Abstract
Two B-type cyclins, B1 and B2, have been identified in mammals. Proliferating cells express both cyclins, which bind to and activate p34cdc2. To test whether the two B-type cyclins have distinct roles, we generated lines of transgenic mice, one lacking cyclin B1 and the other lacking cyclin B2. Cyclin B1 proved to be an essential gene; no homozygous B1-null pups were born. In contrast, nullizygous B2 mice developed normally and did not display any obvious abnormalities. Both male and female cyclin B2-null mice were fertile, which was unexpected in view of the high levels and distinct patterns of expression of cyclin B2 during spermatogenesis. We show that the expression of cyclin B1 overlaps the expression of cyclin B2 in the mature testis, but not vice versa. Cyclin B1 can be found both on intracellular membranes and free in the cytoplasm, in contrast to cyclin B2, which is membrane-associated. These observations suggest that cyclin B1 may compensate for the loss of cyclin B2 in the mutant mice, and implies that cyclin B1 is capable of targeting the p34cdc2 kinase to the essential substrates of cyclin B2.
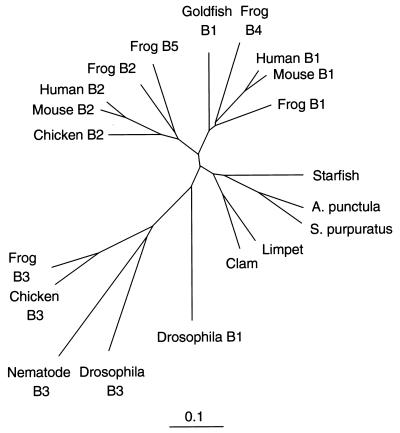
Mitotic B-type cyclins activate the p34cdc2 protein kinase to form maturation promoting factor, which is required for cells to undergo mitosis (1–3). Only two B-type cyclins, B1 (4, 5) and B2 (6, 7), have been identified so far in mammals, although chickens, frogs, flies, and nematode worms possess a third, more distant relative—cyclin B3 (8, 9). The genes of cyclin B1 and B2 show very little similarity in the first 100 residues, and about 57% identity in the remaining 300 residues. The two genes must have diverged early in vertebrate evolution, because frogs have both genes. The family resemblances are strongly conserved: Goldfish B1 is almost 70% identical to mouse B1 in the C-terminal 300 residues, and chicken B2 is 75% identical to mouse B2 in the equivalent region. Fig. 1 shows a dendrogram of the current B-type cyclin family tree, omitting yeast examples that cluster together as a separate branch. Cyclins B1 and B2 are coexpressed in the majority of dividing cells, although their subcellular localization differs, with cyclin B1 usually associated with microtubules, and cyclin B2 with intracellular membranes (10–15). Cyclin B1 enters the nucleus in late G2 phase of the cell cycle, whereas cyclin B2 does not (15). Cyclins B1 and B2 also show different patterns of expression during murine spermatogenesis (6), and in Xenopus oocytes, cyclin B2 protein is already present in unactivated oocytes, whereas cyclin B1 is not synthesized until progesterone induced oocyte maturation (16, 17). These considerations would lead one to suspect that cyclins B1 and B2 have specialized roles in preparing cells for mitosis, but there is no direct evidence for this view from studies in vertebrates.
Figure 1.
Cyclin B1 and B2 diverged early in vertebrate evolution. The protein sequences of cloned cyclins B1, B2 and B3 were compared by using clustal_x (46) and the output displayed by the treeview (47) program, using sequences starting at the beginning of the α–1 helix of the cyclin box, (MRAILIDWLV… ), and finishing at the C terminus. The scale is as calculated by the clustal_x program (46).
