Abstract
Current efforts to recover the Neandertal and mammoth genomes by 454 DNA sequencing demonstrate the sensitivity of this technology. However, routine 454 sequencing applications still require microgram quantities of initial material. This is due to a lack of effective methods for quantifying 454 sequencing libraries, necessitating expensive and labour-intensive procedures when sequencing ancient DNA and other poor DNA samples. Here we report a 454 sequencing library quantification method based on quantitative PCR that effectively eliminates these limitations. We estimated both the molecule numbers and the fragment size distributions in sequencing libraries derived from Neandertal DNA extracts, SAGE ditags and bonobo genomic DNA, obtaining optimal sequencing yields without performing any titration runs. Using this method, 454 sequencing can routinely be performed from as little as 50 pg of initial material without titration runs, thereby drastically reducing costs while increasing the scope of sample throughput and protocol development on the 454 platform. The method should also apply to Illumina/Solexa and ABI/SOLiD sequencing, and should therefore help to widen the accessibility of all three platforms.
INTRODUCTION
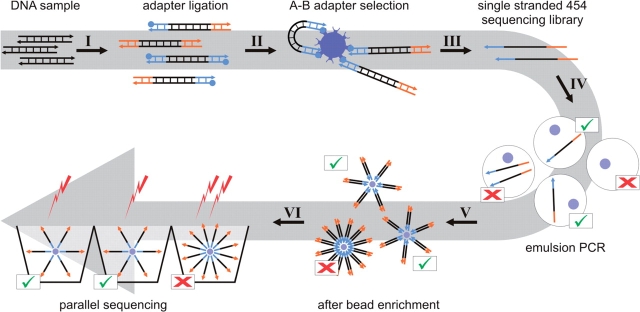
Due to its extremely high throughput, 454 DNA sequencing (1) is currently replacing Sanger capillary sequencing in a wide range of applications (2–5). In theory, any DNA sample can be sequenced with this technology, as long as it can be converted into a 454 sequencing library. A key step in 454 sequencing is emulsion PCR [emPCR, (6)], where numerous DNA templates are clonally amplified in a single reaction vessel containing millions of water-in-oil droplets. In order to obtain universal priming sites for emPCR, two universal oligonucleotide adapters, A and B, are added to the ends of DNA templates during 454 library preparation (see Figure 1 for an overview scheme of 454 sequencing). For complex mixtures of molecules, such as shotgun libraries or pooled PCR products, adapter addition is achieved by ligation. Typically, both adapters are ligated at the same time, and only templates carrying two different adapters are subsequently isolated as a single-stranded sequencing library.
Figure 1.
Overview scheme of 454 sequencing. Double-stranded sequencing templates are blunt end repaired, and two universal adapters, A and B, are ligated to their ends. The B-adapter carries a 5′ biotin (I). Streptavidin beads are used to isolate only molecules carrying an A and a B adapter (II). The single-stranded sequencing library is melted from the beads through alkaline treatment (III). A PCR reaction mix containing 600 000 oligonucleotide-coated sepharose beads and an appropriate number of library molecules is emulsified to produce physically separated droplets as reaction vessels (IV). After amplification, the emulsion is broken while the PCR products remain attached to the beads. Since most beads remain empty in emPCR, amplified beads are isolated through a bead enrichment procedure (V). A total of 50 000 beads are required for loading onto the wells of a 16th 454 FLX picotitre plate region, and the sequencing reaction is performed by flowing nucleotides over the plate and measuring light emissions. Beads carrying multiple amplicons produce mixed signals, which are recognized and filtered out by the run-processing software.
Emulsion PCR involves mixing single-stranded library templates with DNA-capturing sepharose beads in an emulsion containing millions of droplets. Successful sequencing requires single-template-single-bead droplets, allowing clonal amplification of each template onto a bead. Therefore, the success of emPCR and subsequent sequencing reactions is critically dependent on the relative number of template molecules and DNA capture beads (the ‘copy per bead’ ratio) added to the emulsion. Adding either too few or too many template molecules results in poor sequence yields, as beads remain either empty or produce ‘mixed’ signals. Obtaining an optimal copy per bead ratio relies on accurate quantification of the single-stranded 454 sequencing library. Although less than 1 pg of library is eventually used for emPCR, correspondingly low library concentrations are several orders of magnitude below the detection limits of capillary gel electrophoresis (7) and Ribogreen quantification (8), which are the recommended quantification methods in the current 454 library preparation kit (Roche). Since losses occur in 454 library preparation, several micrograms of original DNA sample are usually required to produce nanograms of library suitable for quantification. Even when starting from high amounts of DNA, performing titration runs to obtain an optimal copy per bead ratio is recommended.
