Abstract
Eukaryotic viruses can maintain latency in dividing cells as extrachromosomal nuclear plasmids. Segregation and nuclear retention of DNA is, therefore, a key issue in retaining copy number. The E2 enhancer protein of the papillomaviruses is required for viral DNA replication and transcription. Viral mutants that prevent phosphorylation of the bovine papillomavirus type 1 (BPV) E2 protein are transformation-defective, despite normal viral gene expression and replication function. Cell colonies harboring such mutants show sectoring of viral DNA and are unable to maintain the episome. We find that transforming viral DNA attaches to mitotic chromosomes, in contrast to the mutant genome encoding the E2 phosphorylation mutant. Second-site suppressor mutations were uncovered in both E1 and E2 genes that allow for transformation, maintenance, and chromosomal attachment. E2 protein was also found to colocalize to mitotic chromosomes, whereas the mutant did not, suggesting a direct role for E2 in viral attachment to chromosomes. Such viral hitch-hiking onto cellular chromosomes is likely to provide a general mechanism for maintaining nuclear plasmids.
Viruses must be replicated and effectively partitioned to each daughter cell to be maintained in a population of dividing cells. Covalent integration of viral DNA into cellular chromosomes is the most widely studied and perhaps the most prevalent mechanism used by such episomes in solution of the partitioning and replication problems. However, some eukaryotic viruses do replicate as nuclear plasmids in the cells in which they have established latency. Low-copy-number plasmids in prokaryotes have evolved elaborate mechanisms to facilitate effective segregation (1). In some cases, plasmid-encoded proteins bind to centromere-like sites on free plasmids (2), and from both evolutionary and mechanistic perspectives, it is interesting that the cellular chromosome has homologous genes that play critical roles in partitioning the bacterial chromosome (3, 4). A direct demonstration that eukaryotic plasmids use partitioning strategies for critical maintenance functions has not been documented.
Papillomaviruses have small DNA genomes that are maintained through long latency periods as multicopy plasmids in dividing basal epithelial cell layers. Only after passage through the differentiating layers when the infected cells reach the terminally differentiated keratinocyte level does viral DNA amplification and viral particle production occur. Some forms of human papillomavirus are also associated with cervical and other cancers, so interest in the viral life cycle is high (5, 6). It may be presupposed that high-copy-number plasmids are able to faithfully segregate by random division of the nuclear contents. However if these plasmids are nonrandomly localized (for example, in replication foci) the effective number of units for division would in fact be much lower and additional mechanisms may be required. If a virus is unable to amplify from a low-copy number, accurate segregation is required, particularly, if multiple genomes are required for adequate levels of transforming functions. A particular problem in eukaryotes for maintaining plasmids may be presented by the nuclear membrane and its breakdown and reassembly after mitosis. Thus even high-copy-number plasmids unable to tether to chromosomes may be excluded from the mitotic apparatus and left in the cytoplasm where they are subject to lysosomal degradation.
Bovine papillomavirus (BPV) has been a prototypic virus that has served as a model for studies of papillomavirus replication and transformation. Only two viral proteins are required for DNA replication: E1 is the viraly encoded helicase required for replicational initiation and elongation (7, 8), and E2 is a transcription factor that regulates gene expression from several viral promoters and also facilitates specific E1 binding to the origin (9, 10, 22, 33).
The E2 protein is phosphorylated in its central hinge region and previous studies showed that at least one of the phosphorylation sites was critical for transformation functions (ref. 11 and Fig. 1). A combination of mutations (called A4 because four Ser residues in the central hinge region were mutated to Ala) in the E2 gene could block transformation. Additionally this mutant, in contrast to the A3 (which has three of the four A4 mutations) or wild-type viral DNA was unable to maintain its genome despite high replication and gene expression activities. In transient assays with intact genomes harboring either the A3 or A4 mutations in the E2 gene, replication rates were in fact higher than those of wild type. Moreover, when the E1 and mutant E2 genes were expressed from recombinant vectors, replication reporters showed high levels of transient replication. Cotransfection of the defective A4 genome with a neomycin-resistance marker did not affect the frequency of drug-resistant colonies and adverse effects on cell growth were not detected with overexpression of the A4 protein (11). Thus the transformation defects of the A4 genome could not be ascribed to an apoptotic or cell static effect of the mutant. These transformation defects of the A4 genome could be abrogated by mutation of one of the Ala codons to Asp that when incorporated into the protein mimics phosphorylation at the site by charge and shape, suggesting that phosphorylation per se is the key.
Figure 1.