Yeasts possess an even wider range of B-type cyclins. The Saccharomyces cerevisiae genome encodes six B-type cyclins (Clb 1–6), which display functional redundancy (18–21). Not all of them have identical roles, however, nor are they expressed at the same stage of the cell cycle. Clb1 and 2 are expressed in late S-phase and G2 and clearly regulate mitosis (19), as do Clb3 and 4, which are expressed earlier in the cycle and may also be involved in S phase (21, 22). Clb5 and 6 are expressed at the onset of S phase and clearly play a role in DNA replication (20, 23). While none of these genes is essential, Clb2 seems to play a key role, being able to rescue Clb1, -3, and -4 triple mutants (21). In S. pombe three B-type cyclins are known so far—Cdc13, Cig1, and Cig2 (24). Cdc13 is an essential gene and its deletion prevents cells from entering mitosis (25, 26). Cig1 and cig2 are nonessential genes playing a role in S phase and the control of sporulation, rather than in mitosis (27, 28).
In this study we generated two strains of mice, the first heterozygous for a targeted deletion of the cyclin B1 gene and the second completely lacking cyclin B2, with the aim of shedding light on their specific functions in vertebrate cells. Cyclin B1 proved to be an essential gene and its deletion resulted in embryonic lethality. In contrast, nullizygous B2 mice developed normally and did not display any obvious abnormalities in adult life. Moreover, both male and female cyclin B2-null mice proved to be fertile, which was somewhat unexpected in view of the high levels and distinct patterns of expression of cyclin B2 during spermatogenesis described by Chapman and Wolgemuth (6), and explored further in this paper (see Fig. 5).
Figure 5.
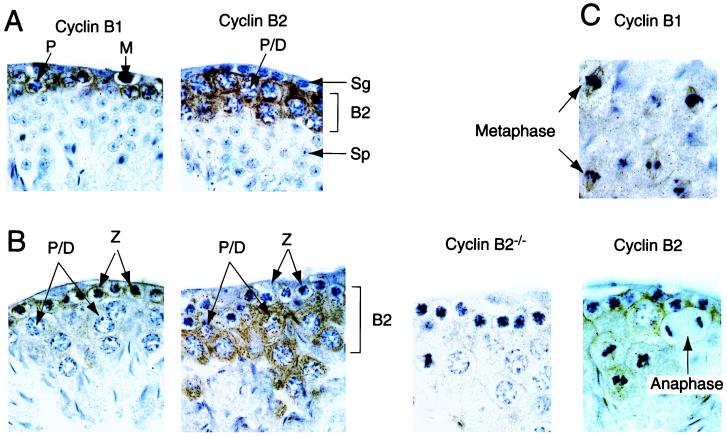
Cyclin B1 and B2 localization in the testis. Sections of testis, prepared from cyclin B2 nullizygous mice or their heterozygous littermates, were analyzed by immunostaining with the indicated antibodies (brown) and counterstained with hematoxylin (blue) The sections in A show mitotic precursors (M, Sg) and postmeiotic spermatocytes (Sp). Cyclin B2 staining is seen in late pachytene and diplotene cells, whereas cyclin B1 staining is most intense at the earlier zygotene stage. The postmeiotic spermatocytes do not stain with either anti-B type cyclin antibody. B represents stages X or XI of spermatogenesis, cell types are: P/D, late pachytene or diplotene; Z, zygotene. A section from a B2 nullizygous animal is shown as a control. C shows stage XII sections with fields of meiotic metaphases and an anaphase from the same series of testis sections (B2+/− heterozygotes), stained with anti-cyclin B1 (Upper) or anti-cyclin B2 (Lower). Note the disappearance of cyclin B2 at anaphase, and the association of both B-type cyclins with mitotic spindles. The upper two metaphase figures in the cyclin B1 panel probably represent meiosis I, and the lower set are probably meiosis II, judged by position in the section and the intensity of chromosome staining.
MATERIALS AND METHODS
Cloning and Construction of the Cyclin B2 Targeting Vector.
