Abstract
Transplanted donor lymphocytes infused during hematopoietic stem cell transplantation (HSCT) have been shown to cure patients with hematological malignancies. However, less is known about the effects of HSCT on metastatic solid tumors. Thus, a better understanding of the immune cells and their target antigens that mediate tumor regression is urgently needed to develop more effective HSCT approaches for solid tumors. Here we report regression of metastatic renal cell carcinoma (RCC) in patients following nonmyeloablative HSCT consistent with a graft-versus-tumor effect. We detected RCC-reactive donor-derived CD8+ T cells in the blood of patients following nonmyeloablative HSCT. Using cDNA expression cloning, we identified a 10-mer peptide (CT-RCC-1) as a target antigen of RCC-specific CD8+ T cells. The genes encoding this antigen were found to be derived from human endogenous retrovirus (HERV) type E and were expressed in RCC cell lines and fresh RCC tissue but not in normal kidney or other tissues. We believe this to be the first solid tumor antigen identified using allogeneic T cells from a patient undergoing HSCT. These data suggest that HERV-E is activated in RCC and that it encodes an overexpressed immunogenic antigen, therefore providing a potential target for cellular immunity.
Introduction
Donor T cells mediating graft-versus-leukemia (GVL) effects can cure patients with a variety of different hematological malignancies. Until recently, little data existed as to whether solid tumors might have similar susceptibility to allogeneic immunotherapy. We and others have recently reported that metastatic renal cell carcinoma (RCC) can also regress following nonmyeloablative allogeneic hematopoietic stem cell transplantation (HSCT) as a consequence of a donor immune-mediated graft-versus-tumor (GVT) effect (1). Although these GVT effects can be durable and sometimes complete, more than half of patients undergoing HSCT fail to achieve a disease response (2). A better understanding of the immune cells and their target antigens that mediate the regression of these tumors is urgently needed to develop more effective HSC transplant approaches for RCC and other solid tumors.
For hematological malignancies, GVL effects appear to be primarily mediated by allogeneic T cells targeting polymorphic peptides expressed on malignant cells of the recipient (i.e., minor histocompatibility antigens [mHas]) (3). Several lines of clinical evidence suggest that donor T cells targeting antigens expressed on the tumor cells mediate regression of metastatic renal cancer (3–5). The majority of patients who achieve a disease response show early tumor growth in the first few months after transplantation, when the newly engrafted donor immune system is still suppressed by cyclosporine or when mixed T cell chimerism leading to T cell tolerance of host tissues (including the tumor) occurs. In responding patients, tumor regression is typically delayed by 4–6 months, occurring after immunosuppression has been withdrawn following the conversion from mixed to full donor T cell chimerism. As observed with hematological malignancies, graft-versus-host disease (GVHD) is also associated with disease regression (2–4). These clinical observations and recent in vitro findings suggest that regression of metastatic RCC may result from alloreactive T cells targeting mHas broadly expressed on both normal tissues and tumor cells of the recipient (4). However, the observation that tumor shrinkage sometimes occurs in the absence of or at a temporal distance from GVHD (5) implies that antigens overexpressed or even selectively expressed on the tumor are also targets for immune cells mediating RCC regression.
Here we show that donor T cells that recognize and kill patient tumor cells in vitro can be isolated from the blood of patients who had regression of metastatic RCC following HSCT. By using PBMCs obtained from one patient who had disease regression associated with prolonged survival, we generated a tumor-specific CTL clone that allowed us to identify a new tumor-associated RCC antigen. Furthermore, the antigen-encoding region, named CT-RCC, was found to be a part of the human endogenous retrovirus (HERV) type E locus highly expressed on RCC but not normal tissues. Here we present the cloning and expression pattern of what we believe to be the first solid tumor-specific antigen identified using donor T cells from a patient following allogeneic HSCT.
Results
Clinical evidence for graft-versus-RCC effect.
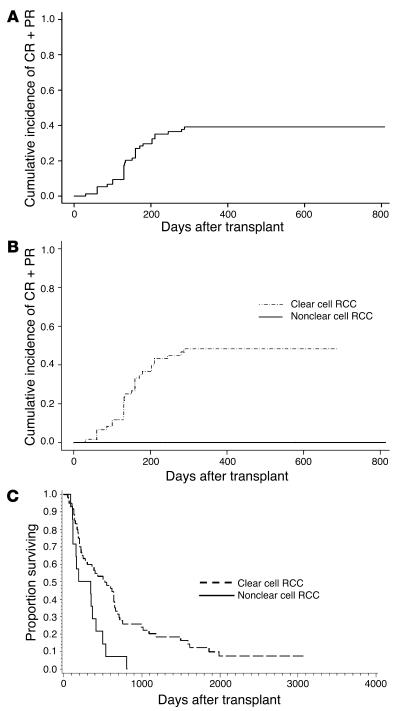
From 1998 through May 2006, 74 patients with metastatic RCC gave written informed consent according to NHLBI protocol 97-H-0196 and then underwent an allogeneic HSCT following nonmyeloablative conditioning with cyclophosphamide and fludarabine (Table 1) (5). Fifty-five percent of patients developed acute grade II–IV GVHD, and 47% developed chronic GVHD. Disease regression consistent with a GVT effect was observed in 29 patients (39.2% cumulative incidence of a complete response plus partial response) and was delayed in onset, occurring at a median 133 (range, 30–287) days following HSCT (Figure 1). RCC patients with clear-cell histology had a higher probability of achieving a GVT effect and had prolonged survival after HSCT compared with those with non-clear-cell histology; the cumulative incidence of a disease response was 48% and 0% (P = 0.0018), and survival was a median 525 versus 273 days; P = 0.003) in patients with clear-cell histologies compared with non-clear-cell histologies, respectively. In 6 patients, tumor regression occurred without any evidence of acute GVHD, suggesting immune responses may have occurred against antigens overexpressed or restricted to the tumor.
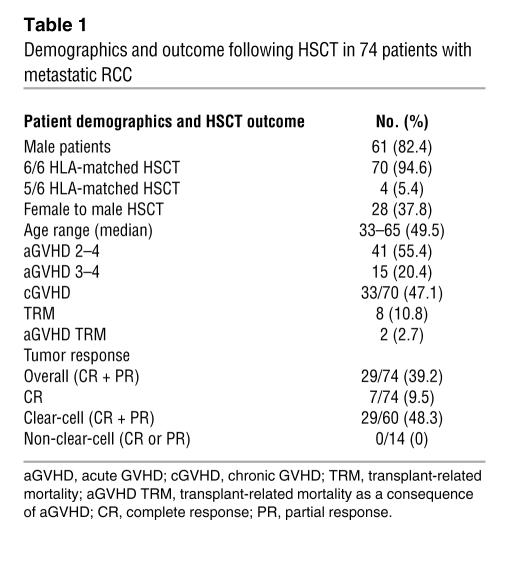
Table 1 .
Demographics and outcome following HSCT in 74 patients with metastatic RCC
Figure 1. Disease response and survival based on RCC histology (clear-cell RCC versus non-clear-cell RCC).