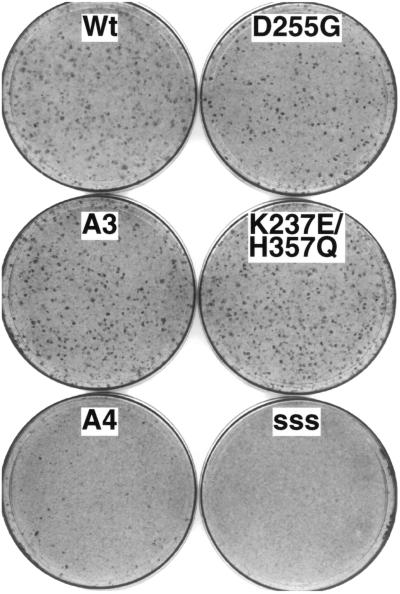
Oncogenic transformation by phosphorylation site mutants of BPV. Mouse fibroblast cells (C127) were transfected with 1 μg of linear BPV DNA and fixed and stained after 2 weeks as described (11). Wt is wild-type BPV, A3 has three Ser residues mutated to Ala at phosphorylation sites in E2 (Ser-290, -298, and -301), A4 has the A3 mutations and one additional Ser → Ala change at amino acid 235, D255G and K237E/H357Q are suppressing mutations in E2 and E1 proteins, respectively, which will be discussed later, and sss is a carrier DNA control.
In this study we have followed the fate of the viral DNA to learn more about long-term maintenance functions and the control of these functions by phosphorylation. Cells containing the A4 genome lose viral DNA in a cell-division-dependent fashion. Both transforming viral DNA and E2 protein were found to colocalize to mitotic chromosomes but A4 mutant DNA and protein do not. Suppressors of this maintenance defect were isolated that map to both E2 and E1, suggesting that E1 may also aid in the chromosomal tethering of the virus.
MATERIALS AND METHODS
Fluorescence in Situ Hybridization (FISH) Analysis.
Mouse mammary fibroblast tumor cells (C127) were transfected with linearized constructs by electroporation as described (11). Colonies were fixed with formaldehyde/acetic acid (4% formaldehyde/5% acetic acid/0.9% NaCl for 30 min), washed with PBS, stored in 70% ethanol until further processed, washed with PBS, treated with RNase A (100 mg/ml in 2× SSC for 1 hr at 37°C), digested with proteinase K (0.5 μg/ml in 20 mM Tris⋅HCl, pH 7.5/2 mM CaCl2 for 10 min at 37°C), postfixed for 10 min (1% formaldehyde/PBS), and dried in 70%, 90%, and 100% ethanol. Random-primed BPV DNA with digoxigenin-dUTP incorporated was used as probe (Boehringer Mannheim). Denaturation of target and probe DNA was at 80°C for 20 min in a hybridization mixture [50% formamide/2× SSC/50 mM NaHPO4, pH 7.2/1 mM EDTA/1× Denhardt’s solution/sheared salmon sperm DNA (1 mg/ml)/tRNA (0.5 mg/ml)/digoxigenin-labeled DNA (∼2 μg/ml)] followed by overnight hybridization at 37°C. After washing, the probe was detected with an anti-digoxigenin antibody coupled to alkaline phosphatase and HNPP and Fast Red as substrates (Boehringer Mannheim), DNA was counterstained with 4′,6-diamidino-2-phenylindole (DAPI; 0.1 μg/ml) in TBS, and slides were mounted with diazabicyclo[2.2.2]octane DABCO (50% glycerol/2% DABCO in PBS). Metaphase arrest was by a 4-hr incubation with Colcemid (0.1 μg/ml), after which mitotic cells were removed by a shakeoff. Chromosomes were isolated by hypotonic swelling followed by fixation (12). Briefly, cells were removed by shakeoff and centrifuged 10 min at 200 × g, all but 0.5 ml of the supernatant was removed, 5 ml 75 mM KCl was added, and the cells were allowed to swell at 37°C for 20 min. One milliliter of fixative (75% methanol/25% acetic acid) was added with mixing and the cells were incubated for 20 min at room temperature. The cells were pelleted, washed with 5 ml of fixative, incubated for 10 min, and centrifuged. This washing was repeated for a total of three times, and then the cells were dropped onto clean slides. The slides were baked at 65°C for 4 hr. Hybridization, detection, counterstaining, and mounting were as for colony studies. Fluorescence microscopy used the DAPI or rhodamine filter sets and ×5 (Fig. 2A) or ×100 (Figs. 3 and 4C) objectives.
Figure 2.
