Abstract
Adipose differentiation is an important part of the energy homeostasis system of higher organisms. Recent data have suggested that this process is controlled by an interplay of transcription factors including PPARγ, the C/EBPs, and ADD1/SREBP1. Although these factors interact functionally to initiate the program of differentiation, there are no data concerning specific mechanisms of interaction. We show here that the expression of ADD1/SREBP1 specifically increases the activity of PPARγ but not other isoforms, PPARα, or PPARδ. This activation occurs through the ligand-binding domain of PPARγ when it is fused to the DNA-binding domain of Gal4. The stimulation of PPARγ by ADD1/SREBP1 does not require coexpression in the same cells; supernatants from cultures that express ADD1/SREBP1 augment the transcriptional activity of PPARγ. Finally, we demonstrate directly that cells expressing ADD1/SREBP1 produce and secrete lipid molecule(s) that bind directly to PPARγ, displacing the binding of radioactive thiazolidinedione ligands. These data establish that ADD1/SREBP1 can control the production of endogenous ligand(s) for PPARγ and suggest a mechanism for coordinating the actions of these adipogenic factors.
Keywords: fatty acid metabolism, nuclear hormone receptor, adipocyte differentiation
The adipocyte is a highly specialized cell type that plays an important role in energy homeostasis. The primary role of fat cells is to store triglycerides in periods of energy excess and to release this energy during nutritional deprivation. Adipocyte differentiation is a complex process accompanied by coordinated changes in cell morphology, hormone sensitivity, and gene expression. These changes result from the action of several transcription factors that influence these processes, including PPARγ, the C/EBPs, and ADD1/SREBP1 (ref. 1 and references therein). These factors do not work completely independently but interact functionally in several important ways.
PPARγ is a member of the nuclear hormone receptor family that is expressed preferentially in adipose tissue (2). It functions as an obligate heterodimer with the retinoid X receptor (RXR) to bind to target sequences termed DR-1 sites (2). Activation of PPARγ by ligands is sufficient to stimulate adipose differentiation in many types of fibroblastic cells (2–4). Ligands for this receptor include the antidiabetic thiazolidinedione (TZD) drugs, 15-deoxy-Δ12,14-prostaglandin J2 (15-deoxy-PGJ2), and certain polyunsaturated fatty acids (5–7). The biological ligands now known bind to PPARγ with relatively low affinity (Kd = 2–50 μM), compared with the affinity (typically Kd < 1 nM) for most bona fide ligands for nuclear receptors. Whether the known PPARγ ligands represent endogenous activators for this receptor is not clear.
The C/EBP family also appears to be very important in adipogenesis. Ectopic expression of C/EBPβ or C/EBPα can promote the differentiation of fibroblastic cells (8–13). There is evidence that C/EBPβ does this by increasing the expression of PPARγ, whereas C/EBPα appears to be able to powerfully synergize with PPARγ to promote differentiation (8, 14).
Another key transcription factor in adipose development is ADD1/SREBP1 (15). As a member of the basic helix–loop–helix transcription factor family, ADD1/SREBP1 has an unusual ability to bind to two distinct DNA sequences (16, 17): E-boxes (CANNTG) and the sterol regulatory element (SRE), a sequence important in the control of genes involved in cholesterol homeostasis (18). This protein and its close homolog SREBP2 (19) are expressed as large precursors that are proteolytically cleaved from the endoplasmic reticulum and nuclear membrane (20, 21). The cleavage of SREBP2 is controlled by cholesterol level in vivo. Although SREBP1 can be controlled by cholesterol levels in cultured cells, this does not seem to occur in vivo, leaving the question of its mechanism of activation unresolved (22).
Ectopic expression of ADD1/SREBP1 is sufficient to increase endogenous expression of fatty acid synthetase and lipoprotein lipase in cultured fibroblasts or in the livers of transgenic animals (23–25). In addition, expression of ADD1/SREBP1 promotes adipogenesis when cells are also stimulated with multiple hormonal inducers (23). Coexpression of ADD1/SREBP1 increases the transcriptional activity of PPARγ, suggesting that this cooperation may be responsible for the effects of ADD1/SREBP1 in differentiation.