(A) The cumulative incidence of disease response (combined complete and partial responses [CR + PR]) following nonmyeloablative allogeneic HSCT. Onset of disease regression was delayed a median 133 days (range, 30–287 days) following HSCT. (B) Disease responses (complete responses plus partial responses) were only observed in RCC patients with clear-cell histology (48% versus 0% cumulative incidence of a disease response; P = 0.0018). (C) Patients with clear-cell histology had longer survival after HSCT compared with those with non-clear-cell RCC (median survival, 525 versus 273 days; P = 0.003).
Assessing for tumor-reactive T cells after transplantation.
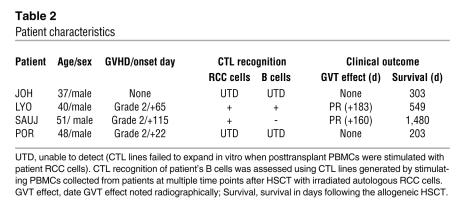
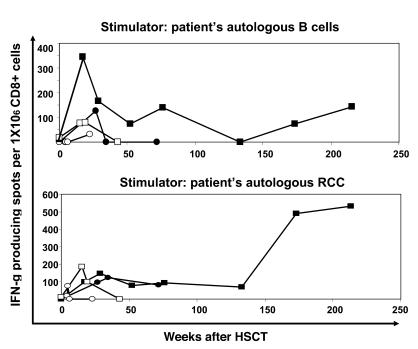
RCC tumor lines were established from the biopsy samples of 4 patients (SAUJ, LYO, JOH, and POR) who underwent HSCT (Table 2). Patients SAUJ and LYO had a partial response with delayed regression of tumor first observed radiographically on post-HSCT days +183 and +160, respectively. Patients JOH and POR had tumor growth after HSCT without evidence for a GVT effect. PCR chimerism analysis of short tandem repeats showed that T cells were 100% donor in origin at least 45 days following HSCT in all 4 patients (data not shown). ELISPOT analysis for interferon (Figure 2) showed that CD8+ T cells that recognized patient B cells were present in the blood of all 4 patients after HSCT, with the highest precursor frequency of alloreactive T cells detected within the first few months following transplantation.
Table 2 .
Patient characteristics
Figure 2. Detection of RCC-reactive CD8+ T cells in PBMCs after transplantation.
The frequency of CD8+ T cells in PBMCs before and at multiple time points after HSCT that recognized patient B cells or patient RCC cells was measured by an IFN-γ ELISPOT analysis; SAUJ (filled squares), LYO (filled circles), JOH (open squares), and POR (open circles). Alloreactive CD8+ T cells that recognized patient B cells were absent at baseline, then became detectable in all 4 patients after HSCT, with the highest precursor frequency measured in the first few months after transplantation. RCC-reactive T cells were absent before HSCT in patients but became detectable after HSCT in 3 of 4 RCC patients. In patient JOH, who did not have a disease response, these populations were detected only transiently. In contrast, patients LYO and SAUJ, who had evidence for a GVT effect, had RCC-reactive T cells detected for more than 1.5 and 4 years, respectively, after HSCT.
RCC-reactive T cells were absent in patients and in their respective donors prior to transplantation. However, shortly following HSCT, RCC-reactive CD8+ T cells became detectable in all 4 RCC patients (Figure 2). In patients JOH and POR, who failed to have a disease response, these populations were detected only transiently, then disappeared. In contrast, patients LYO and SAUJ had a prolonged disease response, with RCC-reactive T cells remaining detectable for 1.5 and more than 4 years, respectively, following HSCT. These data show that donor CD8+ T cells that recognize patient RCC cells expand in vivo in patients with metastatic RCC who have failed to mount an autologous antitumor T cell response.
Expansion of RCC-reactive CTLs from posttransplant PBMCs.
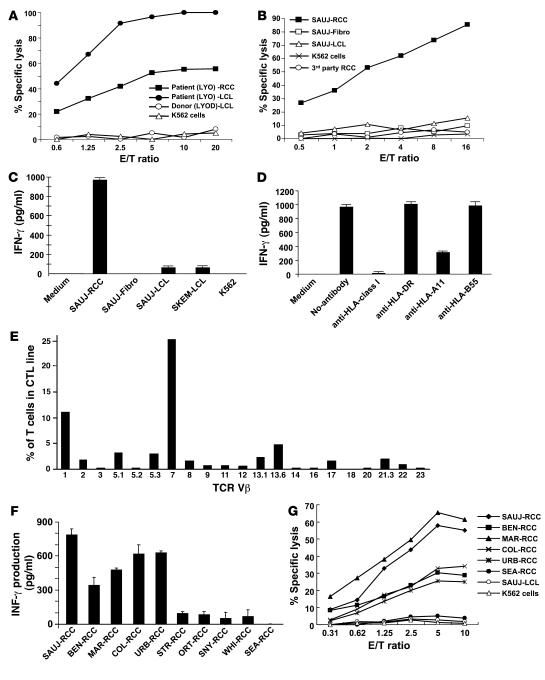
To characterize antigens serving as potential targets for a GVT effect, PBMCs were collected from RCC patients at multiple time points after transplantation and were stimulated in vitro with either irradiated autologous RCC cells or the patient’s EBV-transformed lymphoblastoid cell lines (LCLs). Despite multiple in vitro stimulations, RCC-reactive CTL lines could not be generated from PBMCs collected after HSCT from the 2 nonresponding patients, POR and JOH (Table 2). Furthermore, an mHa-specific CTL line generated by stimulating POR-PBMCs (collected on posttransplant day +96: donor in origin) with patient LCL cells in vitro lysed 98% of patient B cells (20:1 effector/target ratio) but did not kill patient autologous RCC cells (data not shown). In contrast, CTL lines that were cytotoxic to patient tumor cells could be expanded from both patients who had evidence for a GVT effect. Patient LYO was noted to have a delayed regression of lung metastasis by CT scan on posttransplant day +183, shortly following a donor lymphocyte infusion. A CTL line from patient LYO was expanded by stimulating PBMCs obtained during tumor regression (day +211 after transplant; donor in origin) with irradiated patient RCC cells; LYO-CTLs (98% CD3+CD8+) had in vitro cytotoxicity against both patient LYO-LCL and LYO-RCC cells (Figure 3A). Following limiting dilution cloning, an HLA class I–restricted CD8+ T cell clone (LYO–clone 1) was isolated that was cytotoxic to both LYO-LCL and LYO-RCC cells, consistent with recognition of an mHa expressed on both the patient’s hematopoietic cells and RCC cells (Figure 4A).
Figure 3. Characterization and generation of tumor-reactive CTLs.
