Abstract
Gene expression profiling has identified several potentially useful gene signatures for predicting outcome or for selecting targeted therapy. However, these signatures have been developed in fresh or frozen tissue, and there is a need to apply them to routinely processed samples. Here, we demonstrate the feasibility of a potentially high-throughput methodology combining automated in situ hybridization with quantum dot-labeled oligonucleotide probes followed by spectral imaging for the detection and subsequent deconvolution of multiple signals. This method is semiautomated and quantitative and can be applied to formalin-fixed, paraffin-embedded tissues. We have combined dual in situ hybridization with immunohistochemistry, enabling simultaneous measurement of gene expression and cell lineage determination. The technique achieves levels of sensitivity and specificity sufficient for the potential application of known expression signatures to biopsy specimens in a semiquantitative way, and the semiautomated nature of the method enables application to high-throughput studies.
Gene expression profiling experiments have identified several gene signatures as markers either of tumor categories with distinct behaviors or predictive of disease progression and response to therapy.1,2 Application of such signatures to individual patients in a clinical setting holds great potential for improving diagnosis and prognosis and for providing guidance for tailored molecular therapy.1,2,3,4 The generation and use of such signatures in routine clinical practice is presently limited by the need for relatively large amounts of high-quality frozen material for microarray gene expression profiling.2,5 In contrast, nearly all clinical material is routinely formalin-fixed and processed to paraffin,5 a practice that is likely to continue for the foreseeable future, especially outside large academic centers. In addition, gene signatures have largely been derived from tissue homogenates,2 although recent studies have demonstrated the importance of localization of the genes that make up these signatures not only to the tumor cells but also to their neighbors, such as immune and stromal cells.6 There is therefore a need for a generic method for simultaneous visualization of multiple genes in formalin-fixed, paraffin-embedded (FFPE) tissue, ideally by an automated or semiautomated method to enable clinical use and application to high-throughput studies investigating spatial localization of microarray gene signatures.
Currently, gene expression in tissues is determined by immunohistochemistry (IHC) and in situ hybridization (ISH). However, the use of IHC for mapping expression of gene signatures in FFPE tissue is hampered because antibodies effective in FFPE sections are not commonly available for most of the markers composing gene signatures.7 In contrast, probes useful for RNA ISH can be easily constructed for any gene, and in theory, clinical use on FFPE samples could be a reliable method of in situ gene expression detection, although it is methodologically long and complex.8,9,10 We currently obviate the lengthy procedure times with the routine use of automated, high-throughput colorimetric RNA ISH.11
Fluorescence-based detection systems, which would be ideal for visualization of multiple probes, are hampered by low sensitivity (particularly for oligonucleotide probes),8,9 high autofluorescence of paraffin-embedded tissue,12,13 with low resultant signal-to-noise ratio, limited numbers of fluorophores, and overlapping spectra,14 making deconvolution of multiple signals challenging. Furthermore, fluorescence-based techniques are nonpermanent, fading with time,15,16,17 rendering them suboptimal for a clinical test for which a permanent record is best for clinical governance. Quantum dots (QDs) have recently been used for bioimaging by immunofluorescence17 for molecule17,18 and cell labeling,19,20,21 and, more recently, in human clinical material.22 These fluorescent semiconductor nanocrystals possess extremely high fluorescence efficiency and photostability, making them near-optimal for many fluorescent applications.15,16,17,18,19,21,22,23,24,25 In addition, whereas their excitation wavelength is constant, their emission wavelength is sharp, symmetrical, and tunable, dependent on diameter, with the potential for multicolor staining.24 These; properties suggest that QDs might be highly useful in application to clinical specimens.
Simultaneous detection of multiple fluorescent signals requires spectral deconvolution to resolve individual signals. Spectral imaging relies on generation of a complete optical profile for each pixel in the image field from which multiple spectral distributions can be reconstructed via a least-squares fitting linear unmixing approach.26 The spectral information in the acquired datasets can then be used to discriminate between autofluorescence and true fluorescent signal and between different fluorescent signals.19 Such methods could also be highly advantageous in a clinical setting but have yet to be explored and their utility tested in a wide range of tissues. We have demonstrated use of QD-based ISH in human specimens using manual techniques.
Here, we have combined high-throughput, automated ISH, QD-labeling of DNA and RNA probes and spectral imaging to produce a novel technique for ISH that can be used to apply gene expression signatures to FFPE tissue biopsies at diagnosis.
Materials and Methods
Tissues
Archival routinely processed (10%) formalin-fixed, paraffin-embedded human glioma tissue was obtained with Institutional Review Board approval from the archives of the Department of Pathology, Brigham and Women’s Hospital. Mouse lung was harvested at necropsy. Mice were perfused with 4% paraformaldehyde and fixed in the same fixative for 18 hours at 4°C before processing to paraffin blocks.
Cell Culture
LNCaP human prostate cancer cells were grown to 70% confluence in RPMI-1640 (StemCell Technologies, Vancouver, BC, Canada) supplemented with 10% fetal bovine serum (StemCell Technologies) and incubated at 37°C with 5% CO2. The cells were then washed three times in phosphate-buffered saline (PBS) and grown in charcoal-stripped serum for 24 hours before R1881 treatment and 48 hours before harvesting. The cells were treated with 10 nmol/L R1881 in 100% ethanol and harvested at 2, 9.5, and 24 hours. Total RNA was isolated for quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) experiments from one-half of the treated cells, whereas the rest were pelleted, formalin-fixed, and paraffin-embedded. Cultured mouse fibroblast cells, both wild type and cyclin E1-null,27 were a kind gift from Dr. Yan Geng and Peter Sicinski (Dana-Farber Cancer Institute).