We have now investigated in detail the biochemical basis of the cooperation between ADD1/SREBP1 and PPARγ. We find that ADD1/SREBP1 can promote the production of ligand(s) for PPARγ. Expression of various mutant alleles of ADD1/SREBP1 indicates that the production of PPARγ ligand occurs via transcription through E-box motifs.
MATERIALS AND METHODS
Cell Culture.
3T3-L1 preadipocytes were grown in DMEM containing 10% calf serum. Differentiation of 3T3-L1 cells was induced as described previously (11). At confluence, the medium was changed to DMEM supplemented with 10% fetal bovine serum (FBS), 1 μM dexamethasone, 0.5 mM iso-butyl methyl xanthine, and 5 μg/ml of insulin for 2 days in the absence or presence of 5 μM pioglitazone. Thereafter, the medium was replaced every other day with 5 μg/ml of insulin and 10% FBS. Cells were fixed with 10% formaldehyde in PBS and stained with Oil Red O (Sigma). Stable cell lines expressing different ADD1 alleles were derived as described previously (23). NIH 3T3 cells were grown in DMEM containing 10% calf serum (HyClone), and Rat1-IR cells were grown in DMEM/F12 (50:50) medium containing 10% FBS as previously described (26).
Transient Transfection and Enzyme Assays.
NIH 3T3 cells were cultured as described above and transfected 1 day before confluence by the calcium phosphate method as previously described (16). Construction of the 520-bp aP2 enhancer linked to a chloramphenicol acetyltransferase (CAT) reporter was described previously (27). The ADD1 expression vector (ADD1–403) (2 μg) was cotransfected with each CAT reporter plasmid (2 μg) and/or combinations of PPARα/RXRα, PPARδ/RXRα, or PPARγ/RXRα expression vector (1 μg/1 μg). The luciferase reporters such as UASG X4-TK-Luc and PPAR response element (PPRE) X3-TK-Luc, and Gal4 fusion expression vectors including Gal4-DBD, Gal4-PPARα-LBD, and Gal4-PPARγ-LBD have been described (5). The level of CAT expression was determined by measuring CAT enzyme activity. β-Galactosidase (β-gal) assays were performed by a standard colorimetric procedure with chlorophenol red-β-d-galactopyranoside (CPRG) as substrate (Boehringer Mannheim). The relative CAT and luciferase activities were normalized to the β-galactosidase activity. All the transfection experiments were performed in duplicate and repeated at least three times independently.
Ligand-Binding Assays for PPARγ.
Ligand-binding assays were performed as described previously (5, 6). Briefly, BL21(DE3) plysS bacteria expressing a histidine-tagged ligand-binding domain of human PPARγ protein (amino acids 176–477) was prepared by freezing/thawing in a lysis buffer containing 10 mM Tris (pH 8), 50 mM KCl, 10 mM DTT, and 1% Triton X-100. The lysate was cleared by centrifugation at 40,000 × g for 30 min. Competition binding assays were performed with the thiazolidinedione PPARγ ligand [3H]BRL 49653 (10 nM, specific activity 31 Ci/mmol; 1 Ci = 37 GBq), 300 μg of bacterial lysate, with and without addition of extracted conditioned medium (see below), in 100 μl of ligand-binding buffer (same buffer as above without Triton). After incubation at 4°C for 3 hr, bound and free radioactivity was separated by elution through a 1-ml Sephadex G-25 desalting column (Boehringer Mannheim). Nonspecific binding of radiolabeled ligand was assessed by using competition with cold BRL 49653 and was found to be negligible. Radioactivity was quantitated by liquid scintillation counting.