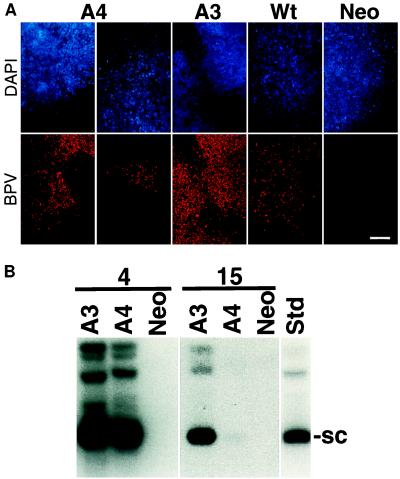
Loss of viral DNA from dividing cells. (A) FISH analysis of colonies of cells cotransfected with BPV DNA and the neomycin-resistance gene. After drug killing of nontransfected cells and growth of colonies for 2 weeks, BPV DNA was detected by the red precipitate and cellular DNA was detected by blue DAPI staining. Wild-type (Wt) and A3 DNAs were detected in nearly all surviving cells. The A3 example shows the edge of two neighboring colonies, all of the rest are from individual colonies. The neor control shows that colonies obtained by single transfection of the drug marker do not give a signal with the BPV probe. A4 DNA was observed in a subset of cells in a range from a few to all within the colony. These examples show sectoring of the colonies. (Bar = 100 μm.) (B) Loss of A4 DNA requires cell division. Cells were transfected, drug-selected, and grown for the same time as in A but were plated under two different conditions. In lanes labeled 4, the cells were plated densely allowing only about four doublings before contact inhibition of further growth. In lanes labeled 15, the cells were plated sparsely and split every 3 days to facilitate logarithmic-phase growth (about 15 doublings). Low molecular weight DNA was isolated from equivalent numbers of cells for each lane and uncut DNA was examined by Southern blotting with random-primed BPV DNA as probe.
Figure 3.
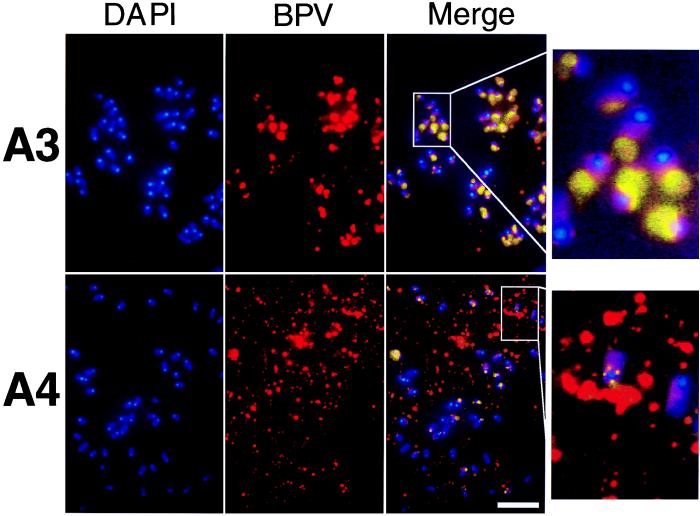
Colocalization of BPV DNA with cellular metaphase chromosomes. DNA staining with DAPI revealed mouse chromosomes that showed predominantly acrocentric organization with brightly staining centromeric regions. BPV DNA was detected by FISH. The chromosomes from one mitotic plate are shown in each panel. Intensely colocalizing BPV and DAPI staining produced magenta, which was pseudocolored yellow, and lower-intensity colocalization produced pink. (Right) Magnifications ×4 of a small region of the merged images. The neor control background levels of staining were similar to those shown in Fig. 2A. (Bar = 5 μm.)
Figure 4.
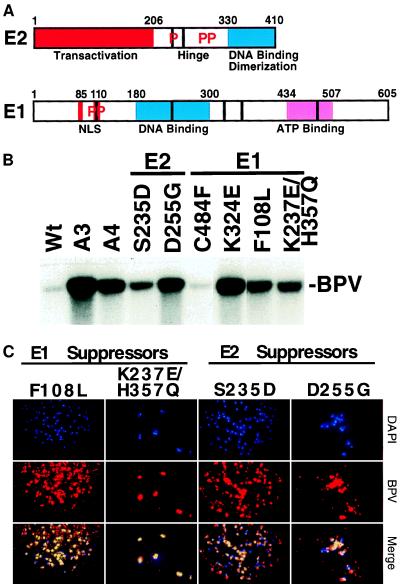
Behavior of suppressors in replication and chromosomal localization assays. (A) Graphical representation of the functional domains of E1 and E2 proteins with the suppressor mutations indicated as vertical bars. The positions of known phosphorylated Ser residues in E2 (11, 13) or Ser and Thr residues in E1 (14, 15) are indicated by Ps. (B) One microgram of the genomes containing the indicated mutation was transfected into C127 cells, low molecular weight DNA was harvested at day 6 and examined by Southern blotting using random-primed BPV as probe. The position of EcoRI-linearized, DpnI-resistant full-length BPV is indicated. (C) Suppressors restored metaphase chromosomal localization of the BPV DNA. FISH analysis was performed as in Fig. 3.
Suppressor Screen.