CTLs and T cell clones that killed patient RCC cells were isolated using PBMCs collected after transplant and stimulated in vitro with irradiated patient tumor cells. (A) A CTL line was expanded from RCC patient LYO by stimulating posttransplant day +211 PBMCs collected following tumor regression with irradiated LYO-RCC cells; a 51Cr release assay showed these CTLs had in vitro cytotoxicity against both patient LYO-LCL and LYO-RCC cells but not donor (LYOD) LCL cells. (B) SAUJ-CTLs generated by stimulating SAUJ day +1,213 PBMCs with irradiated SAUJ RCC cells killed SAUJ-RCC cells but not SAUJ-LCL cells, SAUJ-fibroblasts (SAUJ-Fibro), K562 cells, or a third-party HLA-mismatched RCC cell line. (C) SAUJ-CTLs secreted IFN-γ when cultured with patient SAUJ-RCC cells but not with SAUJ-Fibro, K562 cells, or patient (SAUJ) or donor (SKEM) LCL cells. (D) IFN-γ production by the SAUJ-CTLs following coculture with SAUJ-RCC cells was inhibited by incubation with anti–HLA class I and anti–HLA-A11 mAbs. (E) Flow cytometry revealed CD8+TCR-Vβ7+ cells to be the dominant T cell population in the SAUJ-CTL line. (F) SAUJ-CTL clone BZ-4 was cocultured with various HLA-A11+ RCC cell lines, with tumor recognition assessed by an ELISA measuring IFN-γ secretion; the BZ-4 clone recognized 5 of 10 HLA-A11+ RCC cell lines. (G) A cytotoxicity assay showed that the BZ-4 clone also killed all 5 HLA-A11+ RCC lines recognized in the ELISA assay but not SAUJ-LCL or K562 cells.
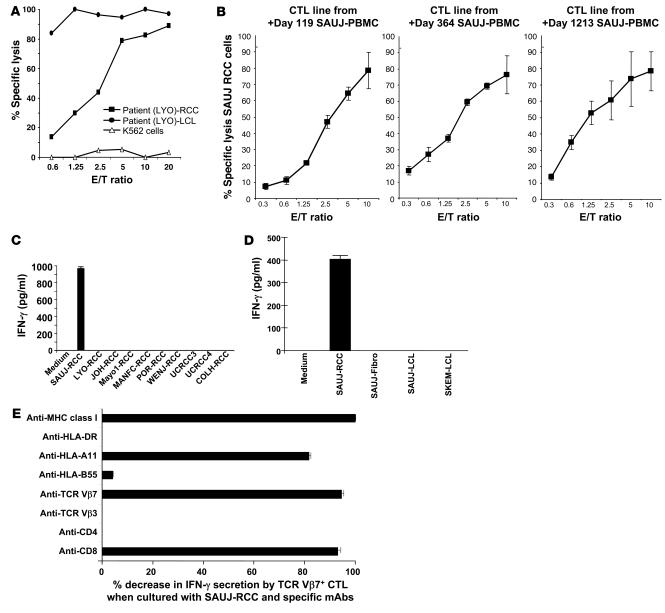
Figure 4. Detection of RCC-reactive CD8+ T cells in PBMCs after transplantation.
(A) A CTL line was expanded from RCC patient LYO by stimulating posttransplant day +211 PBMCs collected following tumor regression with irradiated LYO-RCC cells. Following limiting dilution cloning, an HLA class I–restricted CD8+ T cell clone (LYO-clone 1) was isolated that was highly cytotoxic to both patient autologous LCL and RCC cells. (B) CTL lines were expanded from patient SAUJ by stimulating PBMCs collected on post-HSCT days +119, +364, and +1,213 with irradiated SAUJ-RCC cells; expanded CTLs from all 3 time points were highly cytotoxic to SAUJ-RCC cells. (C) An ELISA showed that day +1,213 SAUJ-CTLs secreted IFN-γ when cultured with SAUJ-RCC cells but not with third-party HLA-mismatched RCC cell lines. (D) An ELISA assay measuring IFN-γ secretion showed that flow-sorted TCR-Vβ7+ SAUJ-CTLs recognized SAUJ-RCC cells but not patient SAUJ-LCL or donor SKEM-LCL cells or fibroblasts (SAUJ-Fibro). (E) Coculture of SAUJ-RCC cells with TCR-Vβ7+ SAUJ-CTLs. IFN-γ secretion by ELISA decreased substantially when SAUJ-CTLs were pretreated with either anti-CD8 or anti–TCR-Vβ7 mAbs or when SAUJ-CTLs were cocultured with SAUJ-RCC cells in the presence of anti–HLA class I or anti–HLA-A11 mAbs.
On posttransplant day +160, RCC patient SAUJ had regression of lung metastasis first noted by CT scan. An ELISPOT assay showed that RCC-reactive CD8+ T cells expanded early following transplantation and persisted in the blood for more than 4 years following HSCT (Figure 2). In contrast, RCC-reactive T cells were undetectable in the PBMCs of both the donor and the patient before transplantation. PBMCs collected from patient SAUJ after transplantation were stimulated in vitro with irradiated SAUJ-RCC cells; CTL lines expanded from PBMCs collected on posttransplant days +119, +364, and +1,213 and were cytotoxic to SAUJ-RCC cells in vitro (Figure 4B). The SAUJ day +1,213 CTL line continued to expand well in vitro and was used for further experiments to identify target antigens expressed on SAUJ-RCC cells. These CTLs consisted mostly of CD8+ T cells and had a cytotoxicity pattern consistent with tumor-restricted recognition, lysing SAUJ-RCC cells but not SAUJ-LCL cells, SAUJ-fibroblasts (SAUJ-Fibro), K562 cells, or a third-party HLA-mismatched RCC cell line (Figure 3B). An ELISA showed that day +1,213 SAUJ-CTLs secreted IFN-γ when cultured with SAUJ-RCC cells but not with SAUJ-Fibro, K562 cells, patient (SAUJ-LCL) or donor (SKEM-LCL) LCL cells (Figure 3C), or HLA-mismatched RCC cell lines (Figure 4C). These results suggested that SAUJ-CTLs, in contrast to the LYO-CTLs, recognized an antigen restricted to RCC. IFN-γ production by SAUJ-CTLs dropped substantially when SAUJ-RCC targets were pretreated with anti–HLA class I and/or HLA-A11 mAbs, suggesting that CTL recognition of the SAUJ-RCC antigen occurred within the context of the MHC class I restricting allele HLA-A11 (Figure 3D).
Flow cytometry revealed TCR-Vb7+CD8+ cells to be the dominant T cell population in the SAUJ-CTL line (Figure 3E). Flow-based intracellular cytokine staining revealed that TCR-Vb7+CD8+ cells secreted IFN-γ when cocultured with SAUJ-RCC cells (data not shown). A greater than 99% pure population of TCR-Vb7+CD8+ SAUJ-CTLs was subsequently isolated from this CTL line by flow sorting. An ELISA measuring IFN-γ secretion showed that Vb7+-sorted SAUJ-CTLs recognized SAUJ-RCC cells but not SAUJ-LCL or donor SKEM-LCL cells or SAUJ-Fibro (Figure 4D). Tumor-reactive TCR-Vb7+T cells within the SAUJ-CTL line were found to have CD8-dependent recognition of an RCC antigen presented in the context of HLA-A11 (Figure 4E).
Limiting dilution T cell cloning was performed on the TCR-Vb7+ SAUJ-CTL line. The HLA-A11–restricted T cell clone BZ-4, confirmed to be TCR-Vb7+ and CD8+ by flow cytometry, expanded well in vitro, allowing further characterization of its antigen specificity. Remarkably, BZ-4 clone secreted IFN-γ and induced high levels of cytotoxicity against 5 of 10 (50%) HLA-A11+ RCC cell lines (Figure 3, F and G). Taken altogether, these data show that the Vb7+CD8+ SAUJ-CTL line and BZ-4 T cell clone isolated from a patient who had regression of RCC after allogeneic HSCT recognized an HLA-A11–restricted tumor antigen that was expressed on a substantial percentage of RCC cell lines.