Despite these limitations, sequences have been obtained directly from ancient samples (9–11). However, the lack of an appropriate library quantification method necessitates both significant efforts and substantial costs to retrieve sequences from such material. In order to find an appropriate concentration for sequencing, extensive titration sequencing from emPCRs with serial dilutions of library must be performed without any a priori knowledge of whether a given library contains sufficient molecules for sequencing (10,11). For example, when sequencing ancient material, many samples must be screened for their endogenous and environmental DNA content, to identify those suitable for a genome sequencing project. In the majority of cases sequencing libraries exhibit DNA concentrations below the detection limits of current quantification methods. A typical example for results we obtained using the titration strategy is given in Table 1. The same problem persists with all other potential applications where only nanogram or picogram amounts of initial material are available, e.g. cDNA libraries derived from a few cells through laser-mediated microdissection (12), or genomic DNA from uncultivated bacteria (13).
Table 1.
Typical result of a sequencing run performed from 454 libraries with low concentrations derived from Neandertal ancient DNA extracts
| Sample | 454 sequencing library dilution | Enriched beads | Filter passed sequences |
|---|---|---|---|
| NT1 | 1:2 | 11 700 | 2448 |
| 1:150 | 8640 | 1180 | |
| 1:450 | 13 860 | 314 | |
| NT2 | 1:2 | 2430 | 267 |
| 1:150 | 1440 | 52 | |
| 1:450 | 1980 | 0 | |
| NT3 | 1:2 | 344 700 | 0 |
| 1:150 | 324 900 | 1247 | |
| 1:450 | 106 740 | 6106 | |
| NT4 | 1:2 | 263 340 | 3805 |
| 1:150 | 29 520 | 3312 | |
| 1:450 | 7110 | 358 | |
| NT5 | 1:2 | 1980 | 36 |
| 1:150 | 1440 | 0 | |
| NT6 | 1:2 | 3420 | 45 |
| 1:150 | 2520 | 4 |
As no copy number estimates could be obtained for the libraries, serial dilutions were used for emulsion PCRs with 600 000 beads each. After bead enrichment, 24 000 or, where applicable, all beads were loaded onto 16th regions of the GS20 picotitre plate. Displayed is the number of sequences obtained after filtering by the run-processing software and removal of artefact sequences. A successful GS20 run usually yields at least 6000 sequences per region.
We have developed a 454 sequencing library quantifica-tion method that eliminates these problems. Our method utilizes the emulsion PCR priming sites for performing a quantitative PCR (qPCR) with SYBR Green dye. As we demonstrate with 15 sequencing libraries constructed from different sources, including ancient DNA extracts from Neandertal bones, SAGE ditags and modern genomic DNA, our method yields accurate estimates for molecular copy numbers and fragment size distributions of 454 libraries without any inherent upper or lower detection limit. Moreover, the precision of our method eliminates the need to perform titration runs, which have previously been a general requirement for 454 sequencing, thereby drastically reducing sequencing costs.
MATERIALS AND METHODS
Constructing and sequencing single-stranded 454 libraries
Several types of material were used for the construction of single-stranded 454 sequencing libraries as part of ongoing research projects. Ancient DNA from twelve Neandertal bones was isolated as described previously (14,15). A pool of PCR products was created by mixing 240 non-purified PCR products in equal volumes. The products were between 130 and 200 bp in size and derived from mammoth DNA using a two-step multiplex approach as described previously (16,17). The PCR product pool was purified using the AMPure PCR purification kit (Agencourt) in order to remove primers and short artefacts. Nebulized bonobo genomic DNA was kindly provided by Anne Fischer. A SAGE ditag library was produced from 10 μg of total RNA from human prefrontal cortex using the I-SAGE long kit (Invitrogen) according to the manufacturer's instructions, but stopping after the first amplification step.
