Abstract
The expression of cPLA2 is critical for transformed growth of non-small cell lung cancer (NSCLC). It is known that phorbol 12-myristate 13-acetate (PMA)-activated signal transduction pathway is thought to be involved in the oncogene action in NSCLC and enzymatic activation of cPLA2. However, the transcriptional regulation of cPLA2α in PMA-activated NSCLC is not clear. In this study, we found that PMA induced the mRNA level and protein expression of cPLA2α. In addition, two Sp1-binding sites of cPLA2α promoter were required for response to PMA and c-Jun overexpression. Small interfering RNA (siRNA) of c-Jun and nucleolin inhibited PMA induced the promoter activity and protein expression of cPLA2α. Furthermore, PMA stimulated the formation of c-Jun/Sp1 and c-Jun/nucleolin complexes as well as the binding of these transcription factor complexes to the cPLA2α promoter. Although Sp1-binding sites were required for the bindings of Sp1 and nucleolin to the promoter, the binding of nucleolin or Sp1 to the promoter was independent of each other. Our results revealed that c-Jun/nucleolin and c-Jun/Sp1 complexes play an important role in PMA-regulated cPLA2α gene expression. It is likely that nucleolin binding at place of Sp1 on gene promoter could also mediate the regulation of c-Jun/Sp1-activated genes.
INTRODUCTION
The production of lipid mediators, the eicosanoids (i.e. prostaglandins and leukotrienes) are derived from metabolism of arachidonic acid, which has been implicated in the regulation of cell growth, inflammation, thrombosis and tumor progression (1–3). There is much evidence indicating that eicosanoids, particularly prostaglandins, are involved in the etiologies of cancer (4). Increased levels of eicosanoids occur in a number of different types of human cancer, including colon, pancreatic, breast and lung. In the case of lung cancer, increased prostaglandin biosynthesis has been found to occur mainly in non-small cell lung cancer (NSCLC), which comprises 80% of lung cancers rather than small cell lung cancer (SCLC) (5,6). In addition, the increase of cPLA2 is correlated with the eicosanoid synthesis that participates in NSCLC transformation (7).
Cytosolic phospholipase A2 (cPLA2) is the major intracellular form of PLA2, which preferentially hydrolyzes membrane phospholipids at the sn-2 position to release arachidonic acid (8). cPLA2 activity is regulated by intracellular Ca2+ and phosphorylation. Increase in Ca2+ results in translocation of cPLA2 to the nuclear envelope and activation (9,10). But it has been shown that phorbol 12-myristate 13-acetate (PMA) provides cPLA2-activating signals without inducing Ca2+ influx (11,12) and by a Rac-p38 kinase-dependent pathway (13). The maximal activation of cPLA2 requires sustained phosphorylation of Ser505, Ser727 and Ser515 by mitogen-activated protein kinases (MAPKs), MAPK-activated protein kinases and by calcium/calmodulin-dependent protein kinase II, respectively (14–16). In addition to acute regulation, expression of cPLA2 through changes in gene transcription is mediated by a number of agents including cytokines, thrombin and growth factors (17–21). The promoter for cPLA2 has been isolated from both human (18) and rat (22). A number of putative binding sites for possible regulatory elements have been identified within the promoter, including AP-1 sites, nuclear factor κB sites and glucocorticoid regulatory elements. Truncation of a 2.4 kb region of the promoter fragment down to the last 58 bp of the 5′-untranslated region has been identified in three regulatory regions and indicates that the transcription factor Sp1 can bind to two of these regions (23,24). In the regulation of cPLA2 promoter, LKLF (lung Krüppel-like factor) as a transcriptional activator also binds to the cPLA2 promoter and may interact with the Sp1 family (25).
Although Sp1 is required for the transcriptional activation of cPLA2 gene, only a limited number of studies have addressed the mechanisms by which cPLA2 gene expression is controlled. In addition to enzymatic activation, in this study, we also clarified that PMA could activate the transcription via the Sp1-binding sites of cPLA2α promoter in NSCLC A549 cells. We demonstrated that transcription factors c-Jun and Sp1 form a complex to regulate PMA-induced gene expression of cPLA2α as the manner as the c-Jun/Sp1-regulated genes including 12(S)-lipoxygenase, keratin 16, p21WAF1/CIP1 and neuronal nicotinic acetylcholine receptor β4 (26–29). We further performed pull-down and protein–DNA interaction assays to show that nucleolin bound to DNA and activated transcription of cPLA2α in PMA-treated cells. Nucleolin is reported to be a ubiquitously expressed multifunctional protein involved in ribosomal biogenesis and the regulation of nucleolar translocation of ribosomal proteins (30). Functional role of nucleolin in the activation and repression of gene transcription as well as in the regulation of RNA metabolism has already been reported (31). Our results revealed a new function for nucleolin as a c-Jun/Sp1-interacting partner and transcription activator in PMA-induced gene expression of cPLA2α.
MATERIALS AND METHODS
Cell culture
Human epidermoid carcinoma A431 and NSCLC A549 cells were grown at 37°C under 5% CO2 in 10 cm plastic dishes containing 10 ml of Dulbecco's modified Eagle's medium (Invitrogen-GIBCO) and nutrition mixture F12 Kaighn's modification medium (Biowest), respectively and supplemented with 10% fetal bovine serum, 100 μg/ml streptomycin and 100 U/ml penicillin. In this series of experiments, cells were treated with 5 nM PMA (Sigma, St Louis, MO) in culture medium supplemented with 10% fetal bovine serum, unless stated otherwise.
Reverse transcription-PCR (RT-PCR)
Total RNA was isolated using the TRIzol RNA extraction kit (Invitrogen, Grand Island, NY), and 5 μg of RNA were subjected to RT-PCR with SuperScript™II (Invitrogen, Grand Island, NY, USA). The cPLA2α-specific primers (sense, 5′-CTG ATG TTT GCA GAT TGG GTT G-3′; antisense, 5′-AAA GGA GAC AGT GGA TAA GAT G-3′), nucleolin-specific primers (sense, 5′- AGA AGG GAG CCA CAC CAG GC-3′; antisense, 5′- AGC TGC TGC TTT CAT CGC TG-3′) and GAPDH primers were used. The PCR products were separated by 1% agarose-gel electrophoresis and visualized with ethidium bromide staining.
Northern blot analysis
Total RNA was isolated using the TRIzol RNA extraction kit (Invitrogen, Grand Island, NY), and 20 μg of RNA were used for electrophoresis and transferred to a nylon membrane as previously described (32). The cDNA probes used were prepared from RT-PCR. Probes were labeled with [α-32P]dCTP by using a Rediprime™II random prime labeling system (Amershan, Bucks, UK). The nylon membranes were washed three times at room temperature in 2× standard saline/phosphate/EDTA buffer (300 mM NaCl, 20 mM NaH2PO4 and 2 mM EDTA) containing 0.1% SDS. Each wash was carried out for 15 min. Autoradiography was then performed.