Identification of a cDNA clone.
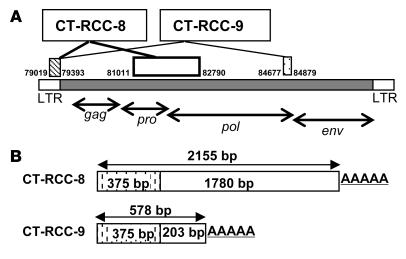
A cDNA library was constructed from the SAUJ-RCC line. SAUJ BZ-4 T cells and TCR-Vb7+CD8+ SAUJ-CTLs were used in an ELISA to screen for recognition of SAUJ-RCC tumor cDNAs transfected into COS7 cells stably expressing the HLA-A11–restricted allele transgene. After repeated screening, 2,155-bp- and 578-bp-long cDNAs were identified to be recognized by SAUJ-CTL clones. Each cDNA shared a common sequence of 375 bp, and each had a unique region spliced to the common region (Figure 5). These sequences were found to have 100% homology with the HERV-E locus on chromosome 6q and were subsequently named CT-RCC-8 and CT-RCC-9. The spliced nonshared regions of CT-RCC-8 and CT-RCC-9 were predicted to be derived from the protease and polymerase regions, respectively, of that HERV-E.
Figure 5. Schematic drawing of the identified clones CT-RCC-8 and CT-RCC-9 and their localization on chromosome 6q.
(A) CT-RCC-8 and CT-RCC-9 are shown with the relative positions of the common region, exons, and the nucleotide numbers corresponding to those of the clone RP3-488C13 on chromosome 6q (GenBank accession number AL133408). gag, capsid region; pro, protease region; pol, polymerase region; env, envelope region. (B) The striped boxes represent the 375-bp common region, and the open and dotted boxes represent the unique regions of CT-RCC-8 (1,780 bp) and CT-RCC-9 (203 bp), respectively.
Identification of the antigenic peptide encoded by CT-RCC-8 and CT-RCC-9.
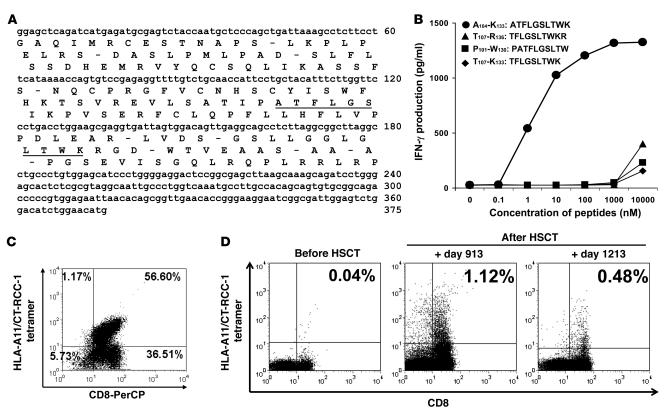
To identify the RCC antigen recognized by tumor-reactive CTLs, plasmids carrying progressively shorter lengths of the common sequence region of CT-RCC-8 and -9 were constructed and transfected into COS7-A11 cells and then screened with SAUJ-CTL lines by an ELISA assay. Four minigenes encoding 4 different candidate 9- or 10-amino-acid peptides were recognized by SAUJ-CTLs (data not shown). The 4 candidate peptides were subsequently predicted and synthesized from the same open reading frame of the common region of CT-RCC-8 and -9 (Figure 6A); the 10-mer peptide ATFLGSLTWK (named CT-RCC-1) induced dose-dependent CTL secretion of IFN-γ at concentrations of at least 1 nM, consistent with this peptide being the HLA-A11–restricted antigen recognized by the RCC-reactive SAUJ-CTLs (Figure 6B).
Figure 6. Identification of the peptide recognized by CTL expanded from a responding patient and detection of circulating CT-RCC-1 peptide–specific T cells after HSCT.
A 10-amino-acid HERV-E–derived peptide (CT-RCC-1) expressed on RCC was identified to be the target antigen of tumor-reactive CTL. (A) The position of the peptide was identified to be located in frame 2 of the 375-bp CT-RCC common region. (B) Four candidate peptides were synthesized from the predicted amino acid translations of these minigenes; only the 10-mer peptide ATFLGSLTWK induced dose-dependent IFN-γ production by SAUJ-RCC–reactive CTLs. (C) RCC-reactive SAUJ-CTL was generated by stimulating SAUJ-PBMCs (day +1,213) with irradiated SAUJ-RCC cells followed by flow sorting for TCR-Vβ7+CD8+ T cells. These CTLs were stained with a PE-conjugated HLA-A*1101/CT-RCC-1 (ATFLGSLTWK) tetramer; 56.6% of the CD3+CD8+ cells in this CTL line had antigen specificity for the CT-RCC-1 peptide. (D) PBMCs collected from SAUJ before HSCT did not bind to the CT-RCC-1 tetramer. CT-RCC-1–specific T cells were detected by tetramer analysis after HSCT in patient SAUJ on days +913 and +1,213 following tumor regression, constituting 1.12% and 0.48% of the CD3+CD8+ T cell repertoire, respectively.
Next, a PE-conjugated HLA-A11/CT-RCC-1 tetramer was synthesized to evaluate for CT-RCC-1–reactive T cells in vivo; 56.6% of the TCR-Vb7+CD8+ SAUJ-CTL line established from SAUJ-PBMCs (day +1,213) bound to the CT-RCC-1, confirming the specificity of the tetramer for CT-RCC-1–reactive T cells (Figure 6C). Tetramer analysis showed that CT-RCC-1–specific T cells were absent in the blood of patient SAUJ before HSCT but became detectable in the posttransplant setting (Figure 6D), constituting 1.12% and 0.48% of the CD3+CD8+ T cell repertoire of PBMCs collected on posttransplant days +913 and +1,213, respectively.
Expression of HERV-E–derived CT-RCC-8 and CT-RCC-9 in RCC and other tissues.