Three λ phages containing partially overlapping cyclin B2 sequences were initially isolated from a 3T3K genomic library screened with labelled mouse cyclin B2 cDNA (a gift of D. Wolgemuth, Columbia University College of Physicians and Surgeons, New York). Exon-intron boundaries were mapped and sequenced. Subsequently a λ phage encompassing most of the cyclin B2 gene was cloned from a 129/SV library (29) and used to construct the targeting vector as follows: A 2 kb XbaI (blunted)-ApaI fragment spanning intron 5 to exon 7 was cloned into the XhoI (blunted) and ApaI sites of pSK-MC1-Neo-PolyA (prepared by cloning the BamHI-XhoI TK-Neo-polyA gene from pMC1-Neo-polyA (Stratagene) into the same sites of Bluescript SKII+). Subsequently, a genomic XbaI fragment of 6 kb, spanning introns 1 to 3, was cloned from the phage into Bluescript, recovered by NotI-SpeI digestion and cloned into the NotI and SpeI sites of the targeting vector.
Cloning and Construction of the Cyclin B1 Targeting Vector.
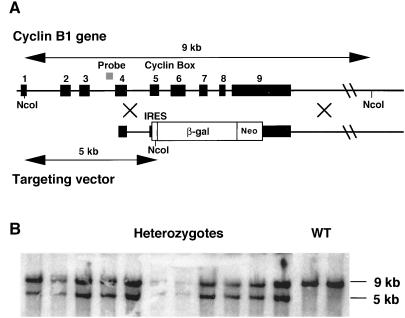
Several λ phages containing genomic cyclin B1 inserts were isolated from a 3T3K genomic library that was screened with labeled mouse cyclin B1 cDNA (a gift of D. Wolgemuth). At least two of these isolates encoded distinct spliced pseudogenes, which confirms reports that the mouse genome harbors several copies of cyclin B1 (30, 31). One clone, which encoded a version of the cyclin B1 gene that contained introns as well as exons, was assumed to be the genuine cyclin B1. We sequenced part of the third intron of cyclin B1 from this phage and used PCR to synthesize a probe. This intron 3 probe was used to clone λ phages coding for cyclin B1 from a 129/SV library (29) that were used to construct the targeting vector as follows: A polylinker (SfiI-EcoRV-XbaI-BamHI-SfiI) was cloned into the XbaI site of Bluescript SKII, making Bluescript SKII-Sfi. A fragment of about 1.5 kb spanning exon 4, intron 4, and the 5′ end of exon 5 (before the cyclin box) was amplified by PCR, digested with EcoRV and XbaI and subcloned into the corresponding sites of Bluescript SKII-Sfi. Next a 6-kb BamHI fragment spanning ≈1 kb of the 3′ untranslated region of the gene and 5 kb of downstream sequences was cloned into the BamHI site of this plasmid. Finally a 4.5-kb XbaI fragment of pGT1.8IresBgeo (a gift of I. Chambers, ref. 32) encoding an internal ribosome entry site and a β-galactosidase-neo fusion protein was ligated into the XbaI site of the vector (see Fig. 3).
Figure 3.
Cyclin B1 targeting in ES cells. (A) Map of the mouse cyclin B1 gene and of the targeting vector used for homologous recombination. (B) Southern blot analysis of the NcoI-digested genomic DNA from G418-resistant colonies with an intron 3 probe (see map).
Generation of Cyclin B2 and B1-Null Mice.
The targeting vectors (20 μg) for disruption of cyclins B1 or B2 were linearized with NotI and separately electroporated into GK129 ES cells from 129/Ola mice and selected as described by Fantl et al. (33). G418-resistant colonies were picked and grown in duplicate 96 well plates. Genomic DNA for screening was prepared from one set of plates while the cells in the other set were stored as frozen stocks. DNA was prepared (34) and screened for homologous recombination as described in the text and in Figs. 2 and 3. Cells from colonies that underwent homologous recombination were thawed, expanded and injected into blastocysts as described by Fantl et al. (33). The chimeras that developed from these blastocysts were mated to C57BL/6 mice and agouti offspring were screened by Southern blotting with the same intron 7 probe used for the screen of the neomycin-resistant colonies for the cyclin B2 gene (Fig. 2A) and intron 3 probe for the cyclin B1 gene.
Figure 2.