Plasmid construction
A 619 bp of human cPLA2α promoter region was PCR-amplified from human genomic DNA and subcloned into luciferase plasmid pXP1 as the pPLA599 plasmid. Respective additions of KpnI to forward primer 5′-GAAATTCAAACCTGAATTCAATTTTCTTCCCT-3′ and of HindIII to reverse primer 5′-GATCCTTTTTCAGCTCCGGA-3′ facilitated subcloning. 5′-Deletions of various lengths, pPLA393, pPLA241, pPLA140, pPLA53, pPLA35 and pPLA27 were generated using reverse primer 5′-GATCCTTTTTCAGCTCCGGA-3′ and the forward primers 5′-AACTTTGCCTTTTTAAATAATGCA-3′, 5′-ACAGAAATCCGCAACAGCACTC-3′, 5′-CATTTACATTTACAATATTAGC-3′, 5′-GGAGACCAGCCCACATTTTAG-3′, 5′-TAGCCCCTCCTACTCAGG-3′ and 5′-CCTACTCAGGATAAGACT-3′, respectively. The mutants at Sp1 site (pLAm) were constructed by the site-directed mutagenesis method. Synthetic primers were shown in the following: pLAm1 mutant primer 5′- GGAGACCAGTTCACATTTTAG -3′, and pLAm2 mutant primer 5′- TAGTTCTTCCTACTCAGG -3′. Mutated positions in the sequence of the primers were underlined. All DNA fragments were directly subcloned into pXP1 using KpnI and HindIII. The vector sequence was confirmed by DNA sequencing. To generate the luciferase plasmid UTR-1-446, a 447 bp of human cPLA2α mRNA 3′UTR was PCR-amplified from cDNA and inserted between luciferase gene and SV40 late poly(A) signal coding regions of luciferase plasmid pGL3. Respective additions of XbaI to forward primer 5′-TTCATGTACTGGAAATGGCAGC-3′ and of FseI to reverse primer 5′-CATGTATGTATATATATGC-3′ facilitated subcloning. The nucleotide sequences of constructs were confirmed by automatic DNA sequencing.
Transfection of cells with plasmids
Cells were transfected with plasmids by lipofection using Lipofectamine 2000 (Invitrogen, Grand Island, NY, USA) according to the manufacturer's instruction with a slight modification. Cells were replated 24 h before transfection at an optimal cell density in 2 ml of fresh culture medium in a 3.5 cm plastic dish. For use in transfection, 0.5 μl of Lipofectamine 2000 was incubated with 0.5 μg of pPLA, pXLO-7-1 of human 12(S)-lipoxygenase (33) or pXC918 of cyclooxygenase-2 (34) luciferase plasmid or the indicated plasmids as described in each experiment, in 1 ml of Opti-MEM medium for 30 min at room temperature. Total DNA concentration for each transfection was normalized with pcDNA3.1. Cells were transfected by changing the medium with 1 ml of Opti-MEM medium containing the plasmids and Lipofectamine 2000, followed by incubation at 37°C in a humidified atmosphere of 5% CO2 for 24 h. After a change of Opti-MEM medium to 2 ml of fresh culture medium, cells were stimulated with PMA if necessary and then incubated for an additional 18 h, unless stated otherwise. The luciferase activity in cell lysate was determined as described previously (33).
DNA affinity precipitation assay (DAPA)
This assay was performed according to the method reported previously (35) with a slight modification. The binding assay was performed by mixing 200 μg of nuclear extract proteins, 2 μg of biotinylated specific wild-type and mutated Sp1 oligonucleotides of cPLA2α, 12(S)-lipoxygenase and gastrin promoters 5′-CAA GGA GAC CAG CCC ACA TTT TAG CCC CTC CTA CTC AGG-3′, 5′-CAA GGA GAC CAG CTT ACA TTT TAT CCC TTC CTA CTC AGG-3′, 5′- TAA AAC TTG CGA GGA GGG CGG GGC CGC AG-3′ and 5′-CAG GGT AGG GGC GGG GTG GGG GGA CA-3′, respectively, and Sp1 and NFκB consensus oligonucleotides 5′-ATT CGA TCG GGG CGG GGC GAG C-3′ and 5′-AGT TGA GGG GAC TTT CCC AGG C-3′, respectively, and 20 μl of streptavidin-agarose beads (4%) with 50% slurry. Mutated positions in the sequence were underlined. The mixture was incubated at room temperature for 1 h with rotating. Beads were pelleted and washed three times with PBS. The binding proteins were eluted by loading buffer and separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), followed by western blot analysis probed with specific antibodies.
Chromatin immunoprecipitation (ChIP) assay
Chromatin immunoprecipitation assay was done according to previous report (36) with minor modification. Briefly, A549 cells were treated with 1% formaldehyde for 15 min. The cross-linked chromatin was sonicated to 400–500 bp fragments. Lysates were precleared with protein A beads and incubated overnight at 4°C with antibodies specific to nucleolin (Santa Cruz Biotechology, Santa Cruz, CA, USA), c-Jun (Santa Cruz, CA, USA) and Sp1 (Santa Cruz, CA, USA). Immune complex was precipitated with protein A beads preabsorbed with sonicated salmon sperm DNA and BSA. After reversal of cross-linking, levels of precipitated cPLA2α promoter DNA were determined by PCR using oligonucleotides spanning the Sp1-binding sites (sense, 5′-ACA GAA ATC CGC AAC AGC ACT C-3′; antisense, 5′-GAT CCT TTT TCA GCT CCG GA-3′). The PCR products were separated by 1% agarose-gel electrophoresis and visualized with ethidium bromide staining.
Western blotting
The cytoplasmic fractions and nuclear extracts of cells were prepared for western blot analysis according to the method described (37). An analytical 10% SDS–PAGE was performed, and 30 μg of protein of each sample were analyzed, unless stated otherwise. For immunoblotting, proteins in the SDS gels were transferred to a polyvinylidene difluoride membrane by an electroblot apparatus. Antibodies against human Sp1 (Santa Cruz, CA, USA), c-Jun (Santa Cruz, CA, USA), nucleolin (Santa Cruz, CA, USA) and β-actin (Santa Cruz, CA, USA) were used as the primary antibodies. Mouse or rabbit IgG antibodies coupled to horseradish peroxidase were used as secondary antibodies. An enhanced chemiluminescence kit (Pierce, Rockford, IL, USA) was used for detection.
Coimmunoprecipitation
Two hundred micrograms protein of nuclear extracts and lysate were incubated under gentle shaking at 4°C overnight with a mixture of anti-c-Jun or anti-Sp1 antibodies and protein A agarose in 300 μl of immunoprecipitation buffer [20 mM HEPES, pH 7.9, 420 mM NaCl, 1.5 mM MgCl2 and 25% glycerol (v/v), 0.5 mM phenylmethylsulfonyl fluoride, 1 mM orthovanadate, 2 μg/ml pepstatin A and 2 μg/ml leupeptin]. Beads were pelleted at 7500g for 2 min and washed three times with RIPA buffer [50 mM Tris–HCl, pH 7.5, 1% IGEPAL CA-630 (v/v), 150 mM NaCl, and 0.5% sodium deoxycholate]. Protein was removed from the beads by boiling in sample buffer (120 mM Tris–HCl, pH 6.8, 10% glycerol, 3% SDS, 20 mM DTT, and 0.4% bromophenol blue) for 5 min and subjected to 10% SDS–PAGE. Western blot analysis was carried out as described above.
