCUSSION
The purpose of the current study was to elucidate the mechanisms of TGF-beta-induced, SMAD3-mediated SpB gene repression. We found robust interactions between SMAD3 and two lung-enriched transcription factors, NKX2.1 and FOXA1, both of which are known positive regulators of SpB gene expression (12). The sites of interactions with SMAD3 were identified as the NH2- and COOH-terminal domains of NKX2.1 and principally the winged helix DNA-binding domain of FOXA1. SMAD3 interacted with these proteins through its MAD homology domains MH1 and MH2. Although interactions between SMADs and homeodomain proteins of Drosophila and Xenopus have been reported (19), NKX2.1 is the first mammalian member of this family whose interactions with SMAD3 result in repression of gene expression.
SMADs are intracellular effectors of TGF-beta signaling (1,20). In the lung, TGF-beta/SMAD signaling figures importantly in normal development as well as disease (5,7,21). In addition to activating batteries of genes (e.g. fibronectin, collagen), TGF-beta also represses a number of genes including SpB (19). Although TGF-beta signaling is mediated through both Smad2 and Smad3, to date, in the handful of reported studies in which TGF-beta acts as an inhibitor, Smad3 has been identified as the key mediating mechanism (19). Consistent with these studies, we found SMAD3 to mediate TGF-beta-induced SpB gene repression in lung epithelial cell lines (14).
SMAD3 is composed of two conserved functional domains, the NH2-terminal MH1 and the COOH-terminal MH2 domains, separated by a non-conserved linker domain (LNKR). In the current study, the two-hybrid assays in Figures 5 and 6 identified both MH1 and MH2 domains of SMAD3 as sites of interaction with NKX2.1 and FOXA1, respectively. These results were further verified by the EMSA studies shown in the same Figures (Panels B). Consistent with the two-hybrid assay results, only GST-fused SMAD3, MH1 and MH2 domains and not LNKR inhibited FOXA1 and NKX2.1 DNA binding. The MH1 domain is involved in DNA binding and nuclear translocation but also negatively regulates the functions of the MH2 domain (22,23). The MH2 domain is critical for homo- and hetero-oligomerization of SMADs, as well as the recognition and phosphorylation by type I receptors (1,20). Interactions between transcription factors and SMAD3 through the MH2 or MH1 domains have been reported (19). By far the majority of the reported cases lead to cooperativity and positive regulation of gene expression by TGF-beta. In the few cases in which TGF-beta, through SMAD3 represses gene expression, the sites of interaction were localized to either MH1 or MH2 domains (19).
We also determined the specific domains of NKX2.1 and FOXA1 with which SMAD3 interacts. The sites of interaction with SMAD3 were found to be localized to the NH2- and COOH-terminal domains of NKX2.1. Little to no interaction was observed with the NKX2.1 HD domain. Both NH2- and COOH-terminal domains of NKX2.1 have been found to affect transactivation function of NKX2.1 in target gene transcription (24). The results of the mammalian two-hybrid assays (Figure 4) are consistent with the latter findings. These domains alone were able to render transactivation function to the GAL4 DNA-binding domain and activate transcription from the GAL4-responsive reporter with VP16 (Figure 3). The HD of NKX2.1 had no transactivation function. Despite these observations, interactions of SMAD3 with NKX2.1 resulted in an overall reduction of DNA-binding affinity, as assessed by EMSA in Figure 2. Thus, TGF-beta-induced, SMAD3-mediated repression of SpB may affect not only DNA binding, but also transactivation function of NKX2.1.
FOXA1and FOXA2 proteins share 93% amino acid homology in the winged helix DNA-binding domain and bind to the same DNA consensus sequences (18). The winged helix DNA-binding domain is the site of protein–DNA interactions and may also contain the nuclear localization signal (25). The FOXA proteins also include conserved NH2- and COOH-terminal transcriptional activation domains that are involved in protein–protein interactions (26). Regions II and III sequences within the COOH-terminal domain are essential in transcriptional activation (25,27). In the current study, the DNA binding, winged helix domain of FOXA1 was found to be the principal site of interaction with SMAD3. No statistically significant interaction was measurable between SMAD3 and either the COOH- or NH2-terminal domains of FOXA1. Interaction with SMAD3 reduced the DNA-binding affinity of FOXA1 for its target sequences on the SpB promoter. Interactions between other forkhead/winged helix family members, FOXH1 and FOXO with both SMAD2 and SMAD3 have also been documented, although the sites of interaction remain unknown (28,29).