Conditioned medium from cells transfected with empty vector or vectors expressing ADD1 or C/EBPα was extracted with organic solvents by using Sep-Pak C18 column (Waters) chromatography methods as previously described (28). Briefly, 20 ml of methanol was added to 10 ml of conditioned medium and incubated at −20°C for at least 30 min. The mixture was centrifuged at 500 × g for 5 min, and the supernatant was collected. The supernatant was acidified with 30 ml 0.01% glacial acetic acid and applied to a preconditioned C18 Sep-Pak cartridge. The cartridge was washed with 10 ml water and eluted in sequence with 10 ml hexane, 10 ml methyl formate, and 10 ml methanol. The methyl formate fraction, which contains fatty acids and eicosanoids, was dried down under N2 gas and resuspended in 100 μl methanol. A small aliquot of this extract was utilized in the ligand-binding assay described above.
RESULTS
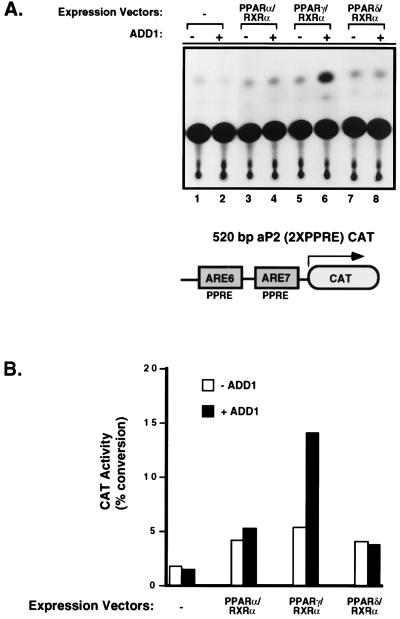
Previous work demonstrated that coexpression of ADD1/SREBP1 with PPARγ could increase the transcriptional activity of PPARγ through its binding site, the PPRE. This enhanced transcriptional activity of PPARγ by ADD1/SREBP1 appears to be indirect because ADD1/SREBP1 alone does not activate transcription of reporter-containing PPREs (23). However, it is not currently known whether this effect is specific for PPARγ among the PPAR isoforms. To investigate this, we cotransfected activated ADD1/SREBP1 (amino acids 1–403), along with each of the three different murine PPAR isoforms and PPAR’s heterodimeric partner, RXRα. The reporter construct used is the adipose-specific enhancer region (520 bp) of the aP2 gene containing two natural PPREs, linked to CAT. As shown in Fig. 1, the transfection of each PPAR form with RXRα caused a small increase in transcriptional activity detected above the control levels. Addition of ADD1/SREBP1 caused an elevation in the transcriptional response seen only from PPARγ, not from PPARα or PPARδ. This indicates that there is selectivity in the responsiveness among the PPARs to ADD1/SREBP1.
Figure 1.
Expression of ADD1/SREBP1 selectively enhances the transcriptional activity of PPARγ. (A) NIH 3T3 cells were cotransfected with the 520-bp enhancer for aP2 gene (520-bp aP2 CAT) and indicated combinations of ADD1 with either PPARα/RXRα, PPARγ/RXRα, or PPARδ/RXRα expression vectors. Transfections were performed in duplicate and repeated at least three times. A representative experiment is shown above. All CAT enzyme activities were normalized to β-gal activity. (B) Quantitation of CAT enzyme activity of A was performed by PhosphorImager analysis (Molecular Dynamics).