Single-stranded 454 sequencing libraries were constructed from between 1 and 20 μl of initial material using the 454 library preparation kit (Roche) according to the manufacturer's instructions, but starting at the blunt end repair step, since nebulization was not required for these samples. The 15 μl of sequencing library obtained from each sample were mixed 1:1 with TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 8.0) for stabilization. Libraries were quantified immediately and stored at −20°C until further use. EmPCR and sequencing were performed according to the standard GS FLX procedure, using 16th regions of the full GS FLX picotitre plate. A total of 50 000 enriched beads were loaded onto each 16th region unless fewer enriched beads were available for a sample, in which case all beads were loaded onto a region.
Constructing a quantification standard
A quantification standard carrying emPCR priming sites with flanking sequences for protection against exonucleolytic degradation was prepared according to the following protocol. Using the emPCR primers (forward primer 5′-CCATCTCATCCCTGCGTGTC-3′; reverse primer 5′-CCTATCCCCTGTGTGCCTTG-3′) a whole library amplicon was generated from a 454 sequencing library produced from nebulized genomic bonobo DNA. One microlitre of a 1:1000 library dilution was used as template for a PCR containing 1× PCR buffer II, 2.5 mM MgCl2, 1.25 U AmpliTaq Gold (all Applied Biosystems), 250 μM each dNTP and 200 nM of each primer. Cycling conditions were comprised of an activation step lasting 10 min at 95°C, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s and elongation at 72°C for 30 s, with a final extension step at 72°C for 10 min. The resulting whole library amplicon was cloned using the TOPO TA cloning kit (Invitrogen). Several colonies were transferred to 50 μl of water and boiled for 10 min at 95°C. One microlitre of lysate from each colony was used as template for colony PCR, using M13 general primers and the PCR conditions described above, except for a lowered annealing temperature of 55°C. PCR products were visualized on an agarose gel for size determination. One colony PCR product with a length of 450 bp was purified using the MinElute PCR purification kit (Qiagen), quantified on a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies) and then used as the qPCR standard with known mass concentration. The molecular concentration (1.36 × 1011 molecules per microlitre) was calculated from the product size and the mass concentration (33 ng/μl). A multiplier of two is included to account for the double-stranded nature of the standard, as opposed to the single-stranded sequencing libraries.
Quantifying 454 sequencing libraries using qPCR
For library quantification, a 10-fold dilution series of the standard was used to obtain a standard curve, ranging from a 1:100 to a 1:109-fold dilution of the standard. Real-time PCRs were performed using a Stratagene MxPro 3005P qPCR System with SYBR Green dye. Duplicate measurements were carried out in 25 μl reactions containing 1 μl template and 1× PCR buffer II, 2.5 mM MgCl2, 1.25 U AmpliTaq Gold (all Applied Biosystems), 1× SYBR Green I, 15 μM ROX reference dye, 3% DMSO, 8% Glycerol (all from the Brilliant SYBR Green QPCR Core Reagent Kit, Stratagene), 250 μM each dNTP, 0.4 mg/ml BSA (Sigma) and 200 nM of each emPCR primer. Cycling was initiated at 95°C for 10 min, followed by 45 cycles of 94°C for 30 s, 60°C for 30 s and 72°C for 45 s. Fluorescence was measured at the end of the elongation phase in each cycle. Amplicons were visualized on 2% agarose gels together with a size marker to infer the average fragment size within the sequencing libraries. The library DNA concentrations were inferred by comparing the measurements to the standard curve, and then corrected for size differences according to the following formula: actual library concentration [molecules/μl] = inferred concentration [molecules/μl] × 450 (length of the standard)/average fragment length in the library.