To examine the in vivo relevance of our findings, we used two approaches. First, using antibodies to each of the three proteins, we showed that complexes consisting of NKX2.1, FOXA1 and SMAD3 can be co-immunoprecipitated (Figure 7). However, the antibodies failed to demonstrate direct NKX2.1–SMAD3 interactions by co-IP. We propose that this may be due to specific intrinsic properties of the antibodies used. An alternative conclusion is that physical interactions between SMAD3 and NKX2.1 occur only through FOXA1. This possibility however is unlikely in the face of the internally consistent data from the stringent analyses conducted in multiple two-hybrid and EMSA assays with full length and fragments of all three proteins (Figures 1, 3 and 5). These latter studies firmly established direct and robust physical interactions between the COOH- and NH2-domains of NKX2.1 and SMAD3.
In a second approach, ChIP analysis showed that TGF-beta decreased promoter occupancy on the SpB gene for both NKX2.1 and FOXA1, suggesting that TGF-beta signaling, through SMAD3 destabilizes the binding of NKX2.1 and FOXA1 to their target DNA sites. A number of mechanisms may be invoked to explain the latter findings. The possibility that SMAD3 may bind and displace NKX2.1 or FOXA1 (competitive inhibition) does not appear likely for two reasons: First, EMSA analysis has failed to identify SMAD3 as a component of nucleoprotein complexes found on the SpB promoter (data not shown). Second, and consistent with the latter, a SMAD3 mutant lacking the DNA-binding domain is equally competent in inhibiting SpB transcription as the wild-type protein (14). Therefore, we favor a model in which interactions between SMAD3 and either NKX2.1 or FOXA1 occurs independently of the DNA thereby preventing the binding of the latter transcription factors to their target site (Figure 9). Although examples of TGF-beta-induced, SMAD3-mediated gene repression are few, a number of findings are consistent with the results of our study in this report. For example, TGF-beta-inhibition of myogenic differentiation is mediated by SMAD3 interaction with MyoD and myogenin. SMAD3 interacts directly with the HLH domains of MyoD or myogenin and interferes with heterodimerization of the latter with their obligatory partner E12/47, thus decreasing their binding to their DNA target site (30).
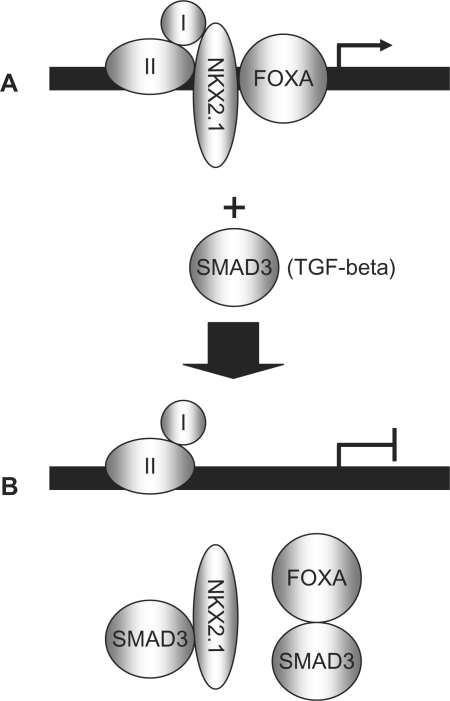
Figure 9.
A hypothetical model of DNA-independent inhibition of NKX2.1 and FOXA1 by SMAD3. (A) In the absence of TGF-beta treatment, NKX2.1 and FOXA1 can readily bind to their cognate sites and activate transcription of the SpB promoter. (B) Treatment with TGF-beta activates SMAD3, which in turn binds through its MH domains to NKX2.1 and FOXA1 and prevents their binding to the SpB promoter. Factors I and II represent other transcription, or co-factors participating in SpB gene activation.
TGF-beta signaling is crucial during normal lung development and maintenance of homeostasis in postnatal life. In addition, many injuries to the lung are characterized by dysregulated TGF-beta signaling. The impact of TGF-beta is manifested in a variety of ways that range from interstitial fibrosis to interruption of alveolar development. Although the overall pathway of TGF-beta signaling may appear simple, physical and functional interactions between its components, such as SMAD3 and other key transcriptional regulators, such as NKX2.1 provide the means for a profound degree of signaling specificity and versatility. Undoubtedly, characterization of the interactions between SMAD3 and NKX2.1 or FOXA1 will aid our understanding of the specificity with which TGF-beta signaling functions in various physiological contexts and therefore its impact on both normal and pathological processes.
ACKNOWLEDGEMENT
Research supported by NIH (NHLBI) and the Hastings Foundation. Funding to pay the Open Access publication charges for this article was provided by NIH (NHLBI).