Low molecular weight DNA was recovered from expanded neomycin-resistant (neor) morphologicaly transformed colonies by a modified HIRT method (11). BPV sequences were recovered by PCR amplification using primers flanking the BamHI site or the AflII and HindIII sites in BPV. A circularly permuted genome was created to allow for linearization of BPV in the late region and for cloning and transfer of fragments that span the early region. The vector was a deleted form of pBluescript SK+, which was cleaved with BamHI and Asp718, blunted with Mung bean nuclease, and religated forming pBX. Circular BPV DNA was cleaved with XbaI and inserted into the XbaI vector site. The pBXBPV DNAs yielded equivalent transformation and replication results to that obtained with pMLBPV (11).
Protein Immunolocalization.
Cells were cotransfected with the neomycin-resistance gene and BPV genomes that were mutant for the splice site for E8/E2 and the initiating methionine for E2C. After neomycin drug selection, the cells were split to slides at day 9 and fixed at day 10 with 50% methanol/50% acetone for 5 min. Fixation and all subsequent steps were performed at room temperature. The slides were washed with PBS and blocked with PBSTB (0.1% Triton X-100/3% BSA in PBS) for 30 min. B201 mAb (provided by Elliot Androphy, Tufts University School of Medicine, Boston) was diluted 1:4 in PBSTB and bound for 1 hr followed by three washes in PBST. Goat anti-mouse secondary antibody coupled to Cy3 (Amersham) was diluted 1:250 in PBSTB and incubated for 30–45 min. Cells were counterstained with DAPI and mounted as for colony staining. Fluorescence microscopy used the DAPI or rhodamine filter sets with a ×100 objective.
RESULTS
To follow the fate of the mutant A4 DNA in dividing cells, we cotransfected the viral DNA with the neomycin-resistance gene and performed FISH with BPV DNA as probe. The cotransformation index was near 100% for wild type and A3 as judged by the frequencies of neor cells and morphological transformation (11) and this point was substantiated by FISH (Fig. 2A) with about 90% of wild-type and A3 colonies staining for BPV DNA. Every cell within these neor colonies contained a bright signal with the BPV probe. However, neor colonies transfected with the A4 mutant yielded a heterogeneous staining pattern for BPV DNA (Fig. 2A). When large numbers of such colonies with the DAPI/FISH comparison were scored, greater than 50% of the colonies showed sectoring with a variety of patterns. In contrast, no sectoring was detected with either wild-type or A3 genomes. The sectoring was not a function of cell plating density because the frequency of sectored colonies was the same at limiting dilution. These staining patterns were reminiscent of bacterial sectoring where a plasmid is periodically lost in a population of nonselected dividing cells. In some cases, only cells in the middle of the colony retained signal, whereas in other colonies stained patches emanated from the center. As cells that have lost viral DNA remain viable and continue to grow, these patterns of sectoring are consistent with the previous results showing that the A4 genome does not cause apoptosis or otherwise block cell growth (11). Moreover, the mutant DNA did not seem to be gradually lost as the colony expanded, either patches of negative or positive signal were observed consistent with a segregation defect rather than a slow reduction in replication rates. These data are consistent with previous results that showed equivalent transient replication for the A3 and A4 genomes. We have also shown that when neor colonies cotransfected with the A4 genome were picked and assayed for viral DNA, none was detectable in five of six such lines, consistent with the rapid loss of FISH signal reported herein (11). In contrast 100% of the neor colonies cotransfected with wild-type or A3 genomes maintained plasmids (11).
We reasoned that if the A4 genome is unable to accurately segregate or be maintained in daughter cells, one would expect that DNA loss would be dependent on cell division rather than time. To test this, viral accumulation was assayed 2 weeks after transfection under two growth conditions. In one culture, cells were allowed to grow until the plates became confluent, thus, arresting further cell growth after limited doublings. In parallel, cells were split repeatedly, facilitating many more cell divisions over the same time course. The level of accumulated A4 genome was comparable to the A3 mutant when the cells had divided about four times over 2 weeks. This indicates that the DNA was not lost by simple degradation or cell killing (Fig. 2B). On the other hand, when the cells had proceeded through about 15 divisions over the same time, the level of DNA maintained by the A4 mutant was only about 8% that of A3. This indicates that the loss of DNA by A4 is dependent on cell division. These results support the contention that the A4 mutant is defective for plasmid segregation.
We sought to follow the BPV DNA localization intracellularly with particular attention to mitosis, when nuclear membranes break down and segregation functions might be observed. In interphase cells, punctate staining was seen, indicating a nonrandom distribution (C.W.L., unpublished results). By blocking cell progression with Colcemid, we were able to enrich the population for mitotic cells. Fig. 3 shows that transforming BPV DNA associated with metaphase chromosomes but the A4 mutant did not. A4 DNA was still apparent in these cells, but staining was diffuse in the nucleoplasm rather than associated with the chromosomes as for A3. In Fig. 3, metaphase chromosomes are stained with DAPI (blue) and the BPV FISH signal is red. The centromeric heterochromatic regions appear as brighter staining dots. When the two signals colocalize, the merge appears as yellow or pink. At this level of resolution, we have detected no specific chromosomal attachment sites and most frequently both sister chromatids had viral DNA. We did notice in some cases one chromatid had a higher concentration of viral DNA than did the other (see enlarged image in Fig. 3). These results suggest an obvious mechanism for effective plasmid segregation and nuclear retention. By attaching to cellular chromosomes before division of sister chromatids in mitosis, the virus can ensure that each progeny cell receives a sufficient complement of viral genomes. The A4 mutant is defective for this association, suggesting a direct role for E2 in this mechanism.