RESULTS
Sequencing library quantification and concentration adjustment
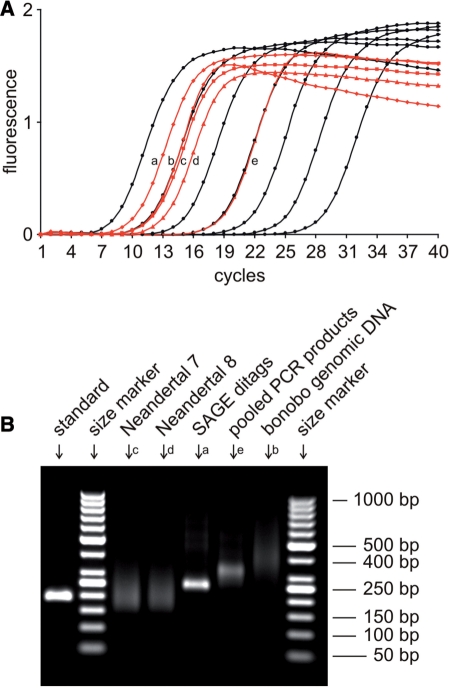
We generated 15 single-stranded 454 sequencing libraries from 12 Neandertal ancient DNA extracts, one SAGE ditag library, one pool of PCR products and one nebulized bonobo genomic DNA sample. The samples used to make these libraries contained much less DNA than the microgram amounts recommended by Roche (Table 2). Except for the bonobo genomic DNA, the sequencing libraries could not be detected by gel electrophoresis on an RNA 6000 pico chip (Agilent, data not shown). However, the libraries could be quantified using qPCR. The slopes of the amplification plots did not noticeably differ between the standard and the sequencing libraries, indicating that the complex mixture of templates in the sequencing libraries does not affect PCR efficiency (Figure 2A). Since SYBR Green fluorescence signals are proportional to the mass of template DNA, qPCR amplification products were visualized on agarose gels together with a size marker to estimate fragment size distributions (Figure 2B). The mean fragment size was subsequently corrected for in the calculation of absolute molecular concentrations for each library (Table 2).
Table 2.
454 library quantification and sequencing results obtained from fifteen samples
| Sample | Library construction | 454 sequencing | ||||||
|---|---|---|---|---|---|---|---|---|
| ID | Initial material (ng) | Mean fragment size (bp) | Concentration (molec./μl) | Recovery (%) | Copies per bead in emPCR | Enriched beads | Mixed sequences (%) | Filter passed sequences |
| Neandertal 1 | n/a | 200 | 2.00 × 106 | n/a | 2.17 | 144 275 | 8.7 | 15 541 |
| Neandertal 2 | n/a | 200 | 1.75 × 107 | n/a | 2.25 | 98 310 | 9.5 | 16 972 |
| Neandertal 3 | n/a | 200 | 1.88 × 107 | n/a | 2.08 | 109 330 | 8.2 | 16 447 |
| Neandertal 4 | n/a | 200 | 1.82 × 107 | n/a | 2.17 | 78 010 | 9.2 | 16 773 |
| Neandertal 5 | n/a | 200 | 5.48 × 106 | n/a | 2.19 | 101 790 | 9.4 | 17 323 |
| Neandertal 6 | n/a | 200 | 7.40 × 107 | n/a | 2.16 | 58 580 | 11.3 | 13 590 |
| Neandertal 7 | n/a | 200 | 9.74 × 107 | n/a | 2.14 | 55 680 | 10.8 | 19 639 |
| Neandertal 8 | n/a | 200 | 4.88 × 107 | n/a | 2.20 | 62 640 | 9.5 | 14 062 |
| Neandertal 9 | n/a | 200 | 3.99 × 107 | n/a | 2.15 | 62 640 | 11.5 | 15 582 |
| Neandertal 10 | n/a | 200 | 1.45 × 108 | n/a | 1.59 | 26 100 | 7.8 | 17 030 |
| Neandertal 11 | n/a | 200 | 7.85 × 107 | n/a | 2.33 | 54 230 | 8.9 | 14 592 |
| Neandertal 12 | n/a | 200 | 4.31 × 107 | n/a | 2.25 | 60 320 | 9.2 | 16 544 |
| SAGE ditags | 32 | 250 | 9.83 × 107 | 0.5 | 1.64 | 76 270 | 3.4 | 21 429 |
| Amplicons | 85 | 320 | 1.72 × 108 | 0.4 | 1.35 | 42 000 | 11.2 | 20 716 |
| Bonobo | 520 | 450 | 1.50 × 109 | 1.7 | 2.33 | 87 800 | 20.3 | 12 232 |
No attempts were made to quantify the Neandertal DNA extracts prior to library preparation in order to preserve precious sample. Library concentrations were inferred by qPCR taking into account the mean fragment size of each library. The percentages of molecules recovered after library preparation are denoted. A narrow range of copy per bead ratios was used in emPCR. The success of emPCR is determined both by the number of enriched beads obtained (50 000 beads required) and the ratio of mixed sequences. Due to the highly fragmented nature of ancient DNA, the run-processing parameters were changed for the Neandertal samples in order to accept shorter reads (see Supplementary Data). Subsequently, emPCR artefact sequences were removed as described in Ref. (11). The other samples were processed with the default run-processing parameters. All plate regions yielded more than 12 000 sequences after filtering.