Reagents
Oligonucleotide probes were high-performance liquid chromotography-purified and biotin-TEG-, digoxigenin (DIG)-, or thiol-modified at the 5′ end (Eurogentec, Seraing, Belgium). Amine-coated QDs (580 nm) (Evident Technologies, Troy, NY) were used for Q-ISH in human brain tissue. QD 605 and 655 streptavidin conjugates (Quantum Dot Corporation, Hayward, CA) were used for conjugation to oligonucleotide/riboprobes for Q-ISH (see below) and QD 655 streptavidin conjugate for IHC. QD 525 anti-DIG antibody conjugates (Quantum Dot Corporation) were used for DIG-labeled riboprobe ISH. Antibodies to mcc10 and CD34 (Abcam, Cambridge, MA) were used for IHC.
Probe Design
Two approaches were used. The probe for cyclin E1 (NM_001238) was designed using a sequence from ENSEMBL (http://www.ensembl.org). Generic sequences from the Illumina Oligator database (San Diego, CA) were used for mcc10 (NM_011681), mammalian achaete-scute homolog 1 (ASCL1) (NM_004316), NKX2-2 (NM_002509), and fatty acid synthase (FAS) (NM_000043). For all genes, 50-mer-long sequences with approximately 50% guanine cytosine content were selected from these areas and specificity checked using BLAST (http://www.ncbi.nlm.nih.gov/blast). Reverse complement sequences were then prepared as oligonucleotide probes (Table 1). RNA probes were transcribed in vitro from DNA templates flanked by phage RNA polymerase promoters, as previously described.11
Table 1.
cDNA Sequences of Probes Used in Study
| Name | Accession no. | cDNA oligonucleotide probe sequence |
|---|---|---|
| ASCL1 | NM_004316 | 5′-TTTCAGCTGTGCGTGTTAGAGGTGATGGGAGTTACTATAACGCGTGTGCT-3′ |
| FAS | NM_000043 | 5′-AAAAACAAACACAGACAAACACCAAACATGGAGTTGGTGCCCGGCGCCGG-3′ |
| MCC10 | NM_011681 | 5′-ATAGACTCCAAATAAACCACATCTACAGACACCAAAGCCTCCAACCTCTA-3′ |
| Cyclin E1 | NM_001238 | 5′-ATCCTCGCTTTTCACAGTCTTGTCAATCTTGGCAATTTCTTCATCTGGGT-3′ |
| Nkx2.2 | NM_00250 | 5′-GGGCAGAGGGCTCCCTGCCTACAGGGTTTTCTTTTCCATATTTGAGAAAT-3′ |
Probe Conjugation
Amine-coated QDs were coupled to probe using a standard protocol (Evident Technologies). In brief, 200 mmol/L N-β-maleimidopropionic acid and 0.1 mmol/L 5′-sulfhydryl-modified oligonucleotide were dissolved in 500 μl of 1× PBS and incubated for 2 hours at room temperature, followed by purification by centrifugation at 5000 × g for 15 minutes to remove excess N-β-maleimidopropionic acid. Seventy microliters of the purified N-β-maleimidopropionic acid-oligo was then added to 270 μl of water, 50 μl of 10× PBS, 100 μl of amine coated QDs (Evident Technologies), and 10 μl of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide-hydrochloride (100 mg/ml), followed by incubation at room temperature for 2 hours, after which the reaction was quenched by addition of 500 μl of 1 mol/L Tris (pH 7.4). The conjugated QDs were then purified by centrifugation in a 100k Omega Nanosep filter (Pall Life Sciences, Hampshire, UK) for 10 minutes at 10,000 × g.
Direct probe conjugation of streptavidin-coated QDs was performed in a QD-to-probe ratio of 1:2 by mixing both in a centrifuge tube and incubating at room temperature for 60 minutes. The resultant conjugate was volume reduced with a 100k Omega Nanosep exclusion filter (Pall Life Sciences).
In Situ Hybridization
Manual in Situ Hybridization
Manual in situ hybridization was performed using standard methods.11 Sections were hybridized with the mcc10 riboprobe for 14 hours at 42°C and washed in 0.1× SSC three times for 20 minutes at 47°C. Digoxigenin-tailed oligonucleotide probe for cyclin E1 was hybridized at 37°C and washed at 42°C.
Semiautomated in Situ Hybridization
DIG-labeled riboprobe ISH using QDs were performed using a Ventana Discovery XT instrument (Ventana Medical Systems, Tucson, AZ). Seven-micrometer-thick tissue sections were deparaffinized (standard xylene and industrial methylated spirits) and fixed in 4% paraformaldehyde for 10 minutes. Tissues were then permeabilized by incubation in 1 mg/ml pepsin in 0.1 mol/L HCl for 20 minutes at 37°C followed by denaturation at 70°C for 10 minutes before hybridization with DIG-labeled riboprobe at 65°C for 6 hours. Washes after hybridization were performed using 0.1× SSC at 75°C twice for 6 minutes followed by PBS at room temperature for 5 minutes. After hybridization, slides were incubated with QDs 605-conjugated anti-DIG antibody (Invitrogen, Carlsbad, CA) for 30 minutes and rinsed thereafter twice in PBS for 5 minutes. All tissue sections were coverslipped using 90% (v/v) glycerol/PBS mounting solution.