To substantiate the role of chromosome attachment in maintenance of a viral transformed state and to potentially learn more about the segregation mechanism, we screened for genetic suppressors of the A4 mutation. The rare morphologically transformed foci obtained with the mutant were isolated and expanded. Low molecular weight DNA was isolated from individual lines and BPV DNA was obtained. The frequency of transformation with the suppressor DNA clones was equivalent to wild type (Table 1). Moreover, the size and appearance of colonies were indistinguishable from wild type (Fig. 1). The positions of the mutations conferring suppression were determined by marker rescue to fully sequenced restriction fragments. Thus far five suppressors have been characterized by this approach. Only one of the suppressors mapped to the E2 hinge region, whereas the larger target seemed to be the E1 gene. These suppressors in E1 mapped throughout the ORF (Fig. 4A). Transient replication by these suppressor variants was equivalent to that detected for the defective A4 genome, implying that suppression was not caused by a hyperreplication phenotype (Fig. 4B and Table 1). Also plasmid copy number and E2 protein levels detected in transformed cell lines ranged between that for wild type and A3 (C.W.L., unpublished results). In the context of a wild-type genome (i.e., without the A4 mutation), these suppressors do not manifest any detectable replication or transformation phenotype consistent with the notion that the selected mutations suppress the A4 defect and do not create defects or new activities in the virus (C.W.L., unpublished results). Most importantly and as anticipated, the suppressor mutations restored colocalization of viral genomes to the mitotic chromosomes (Fig. 4C and Table 1).
Table 1.
Suppressors of the A4 transformation phenotype
| Mutant | Mutation | Transformation | Repl | In situ |
|---|---|---|---|---|
| E1 F108L | T1557C | 64 ± 10 (6) | +++ | + |
| K324E | A1818G | 70 ± 8 (3) | +++ | + |
| C484F | G2299T | 76 ± 13 (6) | + | + |
| K237E/H357Q | A1170G/T1919A | 81 ± 12 (7) | +++ | + |
| E2 D255G | A3371G | 81 ± 20 (4) | +++ | + |
| S235D* | T3310C/G3311A | 76 ± 26 (5) | +++ | + |
Residue changes in E1 and E2 mutants use the single amino acid code. Mutations are in the context of the A4 BPV genome. Transformation is expressed as a percent of wild-type transformation (mean and ± SD), and sample size is in parentheses (n), with A4 being 4 ± 3% (n = 40). Transient replication (Repl): +, equivalent to wild type: +++, equivalent to A3 levels. In situ gives chromosomal localization of DNA by FISH analysis.
*Site-directed mutation.
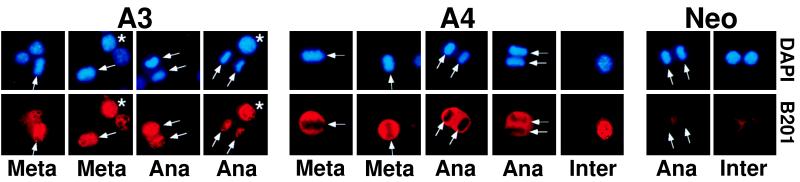
We sought to test directly the hypothesis that the E2 protein itself plays a role in plasmid segregation by attaching to mitotic chromosomes and that this activity is defective in the mutant form. The data obtained by immunofluoresence methods with an E2 mAb are shown in Fig. 5 and establish this point. Whereas the A3 protein appeared to bind chromosomes at both the metaphase and anaphase stages of mitosis, the A4 protein did not. Although the A4 protein appeared nuclear at interphase, it was excluded from the condensed chromatin in mitosis and became cytoplasmic.
Figure 5.
Direct evidence for the role of E2 in chromosomal attachment. E2 protein colocalized with mitotic chromosomes, whereas the A4 mutant protein did not. Cells were cotransfected with the neomycin-resistance gene and BPV genomes that do not express the repressor forms of E2 and fixed after drug selection at day 10 (at which point about half of the cells were neor and contained BPV DNA because of the time course of neomycin killing). E2 protein was detected by B201 monoclonal antibody followed by Cy3-conjugated secondary antibody, and DNA was detected by DAPI staining. The B201 antibody recognizes the wild-type and mutant forms of E2 equally, as demonstrated by the equivalent interphase signals obtained (panels labeled Inter and cells labeled with ∗). Arrows indicate the positions of condensed chromosomes in cells at the metaphase or anaphase stages of mitosis. Two metaphase and two anaphase examples are shown for the A3 and A4 samples. Quantitatively, 14 of 26 mitotic cells transfected with the A3 virus contained chromosomal staining, whereas 0 of 17 mitotic cells transfected with the A4 virus showed this colocalization. Interphase nuclei for both A3 and A4 samples stain for E2. Panels labeled Neo show that the E2 reagents do not stain interphase or anaphase nucleoprotein when BPV genomes were not cotransfected.