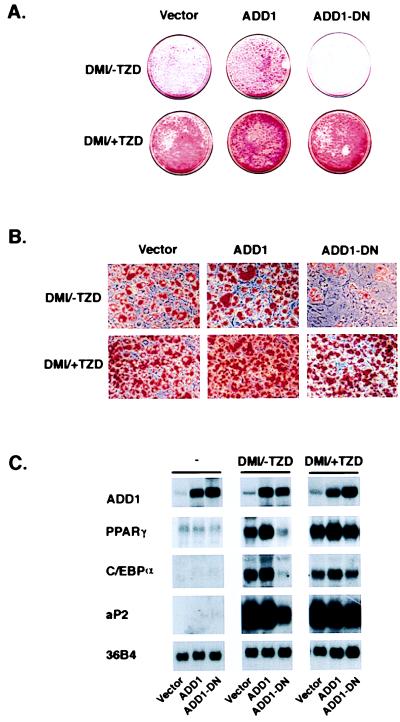
This selectivity of interaction between PPARγ and ADD1/SREBP1 could occur at several levels, such as the transcriptional control of the enzymes responsible for the generation of an endogenous ligand(s) for PPARγ or in the synthesis of other proteins such as coactivators, which are involved in the transcriptional response to PPARγ. To investigate the possibility that ADD1 may be involved in the generation of ligand(s) for PPARγ, we used a previously identified dominant-negative allele of ADD1/SREBP1 to determine whether synthetic ligands for PPARγ could replace the functional role of ADD1/SREBP1 in adipogenesis. This dominant-negative allele of ADD1/SREBP1 contains a substitution of an alanine for an essential tyrosine residue (Y → A, 320) in the basic DNA-binding domain (16). As reported earlier and shown in Fig. 2, the expression of this allele (amino acids 1–403, alanine substitution for tyrosine at 320; termed ADD1-DN) with a retroviral expression system strongly inhibits the differentiation of 3T3-L1 cells treated with the usual differentiation inducers [labeled −TZD/DMI (dexamethasone, methyisobutyl xanthine, and insulin); see Materials and Methods]. Fig. 2A illustrates lipid accumulation in these cells, as shown by staining with Oil Red O. Expression of an activated form of the wild-type allele (amino acids 1–403) caused a small increase in lipid accumulation above the already robust response seen in the control (vector) cells. Addition of pioglitazone, a TZD ligand for PPARγ, completely reverses the ability of the dominant-negative allele of ADD1/SREBP1 to block lipid accumulation and morphological differentiation of the cells (Fig. 2B). At the mRNA level, Fig. 2C shows that although the dominant-negative allele of ADD1 strongly suppresses the expression of genes linked to differentiation, such as PPARγ, C/EBPα, and aP2, this effect is reversed almost completely by exposure of cells to pioglitazone during the differentiation process. These results are consistent with a role for ADD1/SREBP1 in the generation of an endogenous ligand(s) for PPARγ during adipogenesis, though other mechanistic interpretations are also possible.
Figure 2.
A PPARγ ligand rescues the inhibition of adipogenesis by dominant-negative ADD1/SREBP1. 3T3-L1 preadipocytes were infected with retroviruses containing either empty vector, ADD1, or dominant-negative ADD1 (ADD1-DN) as previously described (23). 3T3-L1-vector, -ADD1, and -ADD1-DN cell lines were treated with differentiation inducers in the absence (DMI/-TZD) or presence (DMI/+TZD) of the TZD ligand pioglitazone (5 μM) for 48 hr at confluence and subsequently cultured for 5 days. Cells were fixed and stained with Oil Red O. (A) Macroscopic view of dishes. (B) Microscopic views of dishes shown in A at ×25 original magnification. (C) Total RNA (10 μg per lane) was isolated from each cell lines at confluence (−) and after 5 days of postconfluent culture with treatment of either DMI/-TZD or DMI/+TZD. RNA was separated by electrophoresis, blotted to nylon membrane, and hybridized with the indicated 32P-labeled cDNAs.