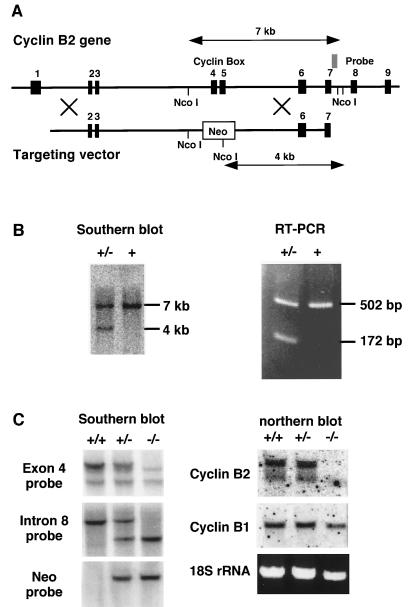
Cyclin B2 targeting in ES cells. (A) Map of the mouse cyclin B2 gene and of the targeting vector used for homologous recombination. (B) Southern blotting of the NcoI-digested genomic DNA from G-418 resistant colonies with an intron 7 probe (see map) and RT-PCR with exon 3 (CTGTGAAACCAGTGCAGATG) and 6 (ACTGGTGTAAGCATTATCTG) specific oligonucleotides, by using cDNA prepared from RNA of heterozygous or wt ES cells. (C) Southern blots of genomic DNA of homozygous, heterozygous, or wt mice were probed with an exon 4 probe, an intron 7 probe (whose location is indicated in A) or a neo probe as indicated. Northern blots of RNA were prepared from the testes of homozygous, heterozygous, or wt mice and were probed with cyclin B1- and B2-specific probes.
Immunostaining of Paraffin Sections.
Slices of decapsulated testis were washed once with saline and transferred to Bouin’s fixative before embedding. Paraffin sections (4 μM) of testis of heterozygous and homozygous littermates were dewaxed with xylene and ethanol and endogenous peroxidase activity quenched in H2O2: methanol (1:1) for 30 min. The sections were boiled in a microwave oven for 10 min in 10 mM Tris, pH 8.0 (35), blocked for 30 min with 10% fetal calf serum in PBS and incubated with anti-cyclin B2 polyclonal antibody 190 or anti-cyclin B1 mAb V152. After three washes in PBS the sections were incubated with the appropriate horseradish peroxidase-conjugated secondary antibody (Dako) and washed again. Horseradish peroxidase-conjugates were visualized with diaminobenzidine and the sections were counterstained for 10 sec with hematoxylin, washed, dehydrated with ethanol followed by xylene, and mounted with DePeX.
Analysis of Thymocytes and Splenocytes.
Fluorescence-activated cell sorter analysis and mAbs were as described (36). Recombinant interleukins were purchased from PharMingen and were used at concentrations recommended by the manufacturers. Dexamethasone and Staphylococcal enterotoxin B were purchased from Sigma and were used at 10−7 M and 10 μg/ml, respectively. Proliferation and viability were measured after 24, 48, and 72 hr by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay as described (37).
RESULTS
Generation of Cyclin B2-Null Mice.
We used homologous recombination in embryonic stem (ES) cells to generate mice lacking functional cyclin B2. Three overlapping λ phages that span the entire 15-kb cyclin B2 gene, which contains nine exons, were isolated from a mouse genomic library, mapped, and partially sequenced (Fig. 2A). The highly conserved “cyclin box” that provides most of the binding site for Cdc2 (38, 39) is encoded by exons 4 and 5. A targeting vector that replaces these two exons with a thymidine kinase promoter-driven neomycin-resistance gene was prepared from a phage isolated from a 129SV library (29) (Fig. 2A) as described in Materials and Methods, and introduced into ES cells by electroporation. The transfected cells were selected with G-418, and resistant colonies screened for homologous recombination. DNA prepared from these colonies was digested with NcoI, transferred to nylon membranes and probed with a labelled DNA fragment within intron 7 (see Fig. 2A). The wild type (wt) allele yielded a 7-kb band and the allele that underwent recombination yielded a 4-kb band (Fig. 2B). About 3% of the screened colonies proved to have undergone homologous recombination, which was confirmed by reverse transcription–PCR of RNA from these cells. When cDNA was amplified with oligonucleotide primers from exons 3 and 6, the wt allele yielded a 502-bp product and the mutated allele a 172-bp product (Fig. 2B). Cells from four of these clones were injected into mouse blastocysts and gave rise to chimeric offspring. One of the chimeras demonstrated germline contribution and transmitted the deleted allele to two offspring, a male and a female, which were successfully mated to provide litters of wt, heterozygous, and homozygous mice (Fig. 2C). These mice were tested by Southern and Northern hybridization (Fig. 2C) and by immunoblotting (Fig. 4). The homozygous mice were completely devoid of detectable cyclin B2 but expressed normal amounts of cyclin B1 (Fig. 2C and Fig. 4). The mRNA and/or protein products of the truncated cyclin B2 are apparently unstable and were not detected in the blots. Both the probe used in the Northern blot and the antibody used in the immunoblot should have been able to identify the 5′ end of the mRNA or the N-terminal end of the protein, respectively.