RESULTS
Effect of PMA on the transcription of cPLA2α gene
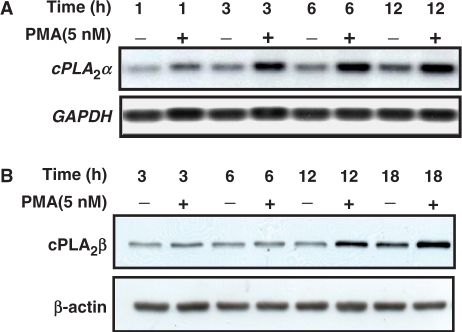
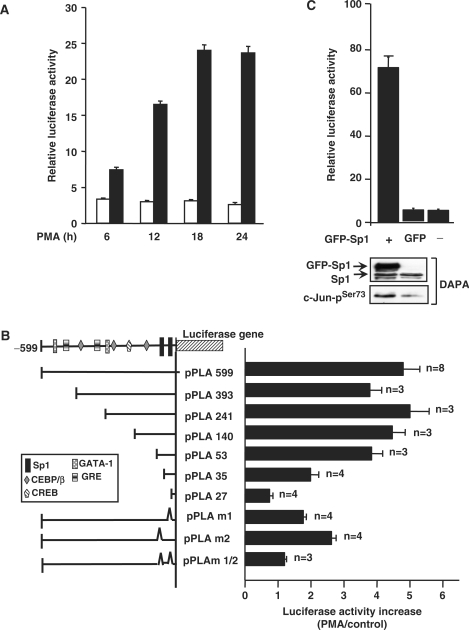
Expression of cPLA2α was detected by northern blot and immunoblot analysis in PMA-treated A549 cells. PMA induced the expression of cPLA2α mRNA (Figure 1A) and protein (Figure 1B) in a time-dependent manner. To confirm that PMA enhanced the expression of cPLA2α through the transcriptional activation of the gene, the vector containing human cPLA2α gene promoter was used in the reporter assay. PMA induced the promoter activity of cPLA2α in a time-dependent manner (Figure 2A). The maximum effect of PMA on the promoter activity was observed in cells treated with PMA for 18 h. To identify the promoter elements responsible for PMA induction, DNA constructs with 5′-deletions of cPLA2α promoter were transiently transfected into cells. The effect of PMA on the luciferase reporter activity was then studied and summarized in Figure 2B. The luciferase activities of vectors pPLA 599, pPLA 393, pPLA 241, pPLA 140 and pPLA 53 were stimulated by PMA treatment. The stimulation was about 5-fold, obtained by comparing the luciferase activity in PMA-treated cells with that in control cells. An apparent decrease in the stimulatory response of PMA was observed in vectors bearing a promoter with a deletion from −53 (pPLA 53) to −27 bp (pPLA 27), indicating that a promoter region ranging from −53 to −27 bp was required for the PMA-induced expression of cPLA2α. To further confirm whether two Sp1-binding sites (−45 to −42 bp and −33 to −29 bp) located in the region were important for PMA response, DNA constructs with mutations on Sp1-binding sites were transfected into cells. Compared to pPLA599, a significant decrease in PMA-stimulated response was observed in pPLA m1 and pPLA m2, and a complete elimination of PMA response was detected in pPLA m1/2. In addition, overexpression of Sp1 also induced the promoter activity of cPLA2α (Figure 2C). These results indicated that two Sp1 sites were essential for PMA-induced transcriptional activation of cPLA2α gene and Sp1 was involved in the regulation of gene expression.
Figure 1.
PMA induces the expression of cPLA2α in A549 cells. (A) A549 cells were starved for 18 h in serum-free culture medium and then treated with 5 nM PMA for a different time period as indicated. Northern blot analysis of mRNA was performed. (B) Cell lysates were subjected to SDS–PAGE and analyzed by western blotting with antibodies against cPLA2α and β-actin.
Figure 2.
Analysis of PMA-responsive element in the promoter region of cPLA2α gene in A549 cells. (A) Cells were transfected with 0.5 μg of luciferase plasmid bearing cPLA2α gene promoter by lipofection, incubated for 24 h and then treated with 5 nM PMA for a different time period as indicated. The luciferase activities and protein concentrations were determined and normalized. The results shown represent the means ± SEM of three determinations. Blank columns: non-treatment; black columns: PMA-treated cells. (B) Truncated promoter fragment was ligated into a luciferase plasmid as described in the Materials and Methods section. Numbers indicate the distance in base pairs from the translation start site. Cells were transfected with 0.5 μg of luciferase plasmid bearing cPLA2α gene promoter by lipofection, incubated for 24 h and then treated with 5 nM PMA for 18 h. The luciferase activities and protein concentrations were determined and normalized. The expression ratio of PMA treated to control cells is indicated. The results shown represent the means ± SEM of 3 to 8 independent experiments in triplicate wells for each construct. (C) Cells were transfected with 0.5 μg of luciferase plasmid bearing cPLA2α gene promoter by lipofection, incubated for 6 h and then infected with adenovirus carrying GFP-Sp1 (Ad-GFP-Sp1) at 50 MOI. Cells infected with Ad-GFP were used as control. The luciferase activities and protein concentrations were determined. The results shown represent the means ± SEM of three determinations. Cell lysates were prepared and DNA affinity precipitation assay was performed as described under Materials and Methods section. Binding of Sp1 and phospho-c-Jun (Ser73) to cPLA2α Sp1 probes was analyzed by western blot.
Effect of c-Jun/Sp1 on the PMA-induced transcriptional activation of cPLA2α gene
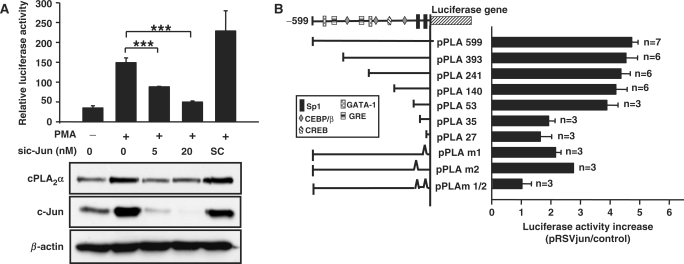
Since overexpressing Ha-Ras leads to induction of c-Jun protein and drives rat cPLA2α promoter activity (23), we then studied the possibility that whether c-Jun also cooperated with Sp1 in the regulation of PMA-induced expression of human cPLA2α gene. To confirm the role of c-Jun in PMA-regulated gene expression, c-Jun siRNA oligonucleotides were transfected into cells. As shown in Figure 3A, c-Jun siRNA inhibited PMA-induced promoter activity and protein expression of cPLA2α. Furthermore, overexpression of c-Jun also induced the promoter activity of cPLA2α gene (Figure 3B). Consistent with the results of Figure 2B, the luciferase activity of vectors pPLA 599, pPLA 393, pPLA 241, pPLA 140 and pPLA 53 but not pPLA 35 and pPLA 27 were stimulated by pRSVjun (Figure 3B). A significant decrease in pRSVjun response was observed in pPLA m1 and pPLA m2, whereas the response was completely abolished in pPLA m1/2. These results indicated that the two Sp1 sites play an important role in c-Jun-induced transcription of cPLA2α gene as well as the PMA response (Figure 2B). Since there is no AP1 site within the promoter region ranging from −53 to −27 bp, we therefore speculated that the effect of c-Jun on gene expression was mediated by cooperation with Sp1.
Figure 3.
Effect of c-Jun on PMA-induced expression of cPLA2α in A549 cells. (A) Cells were transfected with c-Jun siRNA oligonucleotide by lipofection, incubated for 6 h and then transfected with 0.5 μg of pPLA 599. Cells were incubated for 24 h and then treated with 5 nM PMA for 18 h. The luciferase activities and protein concentrations were determined and normalized. The results shown represent the means ± SEM of three determinations. Expressions of c-Jun, cPLA2α and β-actin proteins were analyzed by western blot analysis using anti-c-Jun, cPLA2α and β-actin antibodies, respectively. SC: scramble oligonucleotides. Statistical significance (***P < 0.001) between PMA with c-Jun siRNA and PMA alone was analyzed by Student's t-test. (B) Cells were transfected with 0.5 μg of pRSVjun expression vector and 0.5 μg of luciferase plasmid bearing cPLA2α gene promoter by lipofection, incubated further for 36 h and the luciferase activities and protein concentrations were determined. The expression ratio of pRSVjun treated to control cells is indicated. The results shown represent the means ± SEM of 3 to 7 independent experiments in triplicate wells for each construct.