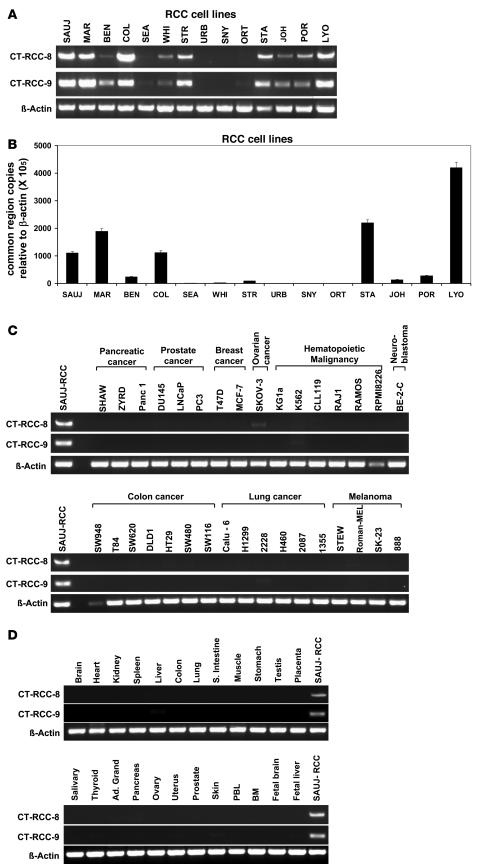
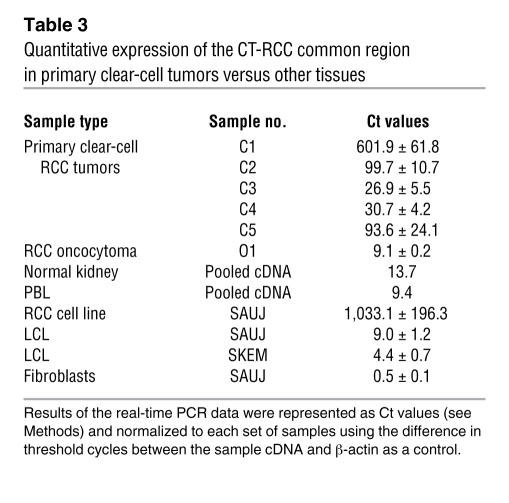
Semiquantitative RT-PCR for expression of CT-RCC-8 and -9 was performed on cDNAs generated from different RCC cell lines and non-RCC tumors, as well as pooled cDNAs from 24 normal human tissues. CT-RCC-8 and -9 were detected in 8 of 14 RCC cell lines, including SAUJ-RCC cells from which the original cDNA library was constructed (Figure 7A). Quantitative real-time PCR using CT-RCC common region–specific primers showed that the transcripts carrying common region were detectable in these same 8 RCC cell lines at varying levels (Figure 7B). Neither CT-RCC-8 nor CT-RCC-9 was detected by RT-PCR in a variety of different non-RCC tumor cell lines (Figure 7C). Neither transcript was detected by RT-PCR in pooled cDNAs obtained from 24 normal tissues, including the kidney and testis (Figure 7D). The CT-RCC common region was also detected by real-time PCR in freshly resected RCC tumors of clear-cell histology (Table 3); in contrast, minimal to no expression of the CT-RCC common region was detected in a fresh oncocytoma specimen (obtained by nephrectomy), SAUJ-fibroblasts, patient (SAUJ) and donor (SKEM) LCL cells, and pooled cDNAs obtained from normal kidneys, PBLs, and 48 different normal human tissue panels.
Figure 7. Expression of CT-RCC-8 and CT-RCC-9 in tumors and nonmalignant tissues.
CT-RCC-8 and CT-RCC -9 were found to be expressed only in RCC cells and not a variety of different non-RCC tumor cell lines or in any normal tissues. (A) Semiquantitative RT-PCR for CT-RCC-8 and -9 was performed using cDNAs prepared from 14 different human RCC tumor lines. Both CT-RCC-8 and -9 were detected in 8 of 14 RCC cell lines. (B) CT-RCC common region transcripts were detectable in the same 8 RCC cell lines at variable levels by quantitative real-time RT-PCR analysis using primers and probes specific to CT-RCC common region. (C) Lack of expression of CT-RCC-8 and -9 transcripts by RT-PCR in a variety of different non-RCC malignant cell lines. (D) Neither transcript was detected by RT-PCR in pooled cDNAs obtained from 24 normal human tissues, including the kidney and testis. β-Actin was used as an internal control, and cDNA from SAUJ-RCC was used as a positive control.
Table 3 .
Quantitative expression of the CT-RCC common region in primary clear-cell tumors versus other tissues
Discussion
In this follow-up to our pilot trial, we observed regression of metastatic RCC in 38% of patients who underwent a nonmyeloablative allogeneic HSCT for metastatic RCC. Although clinical evidence suggests that T cells mediate graft-versus-RCC effects, difficulty in generating tumor lines from patients undergoing transplantation has hampered in vitro studies aimed at characterizing the antigens targeted by donor T cells. In this study, we provide insight into the mechanisms accounting for RCC regression after nonmyeloablative HSCT. We found evidence suggesting that both broadly expressed mHas and antigens restricted to the tumor are targets for donor T cells isolated from patients with evidence for a graft-versus-RCC effect. Using PBMCs collected from both responding patients, we succeeded in expanding the CTLs, which were cytotoxic to patient autologous RCC cells in vitro. In one of these responders (LYO), the CTLs had a cytotoxicity pattern consistent with recognition of an antigen that was not restricted to the tumor, lysing both patient autologous LYO-LCL cells and LYO-RCC cells but not donor LYOD-LCL cells. Following limiting dilution cloning, we identified a CD8+ T cell clone with a similar cytotoxicity pattern that lysed both LYO-RCC cells and LYO-LCL cells but not donor LYOD-LCL cells. These findings are consistent with donor T cell recognition of mHa expressed broadly on both tumor cells and hematopoietic cells of the recipient. Tykodi et al. (4) previously demonstrated that mHa-specific T cell clones cytotoxic to RCC cells in vitro could be isolated from both responding and nonresponding patients with metastatic kidney cancer following an allogeneic HSCT. Such mHas are known to be expressed on a variety of hematological malignancies and are thought to be dominant targets of transplanted donor T cells mediating GVL effects. Our data confirm that mHas absent in the donor but present in the patient are expressed on patient RCC cells. The isolation of an mHa-specific T cell clone from a responding patient that killed patient RCC cells in vitro suggests that donor T cells were primed in vivo to this antigen after HSCT and provide evidence that mHas may be a target for a graft-versus-RCC effect. The detection of mHa-specific CTLs from a responding patient that kill patient RCC cells in vitro is also consistent with the clinical observation that GVT effects occur more frequently in patients who develop GVHD.
In this analysis, we also detected RCC-reactive T cells by ELISPOT in the first several months after HSCT in all 4 patients. However, in the 2 nonresponders, these T cells were detected only transiently. In contrast, in the 2 patients who had tumor regression, RCC-reactive T cells persisted in the blood over a prolonged interval. These findings suggest factors that eliminate or sustain RCC-reactive T cells that expand after HSCT may have a critical impact on the ability to generate graft-versus-RCC tumor effects.
In patient SAUJ, who had a GVT effect against metastatic RCC associated with survival of more than 4 years after HSCT, CD8+ CTL lines and T cell clones were expanded from blood collected after transplantation that had tumor-specific cytotoxicity in vitro. Using cDNA expression cloning, we identified 2 transcripts (CT-RCC-8 and CT-RCC-9) encoding an antigen recognized by HLA-A11–restricted RCC-reactive T cells expanded from a responding patient after HSCT. We subsequently identified a 10-amino-acid peptide antigen (CT-RCC-1) that was encoded from the shared common sequence region of CT-RCC-8 and CT-RCC-9 that was recognized by a T cell clone derived from these RCC-reactive CTLs. Remarkably, CT-RCC-1–specific CTLs recognized approximately 50% of RCC cell lines that expressed HLA-A11 but not patient fibroblasts or patient LCL cells, suggesting this antigen is commonly expressed in RCC tumors at levels that induce CTL killing but not in normal tissues. Semiquantitative RT-PCR for expression of CT-RCC-8 and -9 and real-time PCR for their shared common sequence region showed that these transcripts were expressed at variable levels in fresh kidney cancer samples and in greater than 50% of cultured RCC cell lines but not in pooled cDNA from normal tissues including the kidneys and testis nor in a number of other non-RCC tumor lines.