DISCUSSION
The data presented herein provide support for the notion that BPV plasmid maintenance is dependent on the E2 protein in a manner that is separate from its enhancer role in gene expression or replication. Moreover, this segregation activity appears to be regulated by phosphorylation. The morphological transformation efficiency of the A4 genome is only a few percent of wild type, and as we have shown, the rare drug-resistant colonies that do harbor viral DNA contain second-site suppressor mutations. The inability to establish the A4 genome in a stable cell line seemed paradoxical given the higher than wild-type levels of replication in transient assays. Colonies obtained by coselection showed sectoring of A4-BPV DNA, as monitored by FISH in contrast to a complete absence of sectoring for the wild-type and A3 genomes. The experimental conditions were set so that the cotransformation index (BPV + neor) was close to 100%. Neither the size nor the frequency of neor colonies was affected by the mutant or wild-type BPV DNA. This argues that neither viral genome effects the initial growth rate of the neor recipient. Thus we can reason that the null cells in the A4 sectored colonies arise by loss of viral DNA and that transformation is blocked because the cells lose the viral DNA. This segregation defect, which we suggest below is caused by loss of nuclear retention, may now explain all of the phenotypes associated with the mutant genome, including the higher replication activities. In all cases examined, there is a correlation with chromosomal attachment at mitosis of both the transforming viral DNA and E2 protein and lack thereof for the nontransforming variants. These correlations extend to the second-site suppressors that we have characterized (Table 1). The suppression data coupled with the phenotypes of these mutants suggest that E1 may also be a critical component in the segregation mechanism.
These findings are supported by the recent observations that long-term maintenance of BPV ori constructs depends on both E1 and E2 gene products and significantly multimeric E2 binding sites in cis were critical for such function. In contrast, for transient amplification function, only single E2 binding sites were required, implying that extra E2 sites might be needed for other activities in maintenance (16). Given the results presented herein establishing a genetic role for such activity in maintenance and involvement of a distinct E2 protein activity for such function, it seems likely that the enhancer protein directly functions in this chromosomal tethering.
It is perhaps a bit premature to speculate on the nature of the tethering complex on the viral side and in the chromosome because our data are consistent with a chromosomal target that is generally distributed on all or most chromosomes. Nevertheless, it is interesting to point out that E2 protein binds to the TFIID subunit TATA binding protein and that this interaction for cooperative binding to DNA is at least in part dependent upon the hinge region (17, 18), an interaction that may be regulated by E2 phosphorylation. Furthermore, a hyperphosphorylated form of TFIID has been shown to bind generally to mitotic chromosomes and to thus segregate to daughter cells in this way (19).
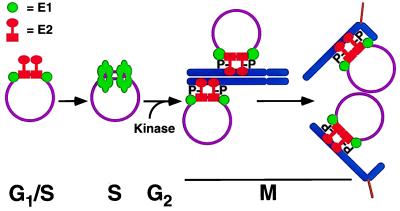
Fig. 6 summarizes the proposed roles of the viral E1 and E2 proteins in the cell cycle. During interphase both proteins have known direct activities in viral gene expression and replication. We suggest that on nuclear membrane breakdown, phosphorylation of E2 becomes critical for segregation and nuclear retention activities. We do not know precisely when this phosphorylation occurs with respect to mitosis but it is known that E2 phosphorylation is highest in the G2/M phase of the cell cycle (20). One of the major phosphorylation residues in the hinge region (position 298) lies within a cdc2p34 kinase consensus site, and it has been reported that in vitro the protein can be modified by this cell cycle kinase only when it is in complex with E1 (14). This would suggest that a preformed E1–E2 complex on the viral DNA might serve as a substrate for such an activity that peaks in early mitotic cells. E1 and E2 are known to associate and to bind cooperatively to DNA; further, this cooperativity can target E1 to multiple sites on the viral chromosome (9, 10, 21–24). However, we must emphasize that phosphorylation within the hinge domain of E2 seems to be redundant because only one major phosphorylation seems to suffice for maintenance activity. Thus other unknown kinases may be of equal importance in regulating this function.
Figure 6.