Figure 2.
Five sequencing libraries were re-quantified in parallel to show the performance of qPCR quantification for libraries derived from different types of initial material. (A) Amplification plots of the standard and the libraries are drawn in black and red, respectively, with duplicates being treated collectively. The bonobo genomic DNA library was measured in a 1:30 dilution to obtain a signal within the range of the standard curve. The library from pooled PCR products was measured in the working dilution (1:100) used for emulsion PCR. (B) qPCR amplicons were size fractionated by agarose gel electrophoresis and visualized by ethidium bromide staining in order to estimate the mean fragment size for each library.
The copy per bead ratio, i.e. the number of library molecules divided by the number of capture beads in emPCR, is the major determinant for sequencing success in the 454 process. EmPCRs with a high copy per bead ratio yield a high proportion of ‘mixed’ sequences, where multiple templates are amplified in the presence of a single bead, leading to nonsense sequencing signals and reducing overall sequence yield. On the other hand, emPCRs with a low copy per bead ratio yield a low proportion of mixed sequences, but also fewer amplified beads for sequencing, either reducing sequence yield or necessitating additional expensive emPCR runs to produce enough beads. Roche recommend titration runs with 1, 4, 16 and 64 copies per bead for each single-stranded sequencing library to determine the copy per bead ratio that maximizes sequence yield. In our previous experience with high concentration, electrophoresis-quantified 454 libraries, copy per bead ratios between one and two usually yielded acceptable sequencing results, if the libraries were freshly prepared. Thus, after quantification of our 15 new libraries by qPCR, we chose copy per bead ratios in this range for emPCR.
Emulsion PCR and 454 sequencing results
We performed emPCRs with 600 000 beads for each of the fifteen 454 libraries. With only two exceptions, emPCRs yielded at least 50 000 beads after enrichment (Table 2), which were then loaded onto one 16th region of a GS FLX picotitre plate and sequenced. The 454 run-processing software provides detailed filtering information for each picotitre plate region (see Supplementary Table). The adequate bead enrichment counts, together with the low percentages of mixed sequences [which were consistently around 10% or lower (Table 2)], demonstrate that the copy per bead ratios used in emPCR were optimal or close to optimal. Crucially, more than 12 000 sequences were obtained for all samples, as can typically be expected from successful runs when using 16th picotitre plate regions. The lowest sequence yield was produced by the bonobo genomic library, which showed a relatively high number of mixed sequences, indicating a slight underestimation of its molecular concentration by qPCR. Since this library contains a substantial fraction of long molecules, one possible explanation may be that the different upper size limit in amplification between emPCR and qPCR led to a drop out of long molecules during quantification. Additional optimizations, e.g. by extending the elongation phase in qPCR, may help to improve the accuracy of quantification for high molecular weight libraries. Nevertheless, the bonobo sample still produced over 12 000 sequences.
Minimal material requirements for 454 sequencing
In order to define the minimal material requirements for 454 sequencing in light of our quantification method, we first estimated the efficiency of the 454 library preparation protocol. For the pooled PCR products, ditag and bonobo libraries, which were constructed from a known amount of initial material, the yield could be calculated by dividing the number of molecules contained in the libraries by the number of initial molecules (Table 2). We observed yields between 0.4% and 1.7%, indicating considerable variation in the efficiency of 454 library preparations.
A single emulsion PCR with a copy per bead ratio of two requires 1.2 × 106 library molecules. Dividing the molecular content of a library by this number yields the number of emPCRs that can be performed from the given amount of initial material. Thus, for a single emulsion PCR, 37 pg of the pool of PCR products, 20 pg of SAGE ditag DNA and 14 pg of bonobo genomic DNA would theoretically have been required for library preparation. Therefore, less than 50 pg of an initial DNA sample should usually be sufficient to produce at least 12 000 sequences on one 16th region of the 454 picotitre plate.