Immunohistochemistry
IHC for CD34 was performed using a rat anti-mouse anti-CD34 antibody (Abcam) with pressure cooker target retrieval in citrate buffer for 2 minutes. Primary anti-CD34 antibody was applied at a dilution of 1:50 in PBS and incubated at room temperature for 60 minutes, followed by detection using a QD-labeled anti-rat IgG antibody (Invitrogen) at a dilution of 1:100 for 30 minutes at room temperature. IHC for mcc10 was performed as previously described28 using a primary rabbit polyclonal antibody against mcc10 (kindly provided by Dr. F.J. De Mayo, Baylor College of Medicine, Houston, TX) at a dilution of 1:100; mcc10 expression is cell specific, being entirely confined to the bronchiolar epithelium.28
Combined Immunohistochemistry and in Situ Hybridization
ISH was performed on a Discovery Machine (Ventana Medical Systems). In brief, paraffin slides were deparaffinized on the machine by heating to 75°C and rinsed with EZ buffer. Slides were then fixed in 4% paraformaldehyde for 10 minutes and digested in 0.5 mg/ml pepsin for 10 minutes at 37°C. DIG-labeled Mcc10 riboprobe (1:50) and biotin-labeled pulmonary surfactant (PS) probe (1:50) were diluted in hybridization buffer and added to the slides. Slides were hybridized at 65°C for 6 hours. After hybridization, slides were rinsed twice in 0.1× SSC buffer at 75°C, each for 6 minutes. Detection of DIG-labeled mCC10 probe and biotin-labeled PS probe and CD34 protein were performed manually. QD 605 anti-DIG (1:100 for mCC10; Invitrogen) and streptavidin QD 525 (1:100 for PS; Invitrogen) and CD34 primary antibody were diluted in PBS buffer and incubated for 1 hour at room temperature. Slides were then rinsed in PBS twice for 5 minutes and incubated with QD 655 anti-rat IgG antibody. Slides were rinsed in phosphate-buffered saline-Tween twice for 5 minutes and coverslipped with Vectashied (Vector Laboratories, Burlingame, CA).
Spectral Imaging and Signal Quantitation
Using a Leitz Diaplan fluorescence microscope and a CRI Nuance spectral analyzer (CRI Inc., Woburn, MA) phosphate-buffered saline-Tween, image files were collected at 5-nm wavelength intervals from 450 to 700 nm. This system uses stacked liquid crystal filters to produce a solid-state tunable Lyot filter. The image files, each comprising the concatenated stack of images at each wavelength interval per pixel, were then used to reconstruct multiple spectral distributions via a maximum likelihood method. Specifically, the maximum likelihood distributions at each pixel were determined for spectral distributions obtained from autofluorescence and for the QDs used in a given experiment. These distributions represent signal intensity at each pixel for the defined spectra and were converted to composite pseudocolorized images to visualize staining distribution and intensity for each QD. The signal intensities in the resultant images were compared numerically using IPLab software (Scanalytics, MD), according to manufacturer’s instructions.
All composite image files were collected from slides at 5-nm wavelength intervals from 450 to 720 nm, with an exposure of 1 second per wavelength interval for ×400 magnification and 0.5 second per wavelength for ×200 magnification, using a CRI Nuance system and analyzed using the spectra of autofluorescence and of the relevant QD emission spectrum using spectral unmixing,26 disclosing signal distribution otherwise obscured by autofluorescence. A long wavelength bypass filter allowing transmission of all emission wavelengths above 450 nm was used, and spectral deconvolution was performed digitally, according to manufacturer’s instructions, on the resultant image stack.
Quantitative Image Analysis
FAS QD ISH images were taken from 500 to 700 nm at 5 nm with a 72.1-millisecond exposure time at ×40 objective magnification. To avoid variations in staining due to heterogeneous cell densities, five images of distinct cell clusters were taken for each time point, and the mean signal intensity was calculated. Image analysis was performed using ImageJ (National Institutes of Health, Bethesda, MD). Mean pixel intensity of the 605-nm QD (FAS) was computed after applying a threshold of 100 to eliminate low-level background and bias due to differing amounts of negative space.
RNA Extraction and Real-Time PCR
LNCAP frozen pellets from single plate were placed in TRIzolR Reagent (Invitrogen), and total RNA was prepared by the acid phenol method. Total RNA was quantified by ND-1000 (NanoDrop Technologies, Wilmington, DE). One microgram of total RNA was subjected to reverse transcription using the SuperScript III First-Strand Synthesis System (Invitrogen) in a total volume of 20 μl according to the manufacturer’s instructions. FAS-specific primers and probe were ordered as ready-to-use primer and probe mix (Assay on Demand, Gene Expression product, Hs00188012_m1; Applied Biosystems, Foster City, CA). The FAS probe was labeled with the FAM reporter at 5′ end and the TAMRA quencher at 3′ end. Human β-glucoronidase (VIC/MGB Probe; Applied Biosystems) was chosen as the endogenous control. In the β-glucoronidase probe labeled with 5′-VIC, the 3′ end quencher has been substituted by the minor groove binder groups that provide a more efficient fluorescence quenching, resulting in an increased sensitivity. Quantitative PCR was performed using an ABI Prism 7500 Sequence Detection System (Applied Biosystems) in a 25-μl final volume containing 2 μl of cDNA and appropriate concentrations of TaqMan Universal Master mix and gene expression assays (Applied Biosystems) according the manufacturer’s instructions. The reactions were performed in two independent analyses, each in duplicate, using MicroAmp Optical 96-well plates (Applied Biosystems). During the PCR operation, the plate was first held at 50°C for 2 minutes and then at 95°C for 10 minutes to denature nucleic acid samples. The amplification process was executed with a thermal cycle of 95°C for 15 seconds and 60°C for 1 minute for 40 cycles. A serial dilution of LNCAP RNA, from 1 μg through 1 ng of total RNA, was analyzed to verify the sensitivity and efficiency of the assays. No template controls and no reverse transcriptase controls were performed in parallel to exclude transcript nonspecific amplification. To determine the quantity of FAS-specific transcripts present in treated cells at different time points relative to untreated ones, the comparative threshold (Ct) method (the 2−ΔΔCt method) (User Bulletin no. 2; Applied Biosystems) was used. The FAS Ct values were first normalized by subtracting the respective Ct value obtained from the β-glucoronidase control (ΔCt = Ct target − Ct control) to compensate for variations in input RNA amounts. The concentration of FAS-specific mRNA in treated cells relative to untreated cells was calculated by subtracting the normalized Ct values obtained for untreated cells from those obtained from treated samples (ΔΔCt = ΔCt treated − ΔCt untreated), and the relative concentration was determined (2−ΔΔCt).