Proposed model for E1 and E2 function in viral replication and maintenance in the cell cycle. Hypophosphorylated E2 loads E1 monomers onto DNA in G1 phase, facilitating E1 multimerization into the active double hexameric E1 helicase and release of E2 from DNA in S phase. After replication, E1 and E2 bind to viral DNA and E2 can be phosphorylated by a G2/M kinase facilitating chromosomal binding. The DNA can then be accurately partitioned to daughter cells and retained in nuclear structures reformed around the cellular chromosomes in telophase. The oligomeric state of E1 and E2 as attached to the chromosome is hypothetical and based on the findings that show a 1:1 complex between an E2 dimer and an E1 monomer. Suppressor mutations in E1 may arise that do not require E2 phosphorylation because these new E1 proteins create a stronger binding surface for either E2 or the chromosome. To start the cycle anew, E2 can be synthesized de novo or a phosphate can be removed from E2. The E2 and E1 functions in regulating transcription are not included here.
The genetic suppression data are only an indirect indication that E1 assists E2 in tethering the viral DNA to chromosomes, and at this point, this part of our model is speculative. We do not believe that it is likely that single point mutations could result in entirely new activities for E1, and the genetic data and biochemical results outlined above are thus consistent with our model. However, we cannot exclude the possibility that E1 normally interferes with hypophosphorylated E2 in tethering to chromosomes and the mutations could relieve such interference. Future experiments aimed at determining what activities are necessary and sufficient for tethering BPV DNA to the chromosome and identification of the chromosomal receptor should clarify these points.
Nuclear retention during or after mitosis may be the critical function in the viral maintenance process emphasized herein. The FISH analysis showed no indication of extreme viral clumping in interphase nuclei and the hypophosphorylated form of E2 was excluded from the mitotic apparatus. By binding to cellular chromosomes, viral DNA would become enclosed within the new nuclear envelopes. Without this tethering mechanism, viral nucleoprotein complexes may remain cytoplasmic and become degraded or be slowly transported back into the nucleus. Therefore, chromosomal binding may serve both partitioning and nuclear retention functions.
A curious feature of both the A3 and A4 mutants is that they both replicate transiently in cells better than does wild-type DNA (Fig. 4B and refs. 11 and 25). Though this in vivo phenotype fits with the fact that E2 phosphorylation levels are lowest in S phase (20) and that phosphorylation would thus negatively regulate DNA replication, we have been unable to detect any differences in vitro for hypophosphorylated E2 (e.g., A4) or hyperphosphorylated wild-type E2 in their abilities to enhance E1 binding or DNA replication (ref. 11 and C.W.L., unpublished results). We suspect that the high-replication phenotypes of the viral mutants that cannot be phosphorylated may be caused by changes in subcellular localization, which might optimize the size of the replication pool. The mutation at Ser-301, for example, may create an E2 more likely to release the viral DNA from chromosomes into a freely replicating pool.
The segregation functions discussed in this report are likely to be homologous to mechanisms used by other eukaryotic viruses. The Epstein–Barr virus has long been known to be associated with metaphase chromosomes (26, 27), and the EBNA1 protein, itself a DNA binding protein, may mediate this attachment (26). Although a clear role for this EBNA1 function in viral maintenance has not been provided, multiple EBNA1 binding sites in tandem can provide for a persistence function when EBNA1 is supplied in trans in cells that cannot replicate such DNA constructs (28, 29). Strikingly, the crystal structure for the E2 and the EBNA1 DNA binding domains reveal great structural homologies, in the absence of sequence homology (30, 31). Both proteins are phosphorylated in regions outside of the DNA binding domain (32) and the gene products play critical roles in both replication and transcription of their respective genomes. It seems reasonable to suggest that chromosomal attachment provides another reason for such convergent evolution.
Acknowledgments
We dedicate this paper to the memory of our friend and colleague Yasha Gluzman. We appreciate technical assistance provided by Anne Fletcher and Sunny Kim. B201 antibody was kindly provided by Elliot Androphy. This work was supported by National Institutes of Health Grants CA42414 and CA30490.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: BPV, bovine papillomavirus; FISH, fluorescence in situ hybridization; neor, neomycin resistance.
A commentary on this article begins on page 4084.