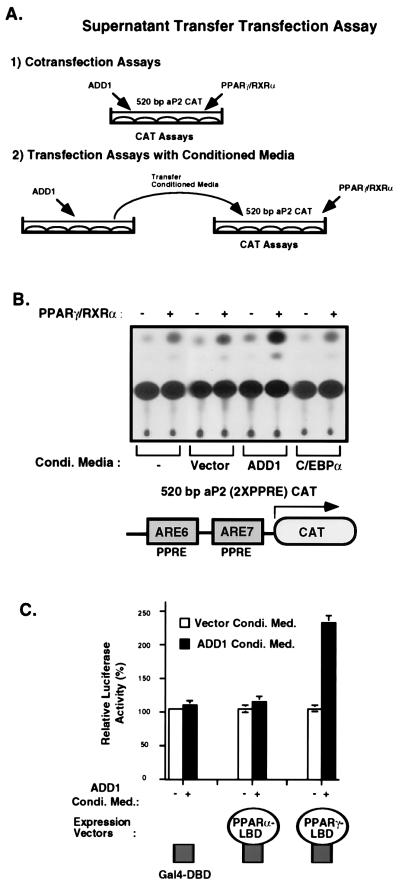
Based on these results, we therefore investigated more directly the notion that ADD1/SREBP1 may generate ligands for PPARγ. Though no molecule has been demonstrated to serve as an endogenous ligand of PPARγ in vivo, several biological ligands or activators of PPARγ and other PPARs have been identified. These are all secreted lipids in the fatty acid family including the prostaglandins, leukotrienes, hydroxy-eicosatetraenoic acids, and polyunsaturated fatty acids (5–7). We therefore have modified the standard cotransfection protocol shown in Fig. 3A, section 1, to a protocol (Fig. 3A, section 2) where the supernatants (conditioned medium) from cells transfected with ADD1 were placed on a second dish of cells transfected with PPARγ. As shown in Fig. 3B, supernatants from cells transfected with ADD1 stimulated a 3-fold increase in transcriptional activity obtained from PPARγ when compared with cells not transfected, transfected with empty vectors, or transfected with C/EBPα.
Figure 3.
Conditioned medium from cells transfected with ADD1/SREBP1 increases the transcriptional activity of PPARγ/RXRα through its ligand-binding domain (LBD). (A) Scheme of supernatant transfer assays. Section 1 shows a cotransfection analysis; section 2 illustrates transient transfection assays with conditioned medium. To collect conditioned medium, transfected cells were incubated with 0.5% BSA in DMEM, and supernatants were isolated after 24 hr. These supernatants were used for the sequential transfection experiments with a 520-bp aP2 CAT reporter in the absence or presence of PPARγ/RXRα. (B) As described above, conditioned medium was prepared by transient transfection of either empty expression vector or vectors expressing ADD1 or C/EBPα. Supernatants collected from different cells were incubated for 24 hr with cells transfected with the 520-bp aP2 CAT reporter and PPARγ/RXRα expression vectors. These transfections were performed in duplicate, and the above is a representative experiment. (C) Rat1-IR cells were transfected with luciferase reporter containing four copies of the UASG and either Gal4 DNA-binding domain (DBD), Gal4-PPARα LBD, or Gal4-PPARγ LBD expression vector. After transfection, cells were incubated with conditioned medium from cells transfected with either vector or ADD1. Normalized luciferase activity was determined and plotted as luciferase activity relative to Gal4 DBD expression vector.
To investigate the domains of PPARγ necessary for this response, we used chimeric molecules where the ligand-binding domains (LBD) of PPARγ or -α were fused to the DNA-binding domain (DBD) of Gal 4. As shown in Fig. 3C, transcriptional activity of the Gal 4 DBD alone or fused to the LBD of PPARα did not respond to these supernatants, whereas Gal4 DBD fused to the LBD of PPARγ showed a marked increase in activity. Supernatants from cells transfected with empty vectors did not show this increase. These data suggest that ADD1/SREBP1 stimulates the production of molecule(s) that can selectively activate PPARγ through its LBD.