Results
Visualization of mRNA Targets by Single Label Quantum Dot-Based in Situ Hybridization
As a first step, we assessed the feasibility of QD conjugation to oligonucleotide cDNA probes and the use of QD-labeled oligonucleotide probes in ISH on paraffin-embedded tissue (Q-ISH), using similar techniques to those we had performed using nonradioactive probes in automated, high-throughput ISH experiments.11 Probes were either labeled by direct conjugation of QD to oligonucleotide or by biotinylation followed by streptavidin-oligonucleotide conjugation in vitro. First, efficient probe-QD conjugation was verified by gel electrophoresis (Figure 1A). Next, we chose a probe that was tissue specific, specific cell type restricted, and abundantly expressed to compare traditional ISH with that performed with QD-labeled probes. The murine mcc10 gene, specifically expressed in the Clara cells of the bronchiolar epithelium,28 was targeted in mouse lung. To first test the effectiveness of the probe sequence, DIG-labeled antisense mcc10 riboprobe was compared with a DIG-labeled mcc10 antisense oligonucleotide probe. Both resulted in specific signal in the mouse bronchial epithelium, albeit with the smaller oligonucleotide probe generated a weaker signal, as expected (Figure 1B, ii and iii). When the same 50-mer-biotinylated oligonucleotide was conjugated with a 605-nm QD, the same specific signal was obtained (Figure 1B, iv). No signal was seen with either scrambled or unrelated oligonucleotide controls (data not shown). The QD-labeled mcc10 riboprobe gave the same bronchiolar signal as the oligonucleotide (Figure 1B, v) and showed similar levels of intensity; IHC for mcc10 demonstrated the same pattern of staining (Figure 1B, vi). In addition, we established feasibility of QD oligonucleotide ISH using a 50-mer 5′ digoxigenin-tailed probe for cyclin E1 in formalin-fixed, paraffin-embedded pellets of mouse fibroblasts, either wild type or null/knockout for cyclin E1, demonstrating strong hybridization signal in the wild type and the absence of signal in the knockout cells (Figure 1C).
Figure 1.
Use of direct quantum dot oligonucleotide conjugates for in situ detection of mcc10 in mouse lung. A: i, Agarose gel (0.8%) electrophoresis showing no migration of unconjugated streptavidin-coated QDs (lane 1) and a differential gel shift with increasing probe-to-QD ratio. The individual samples are, from left to right, 605-nm streptavidin-coated QD alone (lane 1) followed by molar ratios of QDs to probe of 5:1; 1:1, and 1:10 (lanes 2 to 4). ii, Agarose gel (0.8%) electrophoresis showing strongest migration of unconjugated amine coated QDs (lane 1) and reducing gel shift with increasing probe-to-QD ratio. The individual samples are, from left to right, 580-nm amine coated QD alone (lane 1) followed by molar ratios of QDs to probe: 1:20, 1:10, 1:5, 1:2, 1:1, 2:1, and 5:1 (lanes 2 to 8). B: Formalin-fixed, paraffin-embedded mouse (H&E ×20 shown in i) lung hybridized with DIG-labeled antisense mcc10 riboprobe (ii) showing localization of signal to Clara cells located in the bronchiolar epithelium. Magnification, ×20. iii, DIG-labeled mcc10 antisense oligonucleotide showing similar expression distribution to that observed with the antisense mcc10 riboprobe. Magnification, ×20. iv, 5′-biotinylated mcc10 antisense oligonucleotide (sequence the same as in iii) in mouse lung disclosed using a 605-nm streptavidin-coated QD, demonstrating strong signal and localization to the bronchiolar epithelium. Magnification, ×20. v, DIG-labeled antisense mcc10 riboprobe disclosed with a 655-nm QD, demonstrating high signal in the same distribution to that of the QD-disclosed oligonucleotide probe (iv). Magnification, ×20. vi, IHC for mcc10 disclosed with diaminobenzidine demonstrated strong cytoplasmic staining in the same distribution to that seen with either riboprobes or oligonucleotide probes (ii–v). Magnification, ×20. C: i, Formalin-fixed, paraffin-embedded wild-type mouse fibroblasts hybridized with DIG-tailed antisense cyclin E1 oligonucleotide probe showing strong hybridization signal [4,6-diamidino-2-phenylindole (DAPI) counterstained]. Magnification, ×40. ii, Formalin-fixed, paraffin-embedded cyclin E1 knockout mouse fibroblasts hybridized with DIG-tailed antisense cyclin E1 oligonucleotide probe showing the absence of hybridization signal (DAPI counterstained); small amounts of particulate QD are present, but cytoplasmic signal is absent. Magnification, ×40.
Gene Signatures from Expression Profiling Data Can Be Detected in Tumor Tissue by Quantum Dot-Based in Situ Hybridization
To determine whether the technique could be an effective tool in application to a published expression data set, we identified and tested transcription factors ASCL1 and the homeobox-containing gene NKX2-2, which have been recently reported as members of an expression signature set associated with poor prognosis in malignant gliomas.29 Analysis of their expression in two human gliomas using the Q-ISH technique demonstrated abundant expression of both markers in the majority of glioma cells with absent or low-level expression in the normal white matter adjacent to the tumor (Figure 2) similar to results seen by immunohistochemistry (data not shown).
Figure 2.
Spectral analysis of quantum dot-based in situ hybridization to resolve signal against tissue autofluorescence in routinely processed human clinical tissue. i: H&E-stained human glioma is marked by the asterisk, and the arrow points to the tumor normal brain interface. All tissue was routinely formalin fixed, paraffin embedded, and imaged at ×20 magnification. ii: Raw (real color) image of a human glioma with adjacent normal cerebral tissue, hybridized with a 5′ thiol-labeled antisense ASCL1 oligonucleotide probe/amine-coated 580-nm QD conjugate, showing poor signal resolution due to high green autofluorescence of brain tissue. iii and iv: Pseudocolorized images following spectral analysis of image in ii showing signal for ASCL1 (iii) and NKX2-2 (iv) in red and tissue autofluorescence in green, demonstrating ability of spectral imager to resolve hybridization signal against high levels of inherent tissue autofluorescence.