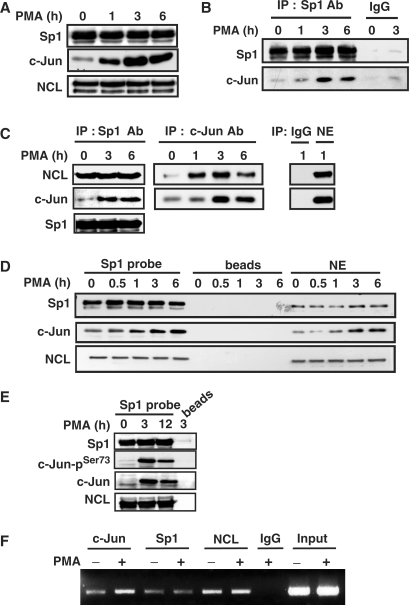
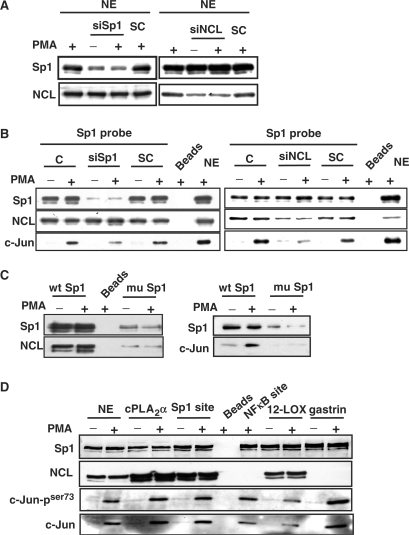
We then studied the functional interaction between c-Jun and Sp1 in the regulation of PMA-induced gene expression of cPLA2α by immunoprecipitation, DAPA and ChIP assay. Expression of Sp1 and c-Jun in nuclear extracts prepared from cells treated with PMA for 1–6 h was studied by using immunoblot analysis. No difference of Sp1 expression between control and PMA-treated cells was observed (Figure 4A). PMA induced the expression of c-Jun (Figure 4A), and enhanced the interaction between c-Jun and Sp1 (Figure 4B). To directly examine whether the c-Jun/Sp1 complex bound to cPLA2 α promoter in PMA-treated cells, the probes containing two Sp1-binding sites of cPLA2α promoter were used in DAPA. The binding of c-Jun and phospho-c-Jun but not Sp1 to the promoter under the PMA stimulation was increased (Figure 4D and E). In addition, phospho-c-Jun binding to the promoter was significantly increased by overexpressed Sp1 in cells (Figure 2C). In ChIP assay, the binding of c-Jun but not Sp1 to the promoter in cells under the PMA treatment was also enhanced (Figure 4F). These results suggested that PMA induced the formation of c-Jun/Sp1 complex, resulting in facilitating the access of activated c-Jun to the promoter and enhanced the transcriptional activation.
Figure 4.
PMA induces the complex formation and the binding of c-Jun, nucleolin and Sp1 to cPLA2α gene promoter in A549 cells. (A) Cells were starved for 18 h in serum-free culture medium and then treated with 5 nM PMA for a different time period as indicated. The Sp1, nucleolin (NCL) and c-Jun proteins were detected by anti-Sp1, anti-nucleolin and anti-c-Jun antibodies, respectively. (B and C) Nuclear extracts were immunoprecipitated (IP) with antibodies (Ab) against Sp1 and c-Jun. The proteins were subjected to SDS–PAGE and analyzed by western blotting with antibodies against c-Jun, NCL and Sp1. IgG: negative control of antibodies. (D and E) Cells were starved for 18 h in serum-free culture medium and then treated with 5 nM PMA for a different time period as indicated. Nuclear extracts were prepared and DNA affinity precipitation assay was performed as described under Materials and Methods section. Binding of Sp1, NCL, c-Jun and phospho-c-Jun (Ser73) to Sp1 probes were analyzed by western blot. The streptavidin-agarose beads were used to serve as a nonspecific binding control. (F) Cross-linked chromatin derived from PMA-treated cells was immunoprecipitated with c-Jun, NCL and Sp1 antibodies and analyzed by PCR with specific primers for the region from −239 to +19 bp of cPLA2α promoter. Input: nonimmunoprecipitated cross-linked chromatin.
Nucleolin mediates the PMA-induced transcriptional activation of cPLA2α gene
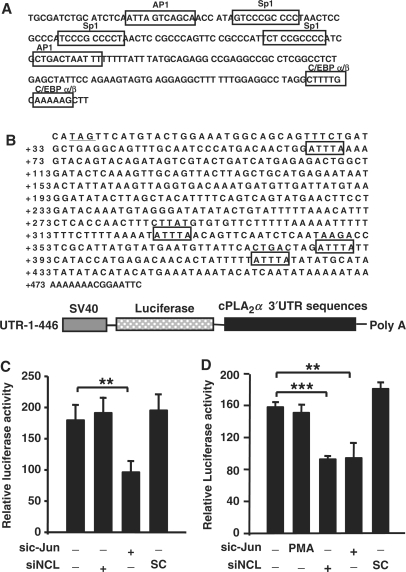
To gain insight into the PMA-regulated transcriptional mechanism of cPLA2α gene, factors bound to Sp1 sties of cPLA2α promoter were analyzed by DAPA. The bound proteins were subsequently analyzed by 2D SDS–PAGE. Proteins on the gel were identified by MALDI-TOF. One of the proteins, nucleolin bound to the probe was then clearly identified (data not shown). As shown in Figure 4D analyzed by DAPA, the binding of nucleolin to the promoter was not changed in PMA-treated cells as well as Sp1 binding. In addition, the constant interaction between nucelolin and Sp1 was observed in control or PMA-treated cells (Figure 4C). However, the interaction of nucleolin with c-Jun was increased in PMA-treated condition (Figure 4C). We further confirmed the binding of nucleolin to cPLA2α promoter by ChIP assay (Figure 4F), which was consistent with the results shown in Figure 4D. Since the formation and binding of c-Jun/nucleolin complex to cPLA2α promoter was increased in PMA-treated cells, it is likely that nucleolin is involved in the regulation of PMA-induced cPLA2α gene expression. We then studied the role of nucleolin in the regulation of PMA-induced gene expression. As shown in Figure 5A, PMA-induced promoter activity and protein expression of cPLA2α were inhibited in nucleolin knockdown cells. Nucleolin siRNA oligonucleotides also significantly inhibited the PMA-induced increase of cPLA2α mRNA (Figure 5B). These results revealed that c-Jun/nucleolin complex played an important role in PMA-induced cPLA2α gene expression. Tay et al. (38) have shown that epidermal growth factor, platelet-derived growth factor, serum and PMA exert their effects on cPLA2 expression through post-translational mechanisms involving stabilization of the mRNA. In order to confirm that whether nucleolin-regulated PMA-induced cPLA2α gene expression through transcriptional activation pathway, pGL3 vector containing SV40 promoter and luciferase cDNA without cPLA2α promoter was used. The sequence of SV40 promoter was analyzed by the computer program ‘TFSEARCH’ and interpreted AP1, Sp1 and C/EBP α/β binding sites on the promoter region (Figure 6A). No change of luciferase activity was observed in nucleolin knockdown cells (Figure 6C), indicating that nucleolin had no effect on the SV40 promoter activity and mRNA stability of the luciferase gene. The expression of luciferase gene was inhibited in c-Jun siRNA oligonucleotides-transfected cells (Figure 6C). Since nucleolin had no effect on the mRNA stability of luciferase gene, together with the reporter assay from Figure 5A, these results indicated that nucleolin was involved in the PMA-induced transcriptional activation of cPLA2α gene. To further study whether nucleolin could also play a function in post-transcriptional activation, we used UTR-1-446 vector, containing SV40 promoter and luciferase cDNA followed by cPLA2α 3′UTR sequence (Figure 6B) to dissect the requirement of 3′UTR sequence responsible for nucleolin-regulated mRNA stability. As shown in Figure 6D, nucleolin siRNA significantly reduced luciferase activity, indicating nucleolin was involved in the regulation of cPLA2α mRNA stability. In addition, PMA had no effect on UTR-1-446 luciferase activity (Figure 6D), indicating that cPLA2α 3′UTR sequence was not essential for PMA-regulated cPLA2α gene expression. These results revealed that PMA regulated cPLA2α expression by favoring transcriptional activation pathway but not through mRNA stabilization. However, nucleolin contributed to not only PMA-induced transcriptional activation of cPLA2α gene but also stabilization of cPLA2α mRNA in cells without PMA treatment.
Figure 5.