Conflict of interest statement. None declared.
REFERENCES
- 1.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. Review. [DOI] [PubMed] [Google Scholar]
- 2.Clark JC, Wert SE, Bachurski CJ, Stahlman MT, Stripp BR, Weaver TE, Whitsett JA. Targeted disruption of the surfactant protein B gene disrupts surfactant homeostasis, causing respiratory failure in newborn mice. Proc. Natl Acad. Sci. USA. 1995;92:7794–7798. doi: 10.1073/pnas.92.17.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nogee LM, Wert SE, Proffit SA, Hull WM, Whitsett JA. Allelic heterogeneity in hereditary surfactant protein B (SP-B) deficiency. Am. J. Respir. Crit. Care Med. 2000;161:973–981. doi: 10.1164/ajrccm.161.3.9903153. [DOI] [PubMed] [Google Scholar]
- 4.Nesslein LL, Melton KR, Ikegami M, Na CL, Wert SE, Rice WR, Whitsett JA, Weaver TE. Partial SP-B deficiency perturbs lung function and causes air space abnormalities. Am. J. Physiol. Lung Cell Mol. Physiol. 2005;288:L1154–L1161. doi: 10.1152/ajplung.00392.2004. [DOI] [PubMed] [Google Scholar]
- 5.Westergren-Thorsson G, Hernnas J, Sarnstrand B, Oldberg A, Heinegard D, Malmstrom A. Altered expression of small proteoglycans, collagen, and transforming growth factor-beta 1 in developing bleomycin-induced pulmonary fibrosis in rats. J. Clin. Invest. 1993;92:632–637. doi: 10.1172/JCI116631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheppard D. Transforming growth factor beta: a central modulator of pulmonary and airway inflammation and fibrosis. Proc. Am. Thorac. Soc. 2006;3:413–417. doi: 10.1513/pats.200601-008AW. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lecart C, Cayabyab R, Buckley S, Morrison J, Kwong KY, Warburton D, Ramanathan R, Jones CA, Minoo P. Bioactive transforming growth factor-beta in the lungs of extremely low birthweight neonates predicts the need for home oxygen supplementation. Biol. Neonate. 2000;77:217–223. doi: 10.1159/000014219. [DOI] [PubMed] [Google Scholar]
- 8.Beers MF, Solarin KO, Guttentag SH, Rosenbloom J, Kormilli A, Gonzales LW, Ballard PL. TGF-beta1 inhibits surfactant component expression and epithelial cell maturation in cultured human fetal lung. Am. J. Physiol. 1998;275:L950–L960. doi: 10.1152/ajplung.1998.275.5.L950. [DOI] [PubMed] [Google Scholar]
- 9.Minoo P, Su G, Drum H, Bringas P, Kimura S. Defects in tracheoesophageal and lung morphogenesis in Nkx2.1(–/–) mouse embryos. Dev. Biol. 1999;209:60–71. doi: 10.1006/dbio.1999.9234. [DOI] [PubMed] [Google Scholar]
- 10.Park KS, Whitsett JA, Di Palma T, Hong JH, Yaffe MB, Zannini M. TAZ interacts with TTF-1 and regulates expression of surfactant protein-C. J. Biol. Chem. 2004;279:17384–17390. doi: 10.1074/jbc.M312569200. [DOI] [PubMed] [Google Scholar]
- 11.Bachurski CJ, Yang GH, Currier TA, Gronostajski RM, Hong D. Nuclear factor I/thyroid transcription factor 1 interactions modulate surfactant protein C transcription. Mol. Cell. Biol. 2003;23:9014–9024. doi: 10.1128/MCB.23.24.9014-9024.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bohinski RJ, Di Lauro R, Whitsett JA. The lung-specific surfactant protein B gene promoter is a target for thyroid transcription factor 1 and hepatocyte nuclear factor 3, indicating common factors for organ-specific gene expression along the foregut axis. Mol. Cell. Biol. 1994;9:5671–5681. doi: 10.1128/mcb.14.9.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wan H, Dingle S, Xu Y, Besnard V, Kaestner KH, Ang SL, Wert S, Stahlman MT, Whitsett JA. Compensatory roles of Foxa1 and Foxa2 during lung morphogenesis. J. Biol. Chem. 2005;280:13809–13816. doi: 10.1074/jbc.M414122200. [DOI] [PubMed] [Google Scholar]
- 14.Li C, Zhu NL, Tan RC, Ballard PL, Derynck R, Minoo P. Transforming growth factor-beta inhibits pulmonary surfactant protein B gene transcription through SMAD3 interactions with NKX2.1 and HNF-3 transcription factors. J. Biol. Chem. 2002;277:38399–38408. doi: 10.1074/jbc.M203188200. [DOI] [PubMed] [Google Scholar]