Quantum Dot-Based in Situ Hybridization Is Photostable, Quantitative, and Reproducible
To demonstrate that the system is durable, quantitative, and reproducible, sections hybridized with mcc10 were analyzed twice at an interval of 18 months, during which time there was no significant reduction in signal intensity (Figure 3A), in contrast to conventional fluorochromes, which are subject to fading over time. Such stability is an important property for use in clinical diagnostics. To test stability of RNA in FFPE using this method, FAS expression was assayed using a directly conjugated oligonucleotide DNA probe in a tissue microarray comprising 30-year-old FFPE benign and malignant prostatic needle cores (Figure 3B, i and ii). Spectral analysis was again used to resolve the hybridization signal from autofluorescence in each core (Figure 3B, iii and iv). After this, the resultant image files were analyzed using IPLab (Scanalytics), a proprietary image analysis software package enabling quantitation of image intensity, demonstrating higher expression of FAS in the malignant cores compared with adjacent normal tissue (Figure 3B, vii) and the ability of the method to quantitate expression differences between samples. This is of particular importance for analysis of microarray identified gene signatures and was capable of quantitation from 10 to nearly 2000 (arbitrary units), demonstrating a wide dynamic range. In addition, fluorescence provides a linear relationship between the amount of probe hybridized and signal intensity not seen with chromogenic methods and is critically important for accurate measurement of relative expression levels. Because oligonucleotides are used and the number of modified nucleotides with attached QDs is identical for each probe, signal intensity is directly proportional to the amount of hybridization present and therefore more precisely representative of the transcript level in the tissue than nonfluorescent methods.
Figure 3.
Quantum dot-based in situ hybridization is photostable and quantifiable. A: Signal intensity for same slide of mouse lung hybridized with DIG-labeled antisense mcc10 riboprobe visualized twice over an 18-month interval, demonstrating stability of QD fluorescence. There was no significant loss of intensity between the first (ii) and second (iii) visualizations. H&E-stained section shown in i. Magnification, ×40. B: FAS hybridization in 30-year benign (i) and malignant (ii) prostatic core biopsies in a tissue microarray demonstrating low signal in benign tissue (iii) and high signal in prostatic adenocarcinoma (iv). QD-disclosed IHC (605 nm) for FAS is shown for benign (v) and malignant (vi) prostatic tissue. Magnification, ×20. Signal intensity was determined by spectral analysis over nine benign (N) and 12 malignant (T) cores (vii) and measured using IPLab, demonstrating higher expression of FAS in the malignant cores.
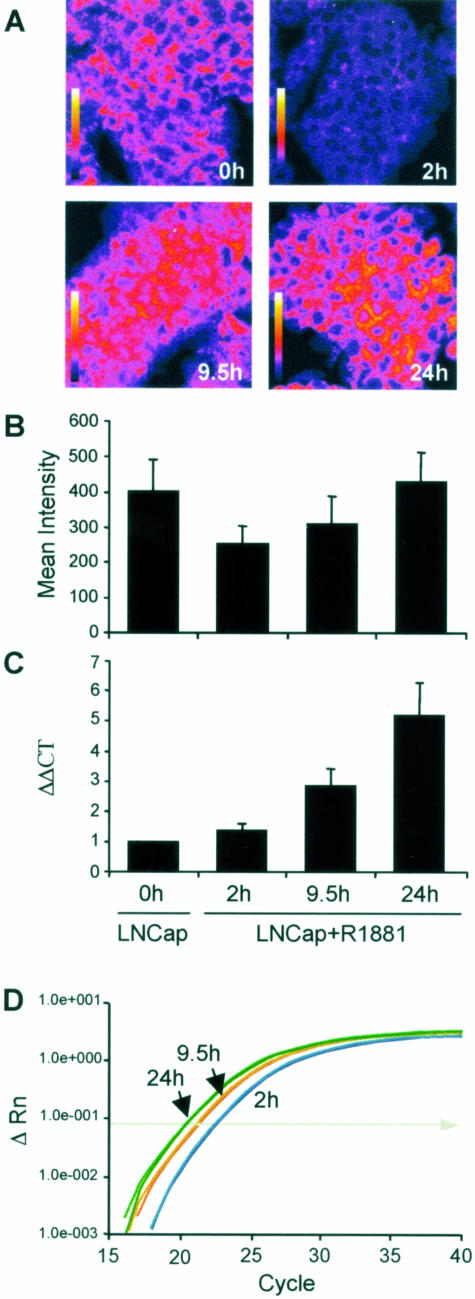
To directly address quantitation, we compared the signal intensity as determined by quantitative spectral imaging to RT-PCR. To do this, LNCAP cells were grown in charcoal-stripped serum, which was subsequently stimulated with the synthetic androgen R1881, and harvested at the times shown in Figure 4. The androgen-regulated gene FAS was evaluated at the transcript level by quantitative RT-PCR on RNA obtained from these cells at time 0 and at various time points and compared with expression levels of FAS as determined by quantitation of spectral images of the same cells, at the same time points, hybridized to FAS after paraffin embedding. FAS mRNA expression as determined by quantitation of fluorescence in tissue sections hybridized with the FAS probe correlated to transcript levels in the same cells determined by quantitative RT-PCR.
Figure 4.
Quantum dot-based in situ hybridization is quantitative. The signal intensity as determined by quantitative spectral imaging (A: magnification, ×40) was compared with that obtained from the same cells by RT-PCR. LNCaP cells were grown in charcoal-stripped serum and subsequently stimulated with the synthetic androgen R1881 and harvested at 2, 9.5, and 24 hours. Transcript levels of the androgen-regulated gene FAS were evaluated both at the tissue level and by quantitative RT-PCR. Quantitation of FAS mRNA by spectral images of FFPE LNCaP cells (A and B) was highly correlated with FAS mRNA expression as determined by quantitation of transcript by quantitative RT-PCR (C and D).