Effect of nucleolin on PMA-induced expression of cPLA2α in A549 cells. (A) Cells were transfected with nucleolin siRNA oligonucleotide (siNCL) by lipofection, incubated for 6 h and then transfected with 0.5 μg of pPLA 599 again. Cells were incubated for 24 h and then treated with 5 nM PMA for 18 h. The luciferase activities and protein concentrations were determined and normalized. The results shown represent the means ± SEM of three determinations. Expressions of nucleolin, cPLA2α and β-actin proteins were analyzed by western blot analysis using anti-nucleolin, cPLA2α and β-actin antibodies, respectively. SC: scramble oligonucleotides. Statistical significance (**P < 0.01 and ***P < 0.001) between PMA with nucleolin siRNA and PMA alone was analyzed by Student's t-test. (B) Cells were transfected with nucleolin siRNA oligonucleotide by lipofection for 36 h and then treated with 5 nM PMA for 18 h. Total RNA was extracted for RT-PCR with nucleolin, cPLA2 and GAPDH primers. The relative density of PCR products was quantified as indicated. SC: scramble oligonucleotides.
Figure 6.
Effect of nucleolin on mRNA stability. (A) Putative transcription factor binding sites in the SV40 promoter of the pGL3-promoter vector were scanned by the computer program available on the Web site (//transfac.gbf.de/TRANSFAC/index.html). Each putative consensus sequence in the SV40 promoter is enclosed in a box. (B) Putative HuR-binding sites in cPLA2α 3′UTR are enclosed in boxes. The sequence of transcription stop site is underlined. Schematic diagram of UTR-1-446 vector was presented. (C and D) A549 Cells were transfected with nucleolin or c-Jun siRNA oligonucleotides by lipofection, incubated for 6 h and then transfected with 1 μg of pGL3 (C) and UTR-1-446 (D) vectors. Cells were incubated for 24 h and then the luciferase activities and protein concentrations were determined and normalized. The results shown represent the means ± SEM of three determinations. SC: scramble oligonucleotides. Statistical significance (**P < 0.01 and ***P < 0.001) between c-Jun siRNA and control or nucleolin siRNA and control was analyzed by Student's t-test.
Nucleolin and Sp1 are individually essential for the recruitment of c-Jun to GC-rich promoter
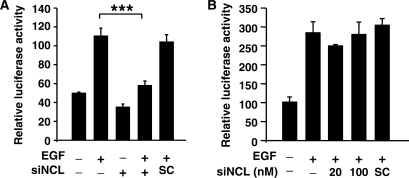
Several lines of evidence reveal that the motif (U/G)CCCG(A/G) is important for nucleolin RNA binding (39). However, it is interesting to note that cPLA2α promoter, containing two potential Sp1-binding sequences, AGCCCA and AGCCCCT might also be considered as the nucleolin recognition element. We then clarified whether the binding of nucleolin to cPLA2α promoter was Sp1-dependent. In Sp1 knockdown cells (Figure 7A), the amount of c-Jun on DNA was reduced to 50% (Figure 7B). However, Sp1 siRNA had no significant effect on the binding of nucleolin to DNA. We further studied whether nucleolin was required for Sp1–DNA interaction. Although the interaction of Sp1 with DNA was not changed in nucleolin siRNA-treated cells, the binding of c-Jun to DNA was almost abolished (Figure 7B). To clarify that whether nucleolin bound to DNA through Sp1-binding sites, we studied the binding of nucleolin to wild-type Sp1 probes, compared to mutated probes. As shown in Figure 7C, mutated Sp1 probes abolished both bindings of Sp1 and nucleolin to DNA, resulting in complete inhibition of the interaction between c-Jun and DNA. These results indicated that the bindings of nucleolin and Sp1 to DNA were independent of each other. Furthermore, in addition to Sp1, nucleolin might also be an anchor protein to recruit c-Jun to Sp1 sites and activated cPLA2α gene expression in PMA-treated cells. In order to further dissect that whether the binding of nucleolin to Sp1 site was mediated by recognizing the canonical or non-canonical Sp1 sequence, the promoters with consensus Sp1 sequence of 12(S)-lipoxygenase (33) and gastrin (40) or consensus Sp1 oligonucelotides were used in DAPA. Although the binding of Sp1 to Sp1-binding site was observed as expectations, the interaction between nucleolin and DNA was only observed in cPLA2α promoter, 12(S)-lipoxygenase promoter and consensus Sp1 oligonucleotide. However, no interaction between nucleolin and DNA was observed in gastrin promoter or consensus NFκB site (Figure 7D). The binding of phospho-c-Jun to DNA was increased in all oligonucleotide probes in PMA-treated cells (Figure 7D). These results revealed that not all the Sp1 or Sp1-like binding sites were in favor of nucleolin binding, indicating that the binding of nucleolin to Sp1 site was not determined by canonical or non-canonical Sp1-binding sequence. To further confirm that nucleolin was essential for c-Jun/Sp1-regulated gene expression resulted from the recruitment of c-Jun to Sp1 sites-dependent transcriptional activation, we studied the effect of nucleolin on the induction of 12(S)-lipoxygenase which is Sp1-dependent, and cyclooxygenase-2 which is Sp1-independent in cells under EGF treatment (33,34). Under these experimental conditions, c-Jun is essential for the EGF-induced expression of 12(S)-lipoxygenase and cyclooxygenase-2. Consistent with that observed in the regulation of cPLA2α gene, nucleolin siRNA abolished the EGF-induced transcriptional activation of 12(S)-lipoxygenase gene (Figure 8A). Contrary to c-Jun/Sp1-regulated genes, nucleolin was not involved in EGF-induced transcriptional activation of cyclooxygenase-2 gene (Figure 8B).
Figure 7.
Effect of nucelolin and Sp1 on the recruitment of c-Jun to Sp1-dependent gene in A549 cells. (A and B) Cells were transfected with nucleolin and Sp1 siRNA oligonucleotides by lipofection, incubated for 30 h and then treated with 5 nM PMA for 3 h. Nuclear extracts (NE) were prepared and DNA affinity precipitation assay was performed as described under Materials and Methods section. Binding of Sp1, nucleolin and c-Jun to Sp1 probes were analyzed by western blot. The streptavidin-agarose beads were used to serve as a nonspecific binding control. SC: scramble oligonucleotides. (C and D) Cells were treated with 5 nM PMA for 3 h. Nuclear extracts (NE) were prepared and DNA affinity precipitation assay was performed as described under Materials and Methods section. Binding of Sp1, nucleolin, phospho-c-Jun (Ser73) and c-Jun to cPLA2α Sp1 (wt Sp1), cPLA2α Sp1 mutant (mu Sp1), 12(S)-lipoxygenase (12-LOX), gastrin, consensus Sp1 site (Sp1 site) and consensus NFκB site probes were analyzed by western blot. The streptavidin-agarose beads were used to serve as a nonspecific binding control.
Figure 8.
Nucelolin is essential for c-Jun/Sp1-regulated gene expression. (A and B) A431 cells were transfected with nucleolin siRNA oligonucleotides by lipofection, incubated for 6 h and then transfected with 0.5 μg of pXLO-7-1 (A) and pXC918 (B) vectors bearing the 12(S)-lipoxygenase and cyclooxygenase-2 promoters, respectively. Cells were incubated for 24 h and pXLO-7-1- and pXC918-transfected cells were treated with EGF (50 ng/ml) for 18 and 6 h, respectively. The luciferase activities and protein concentrations were determined and normalized. The results shown represent the means ± SEM of three determinations. SC: scramble oligonucleotides. Statistical significance (***P < 0.001) between EGF with nucleolin siRNA and EGF alone was analyzed by Student's t-test.