DISCUSSION
Advantages of qPCR quantification for 454 sequencing
The 454 sequencing technology is theoretically well suited for large-scale sequencing from low amounts of biological samples. In practice, however, routine sequencing was previously only possible from microgram amounts of samples due to insufficiently sensitive library quantification methods. Our simple and straightforward quantitative PCR assay complements the high sensitivity of the 454 sequencing technology by shifting the material requirements for routine sequencing almost a million-fold, from micrograms to picograms. Unlike earlier studies (10,11), it allows for retrieval of optimal numbers of sequences from ancient fossils without expensive and time-consuming titration runs. To find the optimal copy per bead ratio of six Neandertal libraries using the titration approach summarized in Table 1, sixteen separate emulsion PCRs and one full 454 sequencing run were performed. The same libraries could have been quantified in a few hours using qPCR and sequenced on only six of the sixteen lanes. Thus, optimizing the copy per bead ratio before emulsion PCR considerably reduces costs and increases output. Furthermore, without an inherent detection limit, the quantification method requires only trace amounts of sequencing library, preserving precious or valuable samples.
Our data indicate that accurate copy number estimates can be obtained for libraries derived from a variety of sources and different amounts of material. Although no comparative data are available, we suggest that our method will also be superior to direct nucleic acid quantification in cases where highly concentrated sequencing libraries are available. By using the same priming sites, the quantitative PCR assay simulates emulsion PCR. Library degradation occurring internally or at the priming sites excludes library molecules from amplification both during quantification and emulsion PCR. Direct nucleic acid quantification methods cannot make this distinction. This may explain the optimal sequence yields we obtained for all our libraries, despite omitting small-scale titration runs as generally recommended by Roche. By providing optimal sequence numbers without prior titration runs, our library quantification method greatly improves the economics of 454 sequencing, particularly in cases where only single or partial runs need to be performed from one sequencing library. Thus, it greatly increases the feasibility of small-scale 454 sequencing projects, such as screening metagenomic samples, sequencing pooled PCR products or sequencing cDNA libraries from a few cells without the need for extensive pre-amplification, thereby retaining quantitative information.
qPCR quantification with other high-throughput sequencing platforms
We propose that sequencing library quantification by qPCR will also be applicable to Illumina/Solexa [www.illumina.com, (18)] and ABI/SOLiD sequencing (www.appliedbiosystems.com), two other massively high-throughput sequencing platforms. Although the sequencing technology is very different among these platforms, all three systems use DNA libraries consisting of templates with two different adapters attached. These libraries must be accurately quantified before picogram or low nanogram quantities are added to emulsion PCR (Roche/454, ABI/SOLiD) or solid-phase bridge amplification (Illumina/Solexa). Therefore, provided that the relevant adapter sequences are made available, our qPCR method should work with Solexa and SOLiD libraries as well as with 454 libraries. If this is the case, then it would also simplify the interchange of library preparation methods between the different technologies. This would be useful for evaluating potential sampling biases associated with the respective methods.
Our method's ability to accurately quantify sequencing libraries produced from nanogram or picogram quantities of initial material will increase the range of sample types and sample sizes available to these sequencing technologies, bridging the current gap between the enormous power of these platforms and the practical and economic limits of sample preparation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors would like to acknowledge Esther Lizano Gonzalez and Anne Fischer for providing SAGE ditag and bonobo genomic DNA. We thank Richard E. Green, Nadin Rohland and Bernd Timmermann for helpful discussions, Christine B. Green for comments on the manuscript, Knut Finstermeier for help with the figures, The Croatian Academy of Sciences and Arts and the Berlin-Brandenburg Academy of Sciences for collaboration on the Neandertal project and the Max Planck Society for financial support. Funding to pay the Open Access publication charges for this article was provided by the Max Planck Society.
Conflict of interest statement. None declared.