Deconvolution by Spectral Imaging
In each of the above experiments, spectral imaging was used to resolve hybridization signal from tissue autofluorescence (Figures 2and 3B). In brief, composite image files were collected from the slides at 5-nm wavelength intervals from 450 to 720 nm using a CRI Nuance system and analyzed using the spectra of autofluorescence and of the relevant QD emission spectrum using spectral unmixing,25 disclosing signal distribution otherwise obscured by autofluorescence (Figure 2).
Multiplex Quantum Dot-Based in Situ Hybridization Is Feasible
Use of QDs for signal disclosure opens the possibility of multiplex detection based on the ability to use QDs with different emission wavelengths.30 Therefore, we next attempted to separate multiple fluorescent signals by spectral deconvolution. Aliquots of QDs were placed on glass slides and coverslipped, and spectral profiles were captured for QDs at each wavelength of emission. The spectral profile of tissue autofluorescence was captured separately, and these profiles were used in a least-squares fitting algorithm to reconstruct autofluorescent and QD fluorescent images.
We then performed ISH using a semiautomated system, because a manual technique is likely to be cumbersome for routine clinical use. Using this semiautomated system, we performed simultaneous riboprobe-ISH for mcc10 and PS disclosed by two different color QDs, both separately and in duplex, and analyzed composite images. Specifically, image stacks were captured, using the spectral analyzer, from sections hybridized simultaneously with a riboprobe to mcc10 and a riboprobe to PS. PS was expressed in both the bronchiolar epithelium and alveolar pneumocytes (Figure 5A, i–v), whereas mcc10 was localized only to the bronchiolar epithelium (Figure 5A, vi–x). Q-ISH for mcc10 and PS were then performed with simultaneous hybridization of a biotinylated mcc10 riboprobe and a DIG-labeled PS riboprobe combined with simultaneous IHC for CD34 (Figure 5B, i). The mcc10 probe was disclosed using streptavidin-coated QDs (605 nm), the PS probe using an anti-DIG antibody directly conjugated to QDs (525 nm), and CD34 antibody staining with a 655-nm QD-conjugated secondary antibody. The signals for mcc10, PS, and CD34 were resolved using spectral deconvolution, demonstrating surfactant expression in type 2 pneumocytes as well as in a subset of Clara cells (Figure 5B, ii), CD34 in alveolar capillaries (Figure 4B, iii), and mcc10 expression in Clara cells (Figure 5B, iv). The resultant triplex signal detection and deconvolution are shown in Figure 5B, v.
Figure 5.
Semiautomated duplex in situ hybridization and immunohistochemistry. A: Single Q-ISH for mcc10 and PS was performed in mouse lung using antisense riboprobes labeled with different color QDs; the probe for mcc10 was labeled with a 605-nm QD and that for PS with a 655-nm QD. Raw images for both were analyzed with the spectral imager using the spectra of both a 605-nm and a 655-nm QD. For the sections hybridized with probe for PS, signal was resolved in the 655-nm channel and was present in both bronchiolar epithelium and the alveolar tissue (ii), whereas no signal was present in the 605-nm channel (i). Conversely, mcc10 was detected in the bronchiolar epithelium in the 605-nm channel only (vi), with no signal in the 655-nm channel (vii). For both, the autofluorescence (AF) was resolved (iii and viii), and composite false color images were generated with AF in green and either PS in blue (iv) or mcc10 in red (ix). DIG-labeled antisense riboprobes were also hybridized for each gene (v and x). Magnification, ×20. B: Duplex Q-ISH was performed in mouse lung for mcc10 and PS, using the same QD-labeled riboprobes as in A (above) with the addition of IHC for CD34, which highlights the alveolar capillary network, although it is not present in the bronchiolar epithelium. The resultant raw image (i) was subjected to spectral analysis using the spectra for 525 nm (PS), 605 nm (mcc10), and 655 nm (CD34) QDs, from which distributions of PS (ii), CD34 (iii), and mcc10 (iv) were resolved. Composite false color image of resolved images for PS, CD34, and mcc10 is shown in v (PS in green, CD34 in red, and mcc10 in blue). Magnification, ×40.
Discussion
The results demonstrate successful generation and use of QD-labeled oligonucleotide and RNA probes for FFPE ISH. This enabled simultaneous detection of multiple mRNA targets by multiplex Q-ISH on FFPE, with deconvolution of individual hybridization signals using quantitative spectral imaging. In addition, multiplex Q-ISH was adapted for semiautomated use. Taken together, these features will enable multiplex in situ transcript detection and quantification in routinely processed human clinical tissue, facilitating translation of gene expression signatures to diagnostic material, potentially in high throughput.
QDs are a novel class of inorganic fluorophores, with near ideal fluorescence properties, that have been used to improve signal detection in immunofluorescence.17 They have been used in detection of FISH after hybridization for 1q1230 and HER2,17 and their use for simultaneous detection of up to two genes has been reported in fresh animal and tissue culture material.31 However, the vast majority of clinical tissue is in the form of formalin-fixed material, and there is therefore a need for application of QDs to such material, not only for analysis of prospective clinical material in a routine diagnostic setting but also for analysis of the existing archive of paraffin-embedded tissue. We have very recently demonstrated use of QD-based ISH in clinical material22 but only in a manual system and only in hemopoietic tissues. To be useful clinically, however, a method must be capable of automation and applicable to all types of tissues. To overcome this limitation of existing methods for clinical use, we have combined high-throughput automated ISH, QD-labeling of DNA and RNA probes, and spectral imaging to produce a novel technique for ISH to enable multiplex mRNA transcript detection in FFPE tissue of both murine and human origin. Specifically, we have combined the extremely high fluorescence of QDs with spectral imaging both to remove autofluorescence digitally, thereby improving signal-to-noise ratio and effectively increasing the sensitivity of the probes used, and to resolve multiple signals in the same tissue section. Importantly for clinical studies, the method used is applicable to paraffin-embedded tissue sections, for which autofluorescence is a particular problem.12,13 The spectral imager also enables quantitation of signal intensity, which is necessary for analysis of microarray-identified genes, many of which show relatively low fold changes in expression level between different groups.1,2 The method described allows precise measurement of signal intensity within the reconstructed images, whereas the linear relationship between probe hybridization and fluorescence also enables semiquantitation by the method described.