DISCUSSION
The expression of cPLA2 can be regulated in NSCLC (7) and is critical for transformed growth of NSCLC. When these cells are treated with a specific inhibitor of the enzyme, or blocking downstream production of prostaglandins with cyclooxygenase inhibitors, resulting in inhibition of anchorage-independent growth of these cells (7). These results suggest that induction of cPLA2 is critical for tumorigenesis. Consistent with this finding, lung tumorigenesis is inhibited in mice that are deficient in cPLA2 (41). In this study, we first found that PMA induced gene expression of cPLA2α in NSCLC. It has been known that PMA-activated PKC signal transduction pathway is thought to be involved in the oncogene action in NSCLC and enzymatic activation of cPLA2. Activation of PKC with PMA impairs progression of lung adenocarcinoma cells from early G1 phase into S phase (42). The broad-range PKC inhibitor staurosporine analog, PKC 412, induces apoptosis in SCLC cells and sensitizes NSCLC cells to apoptosis induced by DNA-damaging agents (43). Taking these results together, we concluded that PMA-induced expression of cPLA2α might be related to either a cause or a consequence of PMA-regulated tumorigenesis of NSCLC.
In the regulation of gene expression, previous studies have reported that c-Jun/Sp1 complex is critical for various different gene expressions, e.g. 12(S)-lipoxygenase, keratin 16, p21WAF1/CIP1 and neuronal nicotinic acetylcholine receptor β4 (26–29). Although overexpression of c-Jun and Sp1 produces a synergistic increase in the rat cPLA2 promoter activity, no evidence shows that the formation of c-Jun/Sp1 complex and the complex binding to the rat cPLA2 promoter are observed (23). However, we first clarified that PMA induced c-Jun/Sp1 interaction and the complex bound to Sp1-binding sites of human cPLA2α promoter, resulting in the induction of transcriptional activation. In addition, we found that nucleolin was also a coactivator interacting with c-Jun to regulate PMA-induced transcription of cPLA2α gene. Nucleolin is a ubiquitous, nonhistone nucleolar phosphoprotein of exponentially growing eukaryotic cells, which is directly involved in the regulation of ribosome biogenesis, the processing of ribosomal RNA, mRNA stability, transcriptional regulation and cell proliferation, and it is also a downstream target of several signal transduction pathways (30). Matching our results, the functional role of nucleolin involved in the transcriptional activation of gene expression has been found. Nucleolin binds acetylated interferon regulatory factor-2 (IRF-2) to enhance H4 promoter activity (44) and plays as a key activator of HPV18 oncogene transcription in cervical cancer (45). Nucleolin also binds transcription factors Myb and tumor suppressor Rb to regulate Myb transcriptional activity and tumor development (46,47). These findings suggest that nucleolin may act as a transcriptional regulator via interacting with activators. Indeed, in the experiments of immunoprecipitation and DAPA, we found that nucleolin bound and bridged c-Jun to Sp1-binding sites of cPLA2α promoter (Figures 4C and 7B). These results suggested the possibility that nucleolin participated in the regulation of c-Jun/Sp1-regulated genes, e.g. 12(S)-lipoxygenase (Figure 7D) and p21WAF1/CIP1 to regulate cell growth (48). From our study, we can conclude that for PMA-induced cPLA2α gene expression, and nucleolin acts as a coactivator via cooperation of c-Jun transcriptional factor. However, the nucleolin/c-Jun complex had no effect on AP1-regulated promoter activation (Figures 6C and 8B). The different binding affinity between nucleolin/c-Jun to Sp1-binding site and to AP1-binding site might be caused by the stereo-recognition of complex to the Sp1 binding but not AP1-binding sequences. Consistent with the role of nucleolin in the regulation of mRNA stability, we found that nucleolin also contributed to the stabilization of cPLA2α mRNA. Since without identical nucleolin-binding site UCCCGA but with HuR-binding element AUUUA is found within the cPLA2α 3′UTR sequence, the effect of nucleolin on the stabilization of cPLA2α mRNA might result from the cooperation of nucleolin with RNA-binding proteins as well as the basal expression of GADD45α mRNA is regulated by association of nucleolin and HuR (49). Indeed, we found that RNA-binding protein HuR bound to AUUUA of cPLA2α 3′UTR sequence and enhanced the mRNA stability (our unpublished data). Although PMA had no effect on the stabilization of cPLA2α mRNA, our data showed that nucleolin was also involved in the regulation of PMA-induced transcriptional activation of cPLA2α gene. These results revealed that nucleolin plays dual functions of transcriptional and post-transcriptional activity in the regulation of cPLA2α gene.
It is known that the motif (U/G)CCCG(A/G) is responsible for the binding of nucleolin to RNA (39). However, we found that the binding of nucleolin to the GC-rich Sp1 site within cPLA2α promoter was in an Sp1-independent manner. Thus, nucleolin might gain access to DNA through two pathways. First, since Sp1 siRNA had no effect on the binding of nucleolin to Sp1 element, indicating that nucleolin might directly bind to DNA and recruit c-Jun transcriptional factor to Sp1 sites and then activate gene expression. Nucleolin is a matrix attachment region (MAR)-binding protein to provide a link between DNA and nuclear matrix scaffolding (50). In addition, nucleolin was also found to bind to the NFκB DNA-binding motif and the KLF2 promoter (51,52). Thus, nucleolin may interact with DNA and other transcriptional factors, such as c-Jun, to regulate transcription. It is very interesting to note that whether the binding of nucleolin to GC-rich Sp1 site has the sequence specificity. Contrary to the binding of nucleolin to cPLA2α promoter, we found that SV40 promoter containing Sp1-binding sites was not regulated by nucleolin. Furthermore, we also found that the interaction between nucleolin and Sp1-binding site was not occurred in all Sp1-regulated promoters such as gastrin. It is possible that the nucleotide sequence surrounding the AGCCC and UCCCGA can modulate the interaction of nucleolin with DNA and RNA (39), respectively. Second, access of nucleolin to cPLA2α promoter may also skip from direct binding but be bridged by other factors to DNA. Although there was no effect of Sp1 siRNA on nucleolin binding, we could not rule out the possibility that the binding of nucleolin to DNA might have occurred concomitant with Sp1-like proteins binding. In addition, in spite of nucleolin and Sp1 bound to DNA was dependent on Sp1-binding sites, the binding of Sp1 to DNA was in a nucleolin-independent manner. These results suggested that Sp1/c-Jun and nucleolin/c-Jun complexes could together or independently bind to Sp1-binding sites and regulate PMA-induced transcriptional activation of cPLA2α gene.
In conclusion, we have identified the functional interaction between c-Jun, nucleolin and Sp1 to mediate cPLA2α expression in PMA signal-activated human tumor cells. Our results demonstrate that the activation of the PMA signaling pathway leads to the binding of c-Jun/nucleolin and c-Jun/Sp1 complexes to the cPLA2α promoter, resulting in the transcriptional activation of cPLA2α gene. Based on the expression of cPLA2α is critical for transformed growth of NSCLC and tumorigenesis (7,41), our study suggested that nucleolin, in addition to contribute to the process of tumor development by linking to major tumor suppressors, Rb and p53 (47,53), could associate with transcription factor c-Jun and up-regulate cPLA2α expression to increase lipid mediators, resulting in the tumorigenesis. Our data also suggested that nucleolin binds to GC-rich elements. It is likely that nucleolin binding at place of Sp1-binding sequences could also mediate the expression of c-Jun/Sp1-activated genes.
ACKNOWLEDGEMENTS
We are greatly indebted to Dr Jan-Jong Hung for providing adenovirus carrying GFP-Sp1 and GFP control. This work was supported in part by grant NSC 95-2320-B-006-071-MY2 from the National Science Council of the Republic of China, and National Cheng Kung University Project of Promoting Academic Excellence and Developing World Class Research Centers. Funding to pay the Open Access publication charges for this article was provided by National Science Council of the Republic of China.
Conflict of interest statement. None declared.