The nucleotide sequences of CT-RCC-8 and -9 matched with genomic sequences of an HERV-E located on chromosome 6q that was not previously known to be expressed in human cells. HERVs exist widely within the human genome as proviruses, with most being transcriptionally inactive (6, 7). Recently, some HERVs were identified to have transcriptionally active components with biological activity expressed widely in human tissues; Seifarth et al. (8) showed that an HERV-E clone, 4-1, had active transcription in many normal tissues. In contrast, the transcriptional products derived from the HERV-E that we identified were selectively expressed in RCC and were not detected in normal tissues.
HERV-K transcripts have been found to be expressed in tissue malignancies including melanoma, teratocarcinoma, and prostate cancer as well as hematological malignancies (9–12). Although more than 50 HERV-Es are estimated to exist in the human genome, this report is, to our knowledge, only the second to identify an HERV-E transcription product expressed in tumor cells (13) and the first to identify an HERV-E expressed in RCC. Furthermore, unlike HERV-K expression, which can be detected not only in tumors but also in normal tissues such as the testis, skin cells (12), and blood cells (10), the sequences derived from this HERV-E appear to be selectively expressed in RCC cells, with virtually undetectable levels of expression in normal tissues, making them potentially ideal targets for tumor immunotherapy.
Recently, CTLs recognizing HERV-K–derived peptides that kill tumor cells in vitro have been identified in a few cancer patients (12, 14). Similarly, we isolated and expanded a T cell clone from the blood of a responding transplant patient that recognized an HERV-E–derived antigen and killed patient RCC cells and HLA-A11+ RCC cells in vitro. The HLA-A11–restricted 10-mer peptide named CT-RCC-1 was identified to be the target antigen of these CTLs that had RCC-specific cytotoxicity. Tetramer analysis showed that CT-RCC-1–specific T cells were absent at baseline but were detected after HSCT following regression of metastatic disease, suggesting that this antigen had immunogenicity in vivo. To the best of our knowledge, this is the first report to identify a T cell population recognizing an HERV-derived antigen with expression restricted to tumor cells.
Because the phenotypic frequency of HLA-A11 expression is low (15), immunotherapy approaches targeting the CT-RCC-1 antigen through tumor peptide vaccination or the adoptive infusion of CT-RCC-1–specific CTLs would be limited to a minority of patients with metastatic kidney cancer. The full protein products translated from CT-RCC-8 and -9 and other genes derived from this HERV-E have not yet been defined. It is possible, therefore, that other immunogenic peptides derived from this HERV could be expressed on more common HLA class I molecules, a finding that potentially would broaden the application of immunotherapy approaches targeting antigens derived from this HERV to a greater percentage of patients with metastatic RCC.
The factors regulating expression of the CT-RCC antigens in RCC are currently unknown. However, the observation that CT-RCC-8 and CT-RCC-9 were not detected in a variety of hematological malignancies and other solid tumors suggests that genetic mutations specific to RCC (i.e., the von Hippel–Lindau [VHL] gene) may in part regulate expression of transcripts derived from this HERV.
Clear-cell RCC is the dominant histological subtype of kidney cancer, accounting for approximately 75% of the cases of spontaneously occurring RCC. Loss of function of the VHL tumor suppressor gene from a mutation or as a result of promoter hypermethylation occurs in nearly 80% of these tumors, which ultimately leads to increased protein expression of HIF-1α and -2α, resulting in overexpression of more than 150 client genes that play a fundamental role in tumor progression, spread, and response to hypoxia (prevalent in tumor tissues) (16). Recent data suggest that clear-cell carcinoma may represent the subtype of kidney cancer that is immunoresponsive, with some studies reporting virtually no responses to IL-2–based immunotherapy occurring in patients with non-clear-cell RCC (17, 18). Although it represented the dominant histological subtype in our transplant trial, it is nonetheless interesting that GVT effects following HSCT were only observed in patients with clear-cell RCC (48% cumulative response rate), with no responses seen in the 14 patients with non-clear-cell tumors. One hypothesis to explain these findings would be that loss of VHL function occurring in clear-cell tumors may result in aberrant expression of CT-RCC HERV-derived transcription products, shown here to be a target for transplanted donor T cells, which theoretically could also be a target for autologous T cells. A histological review of the RCC cell lines and fresh RCC tissues used in experiments presented in this article showed all to be clear-cell carcinomas, with more than half expressing HERV-E transcripts. Furthermore, limited preliminary data from an ongoing study of fresh tumors suggest that this HERV-E may have transcriptional activity limited to the clear-cell variant of kidney cancer (unpublished observations), which is intriguing given the track record for this tumor being the immunoresponsive subtype of RCC.
The unavailability of fresh RCC cells in the majority of patients who received transplants in this study precluded an RT-PCR–based analysis to correlate tumor HERV-E expression with a GVT effect. The development of antibodies that recognize the CT-RCC-1 antigen and/or other peptides and proteins derived from this HERV would permit such an analysis, where immunohistochemistry on paraffin-preserved tumor samples could be used to discern HERV expression. Furthermore, because HLA-A11, the MHC class I restricting allele for the CT-RCC-1 antigen, is present in only a minority of the population, including a small subset of transplanted patients in this transplant series, an analysis correlating response to the expansion of CD8+ CT-RCC-1–specific T cells could not be performed. The identification of other immunogenic peptides derived from CT-RCC-8 and -9 and other gene sequences of this HERV-E that are expressed on more common HLA class I molecules would allow for a study correlating disease response to the generation of CD8+ T cells recognizing HERV-E antigens. Further, it would permit an analysis to discern whether HERV-E antigens represent a target antigen of tumor-infiltrating lymphocytes in fresh kidney cancer tumors.
In conclusion, this study provides insight into the immunological mechanisms accounting for regression of RCC following allogeneic HSCT and provides the first evidence to our knowledge that GVT effects against this malignancy may be associated with tumor-specific immune responses to antigens expressed on RCC cells. The expansion of CT-RCC antigen–specific T cells in a patient who had a GVT effect associated with prolonged regression of metastatic RCC suggests that gene products derived from HERV-E are immunogenic in vivo and may be a novel target for RCC immunotherapy.
Methods
Transplant approach.
Seventy-four patients with metastatic RCC underwent nonmyeloablative transplantation from an HLA-matched or single antigen–mismatched sibling donor at the NHLBI as previously been described (5). Characteristics of the 4 transplanted patients whose tumor and blood samples were used in this study’s in vitro analysis are shown in Table 2.
Cell lines and HLA data.