The use of QDs for biological imaging was first demonstrated by Wu et al,17 who reported their use for antibody detection of HER2. Several reports have underscored their use for protein detection,32 but there have been relatively few reports of their use for detection of in situ hybridization,22,31,32,33,34,35 reflecting the technical difficulty of this endeavor. The specific technical hurdles to their use in ISH, which were also faced and overcome in the present work, include optimization of QD/oligonucleotide conjugation, signal stability (particularly with older sources of QD), tissue autofluorescence (a particular problem in their use in clinical material), and quantitation of signal. The last two are of particular importance in clinical samples and also hamper interpretation of optimization experiments, especially those directed at optimizing tissue permeabilization and conjugation ratio. The use of spectral imaging was critical in overcoming these hurdles.
We have demonstrated use of QD labeling and semiquantitative detection of multiplex ISH in routinely processed clinical tissue samples and laid the groundwork for using this methodology to validate gene expression microarray data. Spectral imaging has been used to reconstruct multiple spectral components from the resultant composite images, enabling sensitive colocalization of multiple genes, whereas use of fluorescence facilitates quantitation. Importantly, the semiautomated nature of the method will facilitate translational application of microarray-identified gene signatures to clinical research and diagnostics, whereas the ability of spectral imaging to resolve multiple signals offers the possibility of multiplexed probe detection.
Acknowledgments
Eyoung Shin and Xingyu Wang provided technical help with in situ hybridization and immunohistochemistry experiments, and Lance Ostrom helped with cell culture.
Footnotes
Related Commentary on page 7
Supported by grants from Dana-Farber/Harvard Cancer Centre, grant for Core Technology Developments on Biological Imaging Spectroscopy, the Howard Hughes Medical Institute/SPORE Biomedical Research Support Program, Novartis, Nuclea, National Institutes of Health [PO1 in Prostate Cancer (2PO1 CA089021-06) and Specialized Programs of Research Excellence 5P50CA90381], Waxmann Foundation and Prostate Cancer Foundation Award (to M.L.), and Research Sabbatical Fellowship from the University of Manchester, United Kingdom (to R.B.).
M.L. is a Novartis investigator.
References
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Ebert BL, Golub TR. Genomic approaches to hematologic malignancies. Blood. 2004;104:923–932. doi: 10.1182/blood-2004-01-0274. [DOI] [PubMed] [Google Scholar]
- Petricoin EF, III, Hackett JL, Lesko LJ, Puri RK, Gutman SI, Chumakov K, Woodcock J, Feigal DW, Jr, Zoon KC, Sistare FD. Medical applications of microarray technologies: a regulatory science perspective. Nat Genet. 2002;32(Suppl):474–479. doi: 10.1038/ng1029. [DOI] [PubMed] [Google Scholar]
- Gerhold DL, Jensen RV, Gullans SR. Better therapeutics through microarrays. Nat Genet. 2002;32(Suppl):547–551. doi: 10.1038/ng1042. [DOI] [PubMed] [Google Scholar]
- Byers R, Roebuck J, Sakhinia E, Hoyland J. PolyA PCR amplification of cDNA from RNA extracted from formalin-fixed paraffin-embedded tissue. Diagn Mol Pathol. 2004;13:144–150. doi: 10.1097/01.pdm.0000133154.73846.66. [DOI] [PubMed] [Google Scholar]
- Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, Fisher RI, Braziel RM, Rimsza LM, Grogan TM, Miller TP, LeBlanc M, Greiner TC, Weisenburger DD, Lynch JC, Vose J, Armitage JO, Smeland EB, Kvaloy S, Holte H, Delabie J, Connors JM, Lansdorp PM, Ouyang Q, Lister TA, Davies AJ, Norton AJ, Muller-Hermelink HK, Ott G, Campo E, Montserrat E, Wilson WH, Jaffe ES, Simon R, Yang L, Powell J, Zhao H, Goldschmidt N, Chiorazzi M, Staudt LM. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351:2159–2169. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RC, Gaasenbeek M, Angelo M, Reich M, Pinkus GS, Ray TS, Koval MA, Last KW, Norton A, Lister TA, Mesirov J, Neuberg DS, Lander ES, Aster JC, Golub TR. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 2002;8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- Zwirglmaier K. Fluorescence in situ hybridisation (FISH): the next generation. FEMS Microbiol Lett. 2005;246:151–158. doi: 10.1016/j.femsle.2005.04.015. [DOI] [PubMed] [Google Scholar]
- Wilcox JN. Fundamental principles of in situ hybridization. J Histochem Cytochem. 1993;41:1725–1733. doi: 10.1177/41.12.8245419. [DOI] [PubMed] [Google Scholar]
- Wilson KH, Schambra UB, Smith MS, Page SO, Richardson CD, Fremeau RT, Schwinn DA. In situ hybridization: identification of rare mRNAs in human tissues. Brain Res Brain Res Protoc. 1997;1:175–185. doi: 10.1016/s1385-299x(96)00028-1. [DOI] [PubMed] [Google Scholar]
- Capodieci P, Magi-Galluzzi C, Moreira G, Jr, Zeheb R, Loda M. Automated in situ hybridization: diagnostic and research applications. Diagn Mol Pathol. 1998;7:69–75. doi: 10.1097/00019606-199804000-00002. [DOI] [PubMed] [Google Scholar]
- Baschong W, Suetterlin R, Laeng RH. Control of autofluorescence of archival formaldehyde-fixed, paraffin-embedded tissue in confocal laser scanning microscopy (CLSM). J Histochem Cytochem. 2001;49:1565–1572. doi: 10.1177/002215540104901210. [DOI] [PubMed] [Google Scholar]