REFERENCES
- 1.Nie D, Honn KV. Cyclooxygenase, lipoxygenase and tumor angiogenesis. Cell. Mol. Life Sci. 2002;59:799–807. doi: 10.1007/s00018-002-8468-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cabral GA. Lipids as bioeffectors in the immune system. Life Sci. 2005;77:1699–1710. doi: 10.1016/j.lfs.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Bogatcheva NV, Sergeeva MG, Dudek SM, Verin AD. Arachidonic acid cascade in endothelial pathobiology. Microvasc. Res. 2005;69:107–127. doi: 10.1016/j.mvr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Xu XC. COX-2 inhibitors in cancer treatment and prevention, a recent development. Anticancer Drugs. 2002;13:127–137. doi: 10.1097/00001813-200202000-00003. [DOI] [PubMed] [Google Scholar]
- 5.Hubbard WC, Alley MC, McLemore TL, Boyd MR. Profiles of prostaglandin biosynthesis in sixteen established cell lines derived from human lung, colon, prostate, and ovarian tumors. Cancer Res. 1988;48:4770–4775. [PubMed] [Google Scholar]
- 6.McLemore TL, Hubbard WC, Litterst CL, Liu MC, Miller S, McMahon NA, Eggleston JC, Boyd MR. Profiles of prostaglandin biosynthesis in normal lung and tumor tissue from lung cancer patients. Cancer Res. 1988;48:3140–3147. [PubMed] [Google Scholar]
- 7.Heasley LE, Thaler S, Nicks M, Price B, Skorecki K, Nemenoff RA. Induction of cytosolic phospholipase A2 by oncogenic Ras in human non-small cell lung cancer. J. Biol. Chem. 1997;272:14501–14504. doi: 10.1074/jbc.272.23.14501. [DOI] [PubMed] [Google Scholar]
- 8.Leslie CC. Properties and regulation of cytosolic phospholipase A2. J. Biol. Chem. 1997;272:16709–16712. doi: 10.1074/jbc.272.27.16709. [DOI] [PubMed] [Google Scholar]
- 9.Evans JH, Spencer DM, Zweifach A, Leslie CC. Intracellular calcium signals regulating cytosolic phospholipase A2 translocation to internal membranes. J. Biol. Chem. 2001;276:30150–30160. doi: 10.1074/jbc.M100943200. [DOI] [PubMed] [Google Scholar]
- 10.Hirabayashi T, Kume K, Hirose K, Yokomizo T, Iino M, Itoh H, Shimizu T. Critical duration of intracellular Ca2+ response required for continuous translocation and activation ofcytosolic phospholipase A2. J. Biol. Chem. 1999;274:5163–5169. doi: 10.1074/jbc.274.8.5163. [DOI] [PubMed] [Google Scholar]
- 11.Gijon MA, Spencer DM, Siddiqi AR, Bonventre JV, Leslie CC. Cytosolic phospholipase A2 is required for macrophage arachidonic acid release by agonists that Do and Do not mobilize calcium. Novel role of mitogen-activated protein kinase pathways in cytosolic phospholipase A2 regulation. J. Biol. Chem. 2000;275:20146–20156. doi: 10.1074/jbc.M908941199. [DOI] [PubMed] [Google Scholar]
- 12.Qiu Z-H, Gijon MA, de Carvalho MS, Spencer DM, Leslie CC. The role of calcium and phosphorylation of cytosolic phospholipase A2 in regulating arachidonic acid release in macrophages. J. Biol. Chem. 1998;273:8203–8211. doi: 10.1074/jbc.273.14.8203. [DOI] [PubMed] [Google Scholar]
- 13.You HJ, Woo C-H, Choi E-Y, Cho S-H, Yoo YJ, Kim J-H. Roles of Rac and p38 kinase in the activation of cytosolic phospholipase A2 in response to PMA. Biochem. J. 2005;388:527–535. doi: 10.1042/BJ20041614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin L-L, Wartmann M, Lin AY, Knopf JL, Seth A, Davis RJ. cPLA2 is phosphorylated and activated by MAP kinase. Cell. 1993;72:269–278. doi: 10.1016/0092-8674(93)90666-e. [DOI] [PubMed] [Google Scholar]
- 15.Hefner Y, Borsch-Haubold AG, Murakami M, Wilde JI, Pasquet S, Schieltz D, Ghomashchi F, Yates J.R., III, Armstrong CG, et al. Serine 727 phosphorylation and activation of cytosolic phospholipase A2 by MNK1-related protein kinases. J. Biol. Chem. 2000;275:37542–37551. doi: 10.1074/jbc.M003395200. [DOI] [PubMed] [Google Scholar]
- 16.Muthalif MM, Hefner Y, Canaan S, Harper J, Zhou H, Parmentier J-H, Aebersold R, Gelb MH, Malik KU. Functional interaction of calcium-/calmodulin-dependent protein kinase II and cytosolic phospholipase A2. J. Biol. Chem. 2001;276:39653–39660. doi: 10.1074/jbc.M103136200. [DOI] [PubMed] [Google Scholar]
- 17.Roshak AK, Jackson JR, McGough K, Chabot-Fletcher M, Mochan E, Marshall LA. Manipulation of distinct NFkappa B proteins alters interleukin-1beta -induced human rheumatoid synovial fibroblast prostaglandin E2 formation. J. Biol. Chem. 1996;271:31496–31501. doi: 10.1074/jbc.271.49.31496. [DOI] [PubMed] [Google Scholar]
- 18.Wu T, lkezono T, Angus CW, Shelhamer JH. Characterization of the promoter for the human 85 kDa cytosolic phospholipase A2 gene. Nucleic Acids Res. 1994;22:5093–5098. doi: 10.1093/nar/22.23.5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dronadula N, Liu Z, Wang C, Cao H, Rao GN. STAT-3-dependent cytosolic phospholipase A2 expression is required for thrombin-induced vascular smooth muscle cell motility. J. Biol. Chem. 2005;280:3112–3120. doi: 10.1074/jbc.M409739200. [DOI] [PubMed] [Google Scholar]
- 20.Neeli I, Liu Z, Dronadula N, Ma ZA, Rao GN. An essential role of the Jak-2/STAT-3/cytosolic phospholipase A2 axis in platelet-derived growth factor BB-induced vascular smooth muscle cell motility. J. Biol. Chem. 2004;279:46122–46128. doi: 10.1074/jbc.M406922200. [DOI] [PubMed] [Google Scholar]
- 21.Dolan-O’Keefe M, Chow V, Monnier J, Visner GA, Nick HS. Transcriptional regulation and structural organization of the human cytosolic phospholipase A2 gene. Am. J. Physiol. Lung Cell Mol. Physiol. 2000;278:L649–L657. doi: 10.1152/ajplung.2000.278.4.L649. [DOI] [PubMed] [Google Scholar]
- 22.Tay A, Maxwell P, Li Z, Goldberg H, Skorecki K. Isolation of promoter for cytosolic phospholipase A2 (cPLA2) Biochim. Biophys. Acta. 1994;1217:345–347. doi: 10.1016/0167-4781(94)90299-2. [DOI] [PubMed] [Google Scholar]
- 23.Blaine SA, Wick M, Dessev C, Nemenoff RA. Induction of cPLA2 in lung epithelial cells and non-small cell lung cancer is mediated by Sp1 and c-Jun. J. Biol. Chem. 2001;276:42737–42743. doi: 10.1074/jbc.M107773200. [DOI] [PubMed] [Google Scholar]
- 24.Van Putten V, Refaat Z, Dessev C, Blaine S, Wick M, Butterfield L, Han S-Y, Heasley LE, Nemenoff RA. Induction of cytosolic phospholipase A2 by oncogenic Ras is mediated through the JNK and ERK pathways in rat epithelial cells. J. Biol. Chem. 2001;276:1226–1232. doi: 10.1074/jbc.M003581200. [DOI] [PubMed] [Google Scholar]
- 25.Wick MJ, Blaine S, Van Putten V, Saavedra M, Nemenoff RA. Lung Kruppel-like factor (LKLF) is a transcriptional activator of the cytosolic phospholipase A2 alpha promoter. Biochem. J. 2005;387:239–246. doi: 10.1042/BJ20041458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen BK, Chang WC. Functional interaction between c-Jun and promoter factor Sp1 in epidermal growth factor-induced gene expression of human 12(S)-lipoxygenase. Proc. Natl Acad. Sci. USA. 2000;97:10406–10411. doi: 10.1073/pnas.180321497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang YN, Chang WC. Induction of disease-associated keratin 16 gene expression by epidermal growth factor is regulated through cooperation of transcription factors Sp1 and c-Jun. J. Biol. Chem. 2003;278:45848–45857. doi: 10.1074/jbc.M302630200. [DOI] [PubMed] [Google Scholar]