The human RCC cell lines SAUJ- (HLA type A11/11, B51/55, Cw1/w7), JOH-, LYO-, POR-, STA-, Mayo1-, MANFC-, WENJ-, MAR-, BEN-, and COLH-RCC, as well as UCRCC3 and UCRCC4 were established from surgically resected tumors procured at the NHLBI and the NCI (samples procured with informed consent of patients according to Institutional Review Board–approved protocols 97-H-0196 and 97-C-0147). Seven RCC cell lines from HLA-A11–negative patients, which were stably transfected to express HLA-A11 (COL-, SEA-, WHI-, STR-, URB-, SNY-, and ORT-RCC) were a gift from Qiong Wang at NCI/NIH. Pathology reports obtained on primary surgical specimens showed that all RCC cell lines were generated from tumors with clear-cell histology. The fibroblast cell line SAUJ-Fibro was established from a skin biopsy sample from patient SAUJ. Expansion of EBV-LCL B cells was performed as previously described (19).
ELISPOT assay.
PBMCs were incubated at 5 × 104 cells/well overnight at 37°C in 5% CO2 in RPMI 1640 (Cellgro) containing 10% FCS in the absence or presence of the same number of stimulator cells. The next day, cells were removed by washing buffer with PBS/0.05% Tween-20 and biotinylated anti–IFN-γ mAb, and after addition of 7-B6-1biotin (1 mg/ml; Mabtech), the cells were left for 2 hours at room temperature, followed by incubation with streptavidin-alkaline phosphatase (Mabtech) for an additional 1 hour. Individual cytokine-producing cells were detected after a 10-minute reaction with 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium alkaline phosphatase substrate (Sigma-Aldrich). Colored spots were counted using a KS-ELISPOT microscopy system (Carl Zeiss).
Generation of CTLs.
PBMCs were prepared by Ficoll density gradient centrifugation and cultured in RPMI 1640 medium supplemented with 10 ng/ml recombinant human IL-15 (rhIL-15) (R&D Systems), 10% human AB serum (Gemini Bio-Products), 25 mM HEPES, 100 U/ml penicillin, and 100 μg/ml streptomycin. CTL lines were established by repeatedly stimulating PBMCs with irradiated (200 Gy) RCC tumor cells at 7- to 10-day intervals. Bulk CTLs were cloned by limiting dilution (0.3, 1, or 3 cells/well) as previously described (19). T cell clones were expanded by resuspending in 25 ml of AIM-V medium (GIBCO; Invitrogen) containing 300 IU/ml rhIL-2, 30 ng/ml OKT3, 2.5 × 106 irradiated (50 Gy) PBMCs as allogeneic feeder cells from a healthy volunteer.
Cytotoxicity and ELISA assays.
IFN-γ or GM-CSF secretion was measured by Human GM-CSF and INF-γ Colorimetric ELISA kit (Pierce) following the manufacturer’s protocol. For the inhibition of IFN-γ secretion assay, the following mAbs were used: anti–MHC class I mAb (DAKO); anti–HLA-A11 and HLA-B55 mAb (One Lambda Inc.); anti–HLA-DR mAb (BD); and anti–TCR-Vβ7, –TCR-Vβ3, and -CD8 (BD).
51Cr release assays were performed as previously described (19).
Construction and screening of SAUJ-RCC cDNA library.
Construction and screening of cDNA library were performed as previously described (20). Briefly, after cDNA synthesis and ligation into the vector pME-SMARTvector, pooled plasmids were transfected into COS7-A11 cells using Lipofectamine 2000 (Invitrogen) in 96-well plates for 24 hours. SAUJ-CTL clone BZ-4 was added and incubated for 24 hours, the supernatants were harvested, and GM-CSF production was measured using an ELISA. Plasmids that carried cDNA clones recognized by tumor-reactive CTLs when transfected into COS-7-A11 were isolated and sequenced using the BigDye Terminator version 3.1 Ready Reaction Kit and ABI Prism 3100 Genetic Analyzer (Applied Biosystems), according to the manufacturer’s protocol. The sequences are deposited in GenBank under accession numbers EU137846 and EU137847.
Identification of the antigenic peptides.
A total of 4 peptides were synthesized and purchased from Genemed Synthesis Inc. The amino acid sequences were as follows: TFLGSLTWK, ATFLGSLTWK, TFLGSLTWKR, PATFLGSLTWK. COS7-A11 cells were loaded with each peptide, and IFN-γ secretion by the SAUJ-CTL clone BZ-4 following coculture with peptide-pulsed COS7-A11 cells was measured by ELISA.
RNA isolation, synthesis of cDNA, and RT-PCR analysis.
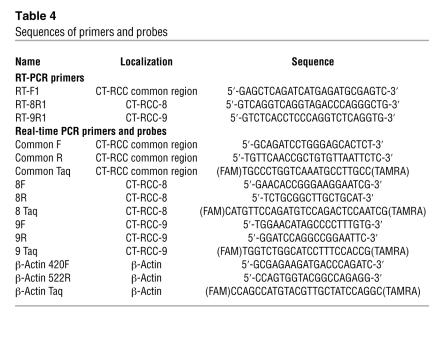
Total cellular RNAs were extracted using the RNeasy Mini RNA Purification Kit (QIAGEN). Kidney tumors obtained from nephrectomy samples frozen in OCT (obtained with informed consent according to protocol 97-C-0147) were used to assess CT-RCC expression in fresh RCC tumors. Slides were made from frozen sections of fresh tumor; tumor cells were microdissected under light microscopy by a pathologist (Maria Merino, NCI/NIH), and total cellular RNAs were obtained using the PicoPure RNA Isolation Kit (Arcturus). cDNAs were synthesized using random primer and the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). For detection of CT-RCC gene expression in human normal tissues, Human 24 Tissue RapidScan gene expression cDNA panels (Oligene) were used. Semiquantitative RT-PCR was done using TaKaRa LA Taq (TaKaRa Bio) and 200 nM of primers (Table 4). PCR conditions were: 95°C, 2 minutes; (95°C, 30 seconds; 55°C or 52°C, 30 seconds; 72°C, 1 minute) × 30 cycles. Quantitative real-time RT-PCR was carried out in 7500 Fast Real-Time PCR System (Applied Biosystem) using primers and probes shown in Table 4. Primers were synthesized by Integrated DNA Technologies (IDT), and TaqMan probes were synthesized by Applied Biosystem or IDT. All data were analyzed using the 7500 System Sequence Detection Software. We normalized each set of samples using the difference in threshold cycles (ΔCt) between the sample cDNA and β-actin. Relative transcripts levels were calculated by the expression 2–ΔΔCt where ΔΔCt = ΔCtsample – ΔCtβ-actin (according to AppliedBiosystems technical protocol).
Table 4 .
Sequences of primers and probes
Flow cytometry and tetramer analysis for CT-RCC-1 peptide–specific T cells.
For the analysis of T cell TCR-Vβ usage, 22 different mAbs of Vβ families (Beckman-Coulter) were used. PE-conjugated HLA-A*1101/CT-RCC-1 (ATFLGSLTWK) tetramer was synthesized by the National Institute of Allergy and Infectious Diseases Tetramer Facility, Emory University Vaccine Center at Yerkes. Cells were stained with the tetramer (1:800 diluted) for 30 minutes at room temperature, then allophycocyanin-conjugated anti-CD3 mAb and peridinin-chlorophyll-protein complex–conjugated (PerCP-conjugated) anti-CD8 mAb were added and incubated for an additional 15 minutes at 4°C. LIVE/DEAD fixable dead cell stain kit (Molecular Probes; Invitrogen) was used for uncultured and frozen PBMCs to analyze only live cells by FACScalibur (BD) or CYAN MLE (Dako-Cytomation). Data analysis was performed using FCS Express V3 (De Novo Software).