- Del Castillo P, Llorente AR, Stockert JC. Influence of fixation, exciting light and section thickness on the primary fluorescence of samples for microfluorometric analysis. Basic Appl Histochem. 1989;33:251–257. [PubMed] [Google Scholar]
- Fu A, Gu W, Larabell C, Alivisatos AP. Semiconductor nanocrystals for biological imaging. Curr Opin Neurobiol. 2005;15:568–575. doi: 10.1016/j.conb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Bruchez M, Jr, Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- Chan WC, Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- Wu X, Liu H, Liu J, Haley KN, Treadway JA, Larson JP, Ge N, Peale F, Bruchez MP. Immunofluorescent labeling of cancer marker Her2 and other cellular targets with semiconductor quantum dots. Nat Biotechnol. 2003;21:41–46. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- Lidke DS, Nagy P, Heintzmann R, Arndt-Jovin DJ, Post JN, Grecco HE, Jares-Erijman EA, Jovin TM. Quantum dot ligands provide new insights into erbB/HER receptor-mediated signal transduction. Nat Biotechnol. 2004;22:198–203. doi: 10.1038/nbt929. [DOI] [PubMed] [Google Scholar]
- Gao X, Cui Y, Levenson RM, Chung LW, Nie S. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- Kim S, Lim YT, Soltesz EG, De Grand AM, Lee J, Nakayama A, Parker JA, Mihaljevic T, Laurence RG, Dor DM, Cohn LH, Bawendi MG, Frangioni JV. Near-infrared fluorescent type II quantum dots for sentinel lymph node mapping. Nat Biotechnol. 2004;22:93–97. doi: 10.1038/nbt920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal JK, Mattoussi H, Mauro JM, Simon SM. Long-term multiple color imaging of live cells using quantum dot bioconjugates. Nat Biotechnol. 2003;21:47–51. doi: 10.1038/nbt767. [DOI] [PubMed] [Google Scholar]
- Tholouli E, Hoyland JA, Di Vizio D, O’Connell F, Macdermott SA, Twomey Levenson R, Yin JA, Golub TR, Loda M, Byers R. Imaging of multiple mRNA targets using quantum dot based in situ hybridization and spectral deconvolution in clinical biopsies. Biochem Biophys Res Comm. 2006;348:628–636. doi: 10.1016/j.bbrc.2006.07.122. [DOI] [PubMed] [Google Scholar]
- Dahan M, Levi S, Luccardini C, Rostaing P, Riveau B, Triller A. Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science. 2003;302:442–445. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- Dubertret B, Skourides P, Norris DJ, Noireaux V, Brivanlou AH, Libchaber A. In vivo imaging of quantum dots encapsulated in phospholipid micelles. Science. 2002;298:1759–1762. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]
- Farkas DL, Du C, Fisher GW, Lau C, Niu W, Wachman ES, Levenson RM. Non-invasive image acquisition and advanced processing in optical bioimaging. Comput Med Imaging Graph. 1998;22:89–102. doi: 10.1016/s0895-6111(98)00011-1. [DOI] [PubMed] [Google Scholar]
- Geng Y, Yu Q, Sicinska E, Das M, Schneider JE, Bhattacharya S, Rideout WM, Bronson RT, Gardner H, Sicinski P. Cyclin E ablation in the mouse. Cell. 2003;114:431–443. doi: 10.1016/s0092-8674(03)00645-7. [DOI] [PubMed] [Google Scholar]
- Ray MK, Wang G, Barrish J, Finegold MJ, DeMayo FJ. Immunohistochemical localization of mouse Clara cell 10-KD protein using antibodies raised against the recombinant protein. Histochem Cytochem. 1996;44:919–927. doi: 10.1177/44.8.8756763. [DOI] [PubMed] [Google Scholar]
- Ray MK, Chen CY, Schwartz RJ, DeMayo FJ. Transcriptional regulation of a mouse Clara cell-specific protein (mCC10) gene by the NKx transcription factor family members thyroid transcription factor 1 and cardiac muscle-specific homeobox protein (CSX). Mol Cell Biol. 1996;16:2056–2064. doi: 10.1128/mcb.16.5.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro JM, Misra A, Zhang L, Smirnov I, Colman H, Griffin C, Ozburn N, Chen M, Pan E, Koul D, Yung WK, Feuerstein BG, Aldape KD. Integrated array-comparative genomic hybridization and expression array profiles identify clinically relevant molecular subtypes of glioblastoma. Cancer Res. 2005;65:1678–1686. doi: 10.1158/0008-5472.CAN-04-2921. [DOI] [PubMed] [Google Scholar]
- Alivisatos AP, Gu W, Larabell C. Quantum dots as cellular probes. Annu Rev Biomed Eng. 2005;7:55–76. doi: 10.1146/annurev.bioeng.7.060804.100432. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Barker PE. Semiconductor nanocrystal probes for human metaphase chromosomes. Nucleic Acids Res. 2004;32:e28. doi: 10.1093/nar/gnh024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P, Yuen T, Ruf F, Gonzalez-Maeso J, Sealfon SC. Method for multiplex cellular detection of mRNAs using quantum dot fluorescent in situ hybridization. Nucleic Acids Res. 2005;33:e161. doi: 10.1093/nar/gni162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountaine TJ, Wincovitch SM, Geho DH, Garfield SH, Pittaluga S. Multispectral imaging of clinically relevant cellular targets in tonsil and lymphoid tissue using semiconductor quantum dots. Mod Pathol. 2000;9:1181–1191. doi: 10.1038/modpathol.3800628. [DOI] [PubMed] [Google Scholar]
- Bentolila LA, Weiss S. Single-step multicolor fluorescence in situ hybridization using semiconductor quantum dot-DNA conjugates. Cell Biochem Biophys. 2006;45:59–70. doi: 10.1385/CBB:45:1:59. [DOI] [PMC free article] [PubMed] [Google Scholar]