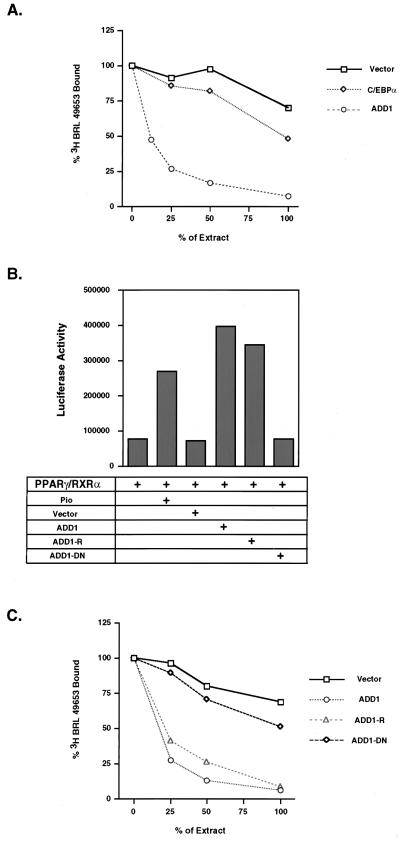
With the availability of synthetic, high-affinity ligands of PPARγ, such as the TZD drugs, it is possible to look directly at ligand-binding activity. The supernatants from cells transfected with ADD1/SREBP1, C/EBPα or empty vectors were extracted with an organic solvent to extract total lipids. They then were resolubilized and put in a competition assay for binding of [3H]BRL 49653 to PPARγ. The supernatants from the cells transfected with ADD1 showed powerful displacement of BRL 49653, with 50% maximal displacement at 12.5% of the neat, extracted materials (Fig. 4). In contrast, cells transfected with empty vectors showed very little displacement, even when the extracted material was used in an undiluted manner. When compared at a point where these two curves overlap, such as the ED25, there appears to be at least 8-fold more ligand activity in cells that were transfected with ADD1. C/EBPα-transfected cells showed a very small amount of displacement, much less than ADD1, but more than the vector-transfected cells. These data illustrate directly that expression of ADD1/SREBP1 triggers the generation of ligand(s) for PPARγ.
Figure 4.
ADD1/SREBP1 induces ligand activity for PPARγ. (A) Conditioned medium from cells transfected with either empty vector, ADD1, or C/EBPα was organically extracted as described in Materials and Methods. Fractions of each extract were diluted in ligand-binding buffer (100% denotes 5 μl of extract in methanol, 50% denotes 2.5 μl, etc.) and used in competition with [3H]BRL 49653 for binding to bacterial-produced PPARγ (see details in Materials and Methods). (B) Cotransfection of several alleles of ADD1 including ADD1, ADD1-R, and ADD1-DN with PPARγ/RXRα to determine transcriptional activity through PPREs with luciferase reporter (PPRE X3-TK-Luc). Luciferase activity was normalized with β-gal activity. (C) Ligand activity from cells transfected with either empty vector, ADD1, ADD1-R, or ADD1-DN. Conditions were the same as in A.
ADD1/SREBP1 has unusual dual DNA-binding specificity, interacting both with E-box motifs and the SRE sequence. This dual DNA-binding specificity is a result of one amino acid, tyrosine 320, at a site where all other basic helix–loop–helix factors have an arginine residue (16). Mutation of tyrosine 320 to arginine converts ADD1/SREBP1 into a pure E-box binding factor and hence allows discrimination of those functions that occur through E-boxes versus those mediated by SRE-like sequences (16). Various alleles of ADD1/SREBP1 were cotransfected with PPARγ, and transcriptional activity was determined through PPRE element. Fig. 4B indicates that the ADD1 320 Y → R mutation is just as effective at promoting transcriptional activity of PPARγ as the wild-type allele. These same alleles were then transfected and ligand activity was assayed from the cellular supernatants. As shown in Fig. 4C, the ADD1-R allele stimulated the production of ligand activity in a manner that was similar to that of the wild-type allele. These data indicate that ADD1/SREBP1 can promote the function of ligand production for PPARγ through E-box motifs of as yet unknown target genes.
DISCUSSION
Cooperation between multiple transcription factors is an important component of most, if not all, differentiation processes, including adipose differentiation. One dramatic example of this has been observed when C/EBPα and PPARγ are coexpressed (14). This yields a powerful adipogenic response under conditions where neither factor alone is sufficient to promote fat cell formation. Another notable example of transcription factor cooperation is that between ADD1/SREBP1 and PPARγ. Both of these factors are induced very early in adipogenesis, and expression of ADD1/SREBP1 enhances the transcriptional activity of PPARγ and increases the percentage of cells undergoing differentiation (23).