Statistics.
Survival distributions were estimated using the Kaplan-Meier method, and comparisons between histology groups were made using a log-rank test. Time to response was estimated using cumulative incidence methods, with death considered to be a competing risk. Comparison of the cumulative incidence of time to response was made using the c2 test. Hypothesis testing was 2-sided; a P value less than 0.05 was considered significant.
Acknowledgments
This work was supported by the intramural program of the NHLBI, NIH. We wish to acknowledge the Dean R. O’Neill Memorial Fellowship for contributions supporting this research and N. Harashima’s postdoctoral fellowship; and the Action to Cure Kidney Cancer (ACKC) for contributions supporting E. Cherkasova’s research fellowship. We also wish to thank Marston Linehan (NCI/NIH), Maria Merino (NCI/NIH), Anthony Suffredini (Clinical Center/Critical Care Medicine Department/NIH), Neal Young (NHLBI), John Barrett (NHLBI), and Maria Berg (NHLBI) for their assistance and invaluable insight related to these translational studies. We also thank the National Institute of Allergy and Infectious Diseases Tetramer Facility, Emory University Vaccine Center at Yerkes, for providing HLA-A*1101/CT-RCC-1 tetramer. We also wish to acknowledge Rose Goodwin, Aleah Smith, Martha Marquesen, Jennifer Karpovich, and many members of the NIH Department of Transfusion Medicine for their contributions to these clinical/translational research studies.
Footnotes
Nonstandard abbreviations used: GVHD, graft-versus-host disease; GVL, graft-versus-leukemia; GVT, graft-versus-tumor; HERV, human endogenous retrovirus; HSCT, hematopoietic stem cell transplantation; LCL, lymphoblastoid cell line; mHa, minor histocompatibility antigen; RCC, renal cell carcinoma; VHL, von Hippel–Lindau.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 118:1099–1109 (2008). doi:10.1172/JCI34409
Yoshiyuki Takahashi and Nanae Harashima contributed equally to this work.
References
- 1.Childs R., et al. Engraftment kinetics after nonmyeloablative allogeneic peripheral blood stem cell transplantation: full donor T-cell chimerism precedes alloimmune responses. Blood. 1999;94:3234–3241. [PubMed] [Google Scholar]
- 2.Bregni M., Ueno N.T., Childs R. The second international meeting on allogeneic transplantation in solid tumors. Bone Marrow Transplant. 2006;38:527–537. doi: 10.1038/sj.bmt.1705479. [DOI] [PubMed] [Google Scholar]
- 3.Sprangers B., Van Wijmeersch B., Fevery S., Waer M., Billiau A.D. Experimental and clinical approaches for optimization of the graft-versus-leukemia effect. Nat. Clin. Pract. Oncol. 2007;4:404–414. doi: 10.1038/ncponc0848. [DOI] [PubMed] [Google Scholar]
- 4.Tykodi S.S., et al. Allogeneic hematopoietic cell transplantation for metastatic renal cell carcinoma after nonmyeloablative conditioning: toxicity, clinical response, and immunological response to minor histocompatibility antigens. Clin. Cancer Res. 2004;10:7799–7811. doi: 10.1158/1078-0432.CCR-04-0072. [DOI] [PubMed] [Google Scholar]
- 5.Childs R., et al. Regression of metastatic renal-cell carcinoma after nonmyeloablative allogeneic peripheral-blood stem-cell transplantation. N. Engl. J. Med. 2000;343:750–758. doi: 10.1056/NEJM200009143431101. [DOI] [PubMed] [Google Scholar]
- 6.Lower R., Lower J., Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl. Acad. Sci. U. S. A. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffiths D.J. Endogenous retroviruses in the human genome sequence. Genome Biol. 2001;2:reviews1017. doi: 10.1186/gb-2001-2-6-reviews1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seifarth W., et al. Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray. J. Virol. 2005;79:341–352. doi: 10.1128/JVI.79.1.341-352.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buscher K., et al. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. 2005;65:4172–4180. doi: 10.1158/0008-5472.CAN-04-2983. [DOI] [PubMed] [Google Scholar]
- 10.Depil S., Roche C., Dussart P., Prin L. Expression of a human endogenous retrovirus, HERV-K, in the blood cells of leukemia patients. Leukemia. 2002;16:254–259. doi: 10.1038/sj.leu.2402355. [DOI] [PubMed] [Google Scholar]
- 11.Muster T., et al. An endogenous retrovirus derived from human melanoma cells. Cancer Res. 2003;63:8735–8741. [PubMed] [Google Scholar]
- 12.Schiavetti F., Thonnard J., Colau D., Boon T., Coulie P.G. A human endogenous retroviral sequence encoding an antigen recognized on melanoma by cytolytic T lymphocytes. Cancer Res. 2002;62:5510–5516. [PubMed] [Google Scholar]
- 13.Wang-Johanning F., et al. Detecting the expression of human endogenous retrovirus E envelop transcripts in human prostate adenocarcinoma. Cancer. 2003;98:187–197. doi: 10.1002/cncr.11451. [DOI] [PubMed] [Google Scholar]
- 14.Rakoff-Nahoum S., et al. Detection of T lymphocytes specific for human endogenous retrovirus K (HERV-K) in patients with seminoma. AIDS Res. Hum. Retroviruses. 2006;22:52–56. doi: 10.1089/aid.2006.22.52. [DOI] [PubMed] [Google Scholar]
- 15.Sidney J., Grey H.M., Kubo R.T., Sette A. Practical, biochemical and evolutionary implications of the discovery of HLA class I supermotifs. Immunol. Today. 1996;17:261–266. doi: 10.1016/0167-5699(96)80542-1. [DOI] [PubMed] [Google Scholar]
- 16.Keith B., Simon M.C. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129:465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrmann E., et al. Histologic subtype of metastatic renal cell carcinoma predicts response to combined immunochemotherapy with interleukin 2, interferon alpha and 5-fluorouracil. Eur. Urol. 2007;51:1625–1631. doi: 10.1016/j.eururo.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Upton M.P., Parker R.A., Youmans A., McDermott D.F., Atkins M.B. Histologic predictors of renal cell carcinoma response to interleukin-2-based therapy. J. Immunother. . 2005;28:488–495. doi: 10.1097/01.cji.0000170357.14962.9b. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi Y., et al. In vitro and in vivo evidence of PNH cell sensitivity to immune attack after nonmyeloablative allogeneic hematopoietic cell transplantation. Blood. 2004;103:1383–1390. doi: 10.1182/blood-2003-04-1281. [DOI] [PubMed] [Google Scholar]
- 20.Hanada K., Perry-Lalley D.M., Ohnmacht G.A., Bettinotti M.P., Yang J.C. Identification of fibroblast growth factor-5 as an overexpressed antigen in multiple human adenocarcinomas. Cancer Res. 2001;61:5511–5516. [PubMed] [Google Scholar]