- 28.Kardassis D, Papakosta P, Pardali K, Moustakas A. c-Jun transactivates the promoter of the human p21WAF1/Cip1 gene by acting as a superactivator of the ubiquitous transcription factor Sp1. J. Biol. Chem. 1999;274:29572–29581. doi: 10.1074/jbc.274.41.29572. [DOI] [PubMed] [Google Scholar]
- 29.Melnikova IN, Gardner PD. The signal transduction pathway underlying ion channel gene regulation by Sp1-c-Jun interactions. J. Biol. Chem. 2001;276:19040–19045. doi: 10.1074/jbc.M010735200. [DOI] [PubMed] [Google Scholar]
- 30.Srivastava M, Pollard HB. Molecular dissection of nucleolin's role in growth and cell proliferation: new insights. FASEB J. 1999;13:1911–1922. [PubMed] [Google Scholar]
- 31.Hanakahi LA, Dempsey LA, Li M-J, Maizels N. Nucleolin is one component of the B cell-specific transcription factor and switch region binding protein, LR1. Proc. Natl Acad. Sci. USA. 1997;94:3605–3610. doi: 10.1073/pnas.94.8.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen BK, Kung HC, Tsai TY, Chang WC. Essential role of mitogen-activated protein kinase pathway and c-Jun induction in epidermal growth factor-induced gene expression of human 12-lipoxygenase. Mol. Pharmacol. 2000;57:153–161. [PubMed] [Google Scholar]
- 33.Liu YW, Arakawa T, Yamamoto S, Chang WC. Transcriptional activation of human 12-lipoxygenase gene promoter is mediated through Sp1 consensus sites in A431 cells. Biochem. J. 1997;324:133–140. doi: 10.1042/bj3240133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen LC, Chen BK, Chang W-C. Activating protein 1-mediated cyclooxygenase-2 expression is independent of N-terminal phosphorylation of c-Jun. Mol. Pharmacol. 2005;67:2057–2069. doi: 10.1124/mol.104.010900. [DOI] [PubMed] [Google Scholar]
- 35.Zhu Y, Saunders MA, Yeh H, Deng W-G, Wu KK. Dynamic regulation of cyclooxygenase-2 promoter activity by isoforms of CCAAT/enhancer-binding proteins. J. Biol. Chem. 2002;277:6923–6928. doi: 10.1074/jbc.M108075200. [DOI] [PubMed] [Google Scholar]
- 36.Saccani S, Pantano S, Natoli G. Two waves of nuclear factor kappaB recruitment to target promoters. J. Exp. Med. 2001;193:1351–1360. doi: 10.1084/jem.193.12.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrews NC, Faller DV. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tay A, Maxwell P, Li Z-G, Goldberg H, Skorecki K. Cytosolic phospholipase A2 gene expression in rat mesangial cells is regulated post-transcriptionally. Biochem. J. 1994;304:417–422. doi: 10.1042/bj3040417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghisolfi-Nieto L, Joseph G, Puvion-Dutilleul F, Amalric F, Bouvet P. Nucleolin is a sequence-specific RNA-binding protein: characterization of targets on pre-ribosomal RNA. J. Mol. Biol. 1996;260:34–53. doi: 10.1006/jmbi.1996.0380. [DOI] [PubMed] [Google Scholar]
- 40.Hansen TVO, Bundgaard JR, Nielsen FC, Rehfeld JF. Composite action of three GC/GT boxes in the proximal promoter region is important for gastrin gene transcription. Mol. Cell. Endocrinol. 1999;155:1–8. doi: 10.1016/s0303-7207(99)00120-3. [DOI] [PubMed] [Google Scholar]
- 41.Meyer AM, Dwyer-Nield LD, Hurteau GJ, Keith RL, O’Leary E, You M, Bonventre JV, Nemenoff RA, Malkinson AM. Decreased lung tumorigenesis in mice genetically deficient in cytosolic phospholipase A2. Carcinogenesis. 2004;25:1517–1524. doi: 10.1093/carcin/bgh150. [DOI] [PubMed] [Google Scholar]
- 42.Nakagawa M, Oliva JL, Kothapalli D, Fournier A, Assoian RK, Kazanietz MG. Phorbol ester-induced G1 phase arrest selectively mediated by protein kinase C-delta-dependent induction of p21. J. Biol. Chem. 2005;280:33926–33934. doi: 10.1074/jbc.M505748200. [DOI] [PubMed] [Google Scholar]
- 43.Hemstrom TH, Joseph B, Schulte G, Lewensohn R, Zhivotovsky B. PKC 412 sensitizes U1810 non-small cell lung cancer cells to DNA damage. Exp. Cell Res. 2005;305:200–213. doi: 10.1016/j.yexcr.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 44.Masumi A, Fukazawa H, Shimazu T, Yoshida M, Ozato K, Komuro K, Yamaguchi K. Nucleolin is involved in interferon regulatory factor-2-dependent transcriptional activation. Oncogene. 2006;25:5113–5124. doi: 10.1038/sj.onc.1209522. [DOI] [PubMed] [Google Scholar]
- 45.Grinstein E, Wernet P, Snijders PJF, Rosl F, Weinert I, Jia W, Kraft R, Schewe C, Schwabe M, et al. Nucleolin as activator of human papillomavirus type 18 oncogene transcription in cervical cancer. J. Exp. Med. 2002;196:1067–1078. doi: 10.1084/jem.20011053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ying G-G, Proost P, van Damme J, Bruschi M, Introna M, Golay J. Nucleolin, a novel partner for the Myb transcription factor family that regulates their activity. J. Biol. Chem. 2000;275:4152–4158. doi: 10.1074/jbc.275.6.4152. [DOI] [PubMed] [Google Scholar]
- 47.Grinstein E, Shan Y, Karawajew L, Snijders PJF, Meijer CJLM, Royer H-D, Wernet P. Cell cycle-controlled interaction of nucleolin with the retinoblastoma protein and cancerous cell transformation. J. Biol. Chem. 2006;281:22223–22235. doi: 10.1074/jbc.M513335200. [DOI] [PubMed] [Google Scholar]
- 48.Saxena A, Rorie CJ, Dimitrova D, Daniely Y, Borowiec JA. Nucleolin inhibits Hdm2 by multiple pathways leading to p53 stabilization. Oncogene. 2006;25:7274–7288. doi: 10.1038/sj.onc.1209714. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Bhatia D, Xia H, Castranova V, Shi X, Chen F. Nucleolin links to arsenic-induced stabilization of GADD45α mRNA. Nucleic Acids Res. 2006;34:485–495. doi: 10.1093/nar/gkj459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dickinson LA, Kohwi-Shigematsu T. Nucleolin is a matrix attachment region DNA-binding protein that specifically recognizes a region with high base-unpairing potential. Mol. Cell. Biol. 1995;15:456–465. doi: 10.1128/mcb.15.1.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Embree-Ku M, Gruppuso PA. The role of nuclear factor kB in late-gestation liver development in the rat. Hepatology. 2005;42:326–334. doi: 10.1002/hep.20796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huddleson JP, Ahmad N, Lingrel JB. Up-regulation of the KLF2 transcription factor by fluid shear stress requires nucleolin. J. Biol. Chem. 2006;281:15121–15128. doi: 10.1074/jbc.M513406200. [DOI] [PubMed] [Google Scholar]
- 53.Daniely Y, Dimitrova DD, Borowiec JA. Stress-dependent nucleolin mobilization mediated by p53-nucleolin complex formation. Mol. Cell. Biol. 2002;22:6014–6022. doi: 10.1128/MCB.22.16.6014-6022.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]