The data presented here indicate clearly that expression of an activated form of ADD1/SREBP1 causes cells to produce ligand(s) for PPARγ. These ligand(s) are likely to be fatty acid derivatives because they can be isolated with organic solvents that extract polar lipids such as eicosanoid derivatives and other fatty acids (Fig. 4). The first biological ligand described for PPARγ was 15-deoxy-Δ12, 14-PGJ2 (5, 6). This molecule, which is produced by the nonenzymatic dehydration of PGD2, has an affinity for PPARγ of approximately 2 μM (6). Most recently, certain polyunsaturated fatty acids such as linoleic acid and arachidonic acid have been shown to bind to and activate PPARγ, although the affinity here is also relatively low (Kd = 10–100 μM) (7). These data raised the possibility that PPARγ ligands may be provided by dietary sources through the circulation, as opposed to being produced endogenously. The experiments described here illustrate that a PPARγ ligand(s) is produced endogenously from cells expressing ADD1/SREBP1, a transcription factor induced early in adipogenesis. Of course, it is quite possible that ligands for this transcription factor come from both endogenous and exogenous sources, dependent on diet, physiological status, and genetic tendency toward obesity and diabetes.
One key issue to be pursued is the target gene(s) for ADD1/SREBP1 related to the biosynthesis of PPARγ ligands. We and others previously have shown that ADD1/SREBP1 can control the expression of fatty acid synthetase and lipoprotein lipase, key genes of fatty acid metabolism (23). Because the endogenous ligand(s) may well be fatty acid derivatives, control of these genes may be rate-limiting. Consistent with this (Fig. 4), we recently have shown that ADD1/SREBP1 can control fatty acid synthetase expression through an E-box motif (29). On the other hand, the most common products of these enzymes, the dietary fatty acids, are likely to have to undergo further enzymatic transformation to function effectively as PPARγ ligands. The genes encoding these modifying enzymes are prime candidates for regulation by ADD1/SREBP1.
Finally, the identification of the endogenous ligand described here will be important to determine. Because PPARγ ligands potentially can be involved in both adipogenesis and insulin resistance/sensitivity, these molecules may open a unique opportunity to understand these problems. Current fractionation studies using HPLC suggest a single peak of activity, indicating one or very few components (not shown). The chromatographic behavior of this activity indicates that it is not equivalent to 15-deoxy-Δ12,14-PGJ2. Structural determination will shed light on the biochemical nature of this molecule and open the door to the study of its biosynthetic regulation.
Acknowledgments
This work was supported by Grant 2R37DK31405 from the National Institutes of Health and a grant from the Sumitomo Chemical Company/J.B.K. was supported by a postdoctoral fellowship from the American Diabetes Association.
ABBREVIATIONS
- PPRE
PPAR response element
- RXR
retinoid X receptor
- TZD
thiazolidinedione
- LBD
ligand-binding domain
- DBD
DNA-binding domain
References
- 1.Spiegelman B M, Flier J S. Cell. 1996;87:377–389. doi: 10.1016/s0092-8674(00)81359-8. [DOI] [PubMed] [Google Scholar]