REFERENCES
- 1.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore MJ, Dhingra A, Soltis PS, Shaw R, Farmerie WG, Folta KM, Soltis DE. Rapid and accurate pyrosequencing of angiosperm plastid genomes. BMC Plant Biol. 2006;6:17. doi: 10.1186/1471-2229-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldberg SM, Johnson J, Busam D, Feldblyum T, Ferriera S, Friedman R, Halpern A, Khouri H, Kravitz SA, et al. A Sanger/pyrosequencing hybrid approach for the generation of high-quality draft assemblies of marine microbial genomes. Proc. Natl Acad. Sci. USA. 2006;103:11240–11245. doi: 10.1073/pnas.0604351103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng P, Tan JJ, Ooi HS, Lee YL, Chiu KP, Fullwood MJ, Srinivasan KG, Perbost C, Du L, et al. Multiplex sequencing of paired-end ditags (MS-PET): a strategy for the ultra-high-throughput analysis of transcriptomes and genomes. Nucleic Acids Res. 2006;34:e84. doi: 10.1093/nar/gkl444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer M, Stenzel U, Myles S, Prufer K, Hofreiter M. Targeted high-throughput sequencing of tagged nucleic acid samples. Nucleic Acids Res. 2007;35:e97. doi: 10.1093/nar/gkm566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tawfik DS, Griffiths AD. Man-made cell-like compartments for molecular evolution. Nat. Biotechnol. 1998;16:652–656. doi: 10.1038/nbt0798-652. [DOI] [PubMed] [Google Scholar]
- 7.Ricicova M, Palkova Z. Comparative analyses of Saccharomyces cerevisiae RNAs using Agilent RNA 6000 Nano Assay and agarose gel electrophoresis. FEMS Yeast Res. 2003;4:119–122. doi: 10.1016/S1567-1356(03)00145-4. [DOI] [PubMed] [Google Scholar]
- 8.Jones LJ, Yue ST, Cheung CY, Singer VL. RNA quantitation by fluorescence-based solution assay: RiboGreen reagent characterization. Anal. Biochem. 1998;265:368–374. doi: 10.1006/abio.1998.2914. [DOI] [PubMed] [Google Scholar]
- 9.Poinar HN, Schwarz C, Qi J, Shapiro B, Macphee RD, Buigues B, Tikhonov A, Huson DH, Tomsho LP, et al. Metagenomics to paleogenomics: large-scale sequencing of mammoth DNA. Science. 2006;311:392–394. doi: 10.1126/science.1123360. [DOI] [PubMed] [Google Scholar]
- 10.Stiller M, Green RE, Ronan M, Simons JF, Du L, He W, Egholm M, Rothberg JM, Keates SG, et al. Patterns of nucleotide misincorporations during enzymatic amplification and direct large-scale sequencing of ancient DNA. Proc. Natl Acad. Sci. USA. 2006;103:13578–13584. doi: 10.1073/pnas.0605327103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green RE, Krause J, Ptak SE, Briggs AW, Ronan MT, Simons JF, Du L, Egholm M, Rothberg JM, et al. Analysis of one million base pairs of Neanderthal DNA. Nature. 2006;444:330–336. doi: 10.1038/nature05336. [DOI] [PubMed] [Google Scholar]
- 12.Schutze K, Lahr G. Identification of expressed genes by laser-mediated manipulation of single cells. Nat. Biotechnol. 1998;16:737–742. doi: 10.1038/nbt0898-737. [DOI] [PubMed] [Google Scholar]
- 13.Rappe MS, Giovannoni SJ. The uncultured microbial majority. Annu. Rev. Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 14.Rohland N, Hofreiter M. Ancient DNA extraction from bones and teeth. Nat. Protoc. 2007;2:1756–1762. doi: 10.1038/nprot.2007.247. [DOI] [PubMed] [Google Scholar]
- 15.Rohland N, Hofreiter M. Comparison and optimization of ancient DNA extraction. Biotechniques. 2007;42:343–352. doi: 10.2144/000112383. [DOI] [PubMed] [Google Scholar]
- 16.Krause J, Dear PH, Pollack JL, Slatkin M, Spriggs H, Barnes I, Lister AM, Ebersberger I, Paabo S, et al. Multiplex amplification of the mammoth mitochondrial genome and the evolution of Elephantidae. Nature. 2006;439:724–727. doi: 10.1038/nature04432. [DOI] [PubMed] [Google Scholar]
- 17.Rompler H, Dear PH, Krause J, Meyer M, Rohland N, Schoneberg T, Spriggs H, Stiller M, Hofreiter M. Multiplex amplification of ancient DNA. Nat. Protoc. 2006;1:720–728. doi: 10.1038/nprot.2006.84. [DOI] [PubMed] [Google Scholar]
- 18.Bentley DR. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 2006;16:545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.