References
- 1.Nordstrom K, Austin S. Annu Rev Genet. 1989;23:37–69. doi: 10.1146/annurev.ge.23.120189.000345. [DOI] [PubMed] [Google Scholar]
- 2.Niki H, Hiraga S. Cell. 1997;90:951–957. doi: 10.1016/s0092-8674(00)80359-1. [DOI] [PubMed] [Google Scholar]
- 3.Mohl D A, Gober J W. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- 4.Webb C D, Teleman A, Gordon S, Straight A, Belmont A, Lin D C, Grossman A D, Wright A, Losick R. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- 5.Zur Hausen H. Virology. 1991;184:9–13. doi: 10.1016/0042-6822(91)90816-t. [DOI] [PubMed] [Google Scholar]
- 6.Howley P M. In: Fundamental Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott-Raven; 1996. pp. 947–978. [Google Scholar]
- 7.Yang L, Mohr I, Fouts E, Lim D A, Nohaile M, Botchan M. Proc Natl Acad Sci USA. 1993;90:5086–5090. doi: 10.1073/pnas.90.11.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seo Y-S, Muller F, Lusky M, Hurwitz J. Proc Natl Acad Sci USA. 1993;90:702–706. doi: 10.1073/pnas.90.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seo Y-S, Muller F, Lusky M, Gibbs E, Kim H-Y, Hurwitz J. Proc Natl Acad Sci USA. 1993;90:2865–2869. doi: 10.1073/pnas.90.7.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sedman J, Stenlund A. EMBO J. 1995;14:6218–6228. doi: 10.1002/j.1460-2075.1995.tb00312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehman C W L, King D M, Botchan M R. J Virol. 1997;71:3652–3665. doi: 10.1128/jvi.71.5.3652-3665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trask B J. Methods Cell Biol. 1991;35:3–35. doi: 10.1016/s0091-679x(08)60567-1. [DOI] [PubMed] [Google Scholar]
- 13.McBride A A, Bolen J B, Howley P M. J Virol. 1989;63:5076–5085. doi: 10.1128/jvi.63.12.5076-5085.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lentz M R, Pak D, Mohr I, Botchan M R. J Virol. 1993;67:1414–1423. doi: 10.1128/jvi.67.3.1414-1423.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zanardi T A, Stanley C M, Saville B M, Spacek S M, Lentz M R. Virology. 1997;228:1–10. doi: 10.1006/viro.1996.8375. [DOI] [PubMed] [Google Scholar]
- 16.Piirsoo M, Ustav E, Mandel T, Stenlund A, Ustav M. EMBO J. 1996;15:1–11. [PMC free article] [PubMed] [Google Scholar]
- 17.Steger G, Ham J, Lefebvre O, Yaniv M. EMBO J. 1995;14:329–340. doi: 10.1002/j.1460-2075.1995.tb07007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rank N M, Lambert P F. J Virol. 1995;69:6323–6334. doi: 10.1128/jvi.69.10.6323-6334.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segil N, Guermah M, Hoffmann A, Roeder R G, Heintz N. Genes Dev. 1996;10:2389–2400. doi: 10.1101/gad.10.19.2389. [DOI] [PubMed] [Google Scholar]
- 20.Notolli T. Ph.D. thesis. Berkeley: University of California; 1992. [Google Scholar]
- 21.Spalholz B A, McBride A A, Sarafi T, Quintero J. Virology. 1993;193:201–212. doi: 10.1006/viro.1993.1116. [DOI] [PubMed] [Google Scholar]
- 22.Yang L, Li R, Mohr I J, Clark R, Botchan M R. Nature (London) 1991;353:628–633. doi: 10.1038/353628a0. [DOI] [PubMed] [Google Scholar]
- 23.Mohr I J, Clark R, Sun S, Androphy E J, MacPherson P, Botchan M R. Science. 1990;250:1694–1699. doi: 10.1126/science.2176744. [DOI] [PubMed] [Google Scholar]
- 24.Mendoza R, Gandhi L, Botchan M R. J Virol. 1995;69:3789–3798. doi: 10.1128/jvi.69.6.3789-3798.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McBride A A, Howley P M. J Virol. 1991;65:6528–6534. doi: 10.1128/jvi.65.12.6528-6534.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grogan E, Summers W, Dowling S, Shedd D, Gradoville L, Miller G. Proc Natl Acad Sci USA. 1983;80:7650–7653. doi: 10.1073/pnas.80.24.7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris A, Young B D, Griffen B E. J Virol. 1985;56:328–332. doi: 10.1128/jvi.56.1.328-332.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reisman D, Sugden B. Mol Cell Biol. 1986;6:3838–3846. doi: 10.1128/mcb.6.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krysan P J, Haase S B, Calos M P. Mol Cell Biol. 1989;9:1026–1033. doi: 10.1128/mcb.9.3.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hegde R S, Grossman S R, Laimins L A, Sigler P B. Nature (London) 1992;359:505–512. doi: 10.1038/359505a0. [DOI] [PubMed] [Google Scholar]
- 31.Bochkarev A, Barwell J, Pfuetzner R, Bochkareva E, Frappier L, Edwards A. Cell. 1996;84:791–800. doi: 10.1016/s0092-8674(00)81056-9. [DOI] [PubMed] [Google Scholar]
- 32.Hearing J C, Levine A J. Virology. 1985;145:105–116. doi: 10.1016/0042-6822(85)90205-3. [DOI] [PubMed] [Google Scholar]
- 33.McBride A A, Romanczulc H, Howley P M. J Biol Chem. 1991;266:18411–18414. [PubMed] [Google Scholar]