- 15.Prokova V, Mavridou S, Papakosta P, Kardassis D. Characterization of a novel transcriptionally active domain in the transforming growth factor beta-regulated Smad3 protein. Nucleic Acids Res. 2005;33:3708–3721. doi: 10.1093/nar/gki679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borok Z, Li X, Fernandes VF, Zhou B, Ann DK, Crandall ED. Differential regulation of rat aquaporin-5 promoter/enhancer activities in lung and salivary epithelial cells. J. Biol. Chem. 2000;275:26507–26514. doi: 10.1074/jbc.M910007199. [DOI] [PubMed] [Google Scholar]
- 17.Yang MC, Guo Y, Liu CC, Weissler JC, Yang YS. The TTF-1/TAP26 complex differentially modulates surfactant protein-B (SP-B) and -C (SP-C) promoters in lung cells. Biochem. Biophys. Res. Commun. 2006;344:484–490. doi: 10.1016/j.bbrc.2006.03.158. [DOI] [PubMed] [Google Scholar]
- 18.Costa RH, Kalinichenko VV, Lim L. Transcription factors in mouse lung development and function. Am. J. Physiol. Lung Cell Mol. Physiol. 2001;280:L823–L838. doi: 10.1152/ajplung.2001.280.5.L823. Review. [DOI] [PubMed] [Google Scholar]
- 19.Feng XH, Derynck R. Specificity and versatility in tgf-beta signaling through Smads. Annu. Rev. Cell Dev. Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. Review. [DOI] [PubMed] [Google Scholar]
- 20.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-β family signaling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 21.Zhou L, Dey CR, Wert SE, Whitsett JA. Arrested lung morphogenesis in transgenic mice bearing an SP-C-TGF-beta 1 chimeric gene. Dev. Biol. 1996;175:227–38. doi: 10.1006/dbio.1996.0110. [DOI] [PubMed] [Google Scholar]
- 22.Kurisaki A, Kose S, Yoneda Y, Heldin C.-H, Moustakas A. Transforming growth factor-beta induces nuclear import of Smad3 in an importin- β1 and Ran-dependent manner. Mol. Biol. Cell. 2001;12:1079–1091. doi: 10.1091/mbc.12.4.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao Z, Liu X, Henis Y.-I, Lodish H.-F. A distinct nuclear localization signal in the N terminus of Smad 3 determines its ligand-induced nuclear translocation. Proc. Natl Acad. Sci. USA. 2000;97:7853–7858. doi: 10.1073/pnas.97.14.7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeFelice M, Damante G, Zannini M, Francis-Lang H, Di Lauro R. Redundant domains contribute to the transcriptional activity of the thyroid transcription factor 1. J. Biol. Chem. 1995;270:26649–26656. doi: 10.1074/jbc.270.44.26649. [DOI] [PubMed] [Google Scholar]
- 25.Qian X, Costa RH. Analysis of HNF-3 protein domains required for transcriptional activation and nuclear targeting. Nucleic Acids Res. 1995;23:1184–1191. doi: 10.1093/nar/23.7.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harnish DC, Malik S, Kilbourne E, Costa R, Karathanasis SK. Control of apolipoprotein AI gene expression through synergistic interactions between hepatocyte nuclear factors 3 and 4. J. Biol. Chem. 1996;271:13621–13628. doi: 10.1074/jbc.271.23.13621. [DOI] [PubMed] [Google Scholar]
- 27.Pani L, Overdier DG, Porcella A, Qian X, Lai E, Costa RH. Hepatocyte nuclear factor 3 contains two transcriptional activation domains, one of which is novel and conserved with the Drosophila fork head protein. Mol. Cell. Biol. 1992;12:3723–3732. doi: 10.1128/mcb.12.9.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seoane J, Le HV, Massague J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- 29.Randall RA, Howell M, Page CS, Daly A, Bates PA, Hill CS. Recognition of phosphorylated-Smad2-containing complexes by a novel Smad interaction motif. Mol. Cell. Biol. 2004;24:1106–1121. doi: 10.1128/MCB.24.3.1106-1121.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu F, Hata A, Baker J.-C, Doody J, Carcamo J, Harland R.-M, Massagué J. A human Mad protein acting as a BMP-regulated transcriptional activator. Nature. 1996;381:620–623. doi: 10.1038/381620a0. [DOI] [PubMed] [Google Scholar]