- 2.Tontonoz P, Hu E, Graves R A, Budavari A I, Spiegelman B M. Genes Dev. 1994;8:1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- 3.Hu E, Tontonoz P, Spiegelman B M. Proc Natl Acad Sci USA. 1995;92:9856–9860. doi: 10.1073/pnas.92.21.9856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lehmann J M, Moore L B, Smith-Oliver T A, Wilkison W O, Willson T M, Kliewer S A. J Biol Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 5.Forman B M, Tontonoz P, Jasmine C, Brun R P, Spiegelman B M, Evans R M. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 6.Kliewer S A, Lenhard J M, Willson T M, Patel I, Morris D C, Lehmann J M. Cell. 1995;83:813–819. doi: 10.1016/0092-8674(95)90194-9. [DOI] [PubMed] [Google Scholar]
- 7.Krey G, Braissant O, L’Horset F, Kalkhoven E, Perroud M, Parker M G, Wahli W. Mol Endocrinol. 1997;11:779–791. doi: 10.1210/mend.11.6.0007. [DOI] [PubMed] [Google Scholar]
- 8.Wu Z, Xie Y, Bucher N, Farmer S R. Genes Dev. 1995;9:2350–2363. doi: 10.1101/gad.9.19.2350. [DOI] [PubMed] [Google Scholar]
- 9.Samuelsson L, Stromberg K, Vikman K, Bjursell G, Enerback S. EMBO J. 1991;10:3787–3793. doi: 10.1002/j.1460-2075.1991.tb04948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Umek R M, Friedman A D, McKnight S L. Science. 1991;251:288–292. doi: 10.1126/science.1987644. [DOI] [PubMed] [Google Scholar]
- 11.Lin F T, Lane M D. Genes Dev. 1992;6:533–544. doi: 10.1101/gad.6.4.533. [DOI] [PubMed] [Google Scholar]
- 12.Lin F T, Lane M D. Proc Natl Acad Sci USA. 1994;91:8757–8761. doi: 10.1073/pnas.91.19.8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freytag S O, Paielli D L, Gilbert J D. Genes Dev. 1994;8:1654–1663. doi: 10.1101/gad.8.14.1654. [DOI] [PubMed] [Google Scholar]
- 14.Tontonoz P, Hu E, Spiegelman B M. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 15.Tontonoz P, Kim J B, Graves R A, Spiegelman B M. Mol Cell Biol. 1993;13:4753–4759. doi: 10.1128/mcb.13.8.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim J B, Spotts G D, Halvorsen Y D, Shih H M, Ellenberger T, Towle H C, Spiegelman B M. Mol Cell Biol. 1995;15:2582–2588. doi: 10.1128/mcb.15.5.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yokoyama C, Wang X, Briggs M R, Admon A, Wu J, Hua X, Goldstein J L, Brown M S. Cell. 1993;75:187–197. [PubMed] [Google Scholar]
- 18.Briggs M R, Yokoyama C, Wang X, Brown M S, Goldstein J L. J Biol Chem. 1993;268:14490–14496. [PubMed] [Google Scholar]
- 19.Hua X, Yokoyama C, Wu J, Briggs M R, Brown M S, Goldstein J L, Wang X. Proc Natl Acad Sci USA. 1993;90:11603–11607. doi: 10.1073/pnas.90.24.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Sato R, Brown M S, Hua X, Goldstein J L. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 21.Sakai J, Duncan E A, Rawaon R B, Hua X, Brown M S, Goldstein J L. Cell. 1996;85:1037–1046. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- 22.Sheng Z, Otani H, Brown M S, Goldstein J L. Proc Natl Acad Sci USA. 1995;92:935–938. doi: 10.1073/pnas.92.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J B, Spiegelman B M. Genes Dev. 1996;10:1096–1107. doi: 10.1101/gad.10.9.1096. [DOI] [PubMed] [Google Scholar]
- 24.Shimano H, Horton J D, Hammer R E, Shimomura I, Brown M S, Goldstein J L. J Clin Invest. 1996;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimano H, Horton J D, Shimomura I, Hammer R E, Brown M S, Goldstein J L. J Clin Invest. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu E, Kim J B, Sarraf P, Spiegelman B M. Science. 1996;274:2100–2103. doi: 10.1126/science.274.5295.2100. [DOI] [PubMed] [Google Scholar]
- 27.Graves R A, Tontonoz P, Spiegelman B M. Mol Cell Biol. 1992;12:1202–1208. doi: 10.1128/mcb.12.3.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serhan C N, Sheppard K A. J Clin Invest. 1990;85:772–780. doi: 10.1172/JCI114503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J B, Pasha S, Wright M, Yao K M, Mueller E, Solanes G, Lowell B B, Spiegelman B M. J Clin Invest. 1998;101:1–9. doi: 10.1172/JCI1411. [DOI] [PMC free article] [PubMed] [Google Scholar]