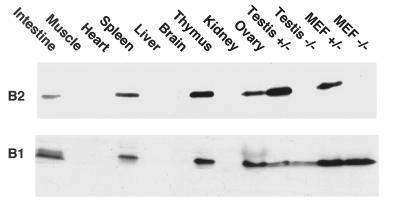
Figure 4.
Total protein was extracted from wt or mutant mouse tissues as indicated, or from fibroblasts prepared from 13-day-old embryos (MEF). Equivalent amounts of protein were analyzed by SDS/PAGE and immunoblotted with the anti-cyclin B1 mouse mAb V143 or by the anti-cyclin B2 rabbit polyclonal antibody 190 directed against the 101 N-terminal residues of cyclin B2. The cyclin B2 immunoblot was exposed about 10× longer than the B1 blot.
The Characteristics of Cyclin B2-Null Mice.
In crosses between heterozygous animals, wt, heterozygous, and homozygous pups were born in the ratio 48:84:28; homozygous B2-null animals were thus somewhat underrepresented. The homozygous B2-null animals did not differ in any obvious way from their littermates, except that in many litters they weighed less than their littermates. The B2-null animals developed normally and a thorough anatomical and histological examination of the mutant mice did not reveal any obvious malformations. To our surprise, matings between male and female homozygous B2-null mice resulted in viable offspring, showing that both the B2-null animals were fertile, although the litters tended to be small (data not shown).
Cyclins B1 and B2 are both expressed in organs that contain dividing cells, such as gut, spleen, thymus, skin, ovaries, and testis as well as in cultured embryonic fibroblasts (Fig. 4), as ws expected from their well-documented role in mitosis (17) and meiosis (6). As previously noted (40), the concentration of cyclin B2 protein was considerably lower than that of cyclin B1 except in the testis, where cyclin B2 was present at comparable levels to those of cyclin B1. Even here, however, the fertility of the nullizygous male mice indicates that cyclin B2 is dispensable. We were unable to detect any obvious differences between wt, heterozygous, and homozygous B2-null mice in histological sections of the testis (see Fig. 5).
Lack of Cyclin B2 Has No Effect on Lymphocyte Proliferation.
Cyclin B2 was also present at relatively high levels in spleen and thymus (Fig. 4), which suggested that it might play a role in proliferation of cells of the immune system. We therefore studied the proliferation of thymocytes and splenocytes in vivo and in vitro. Fluorescence-activated cell sorter analysis showed phenotypically normal thymuses and spleens in cyclin B2-null mutant mice with respect to common cell surface markers, although the thymuses of the mutant mice were somewhat smaller than those of their littermates and contained slightly higher proportion of CD4-CD8 double-negative cells. The distribution of cells in G1, S, and G2/M in in vitro stimulated thymocyte populations was similar in the B2-null animals and their littermates when measured by propidium iodide staining and fluorescence-activated cell sorter analysis of sorted populations. When thymocytes and splenocytes were stimulated in vitro with interleukin-2, -4, and -7; staphylococcal enterotoxin B; or dexamethasone there were no significant differences in survival or proliferation between cells derived from B2-null mutants and their littermates. Propidium iodide staining after two days in culture showed no difference in cell cycle status between mutant, heterozygous and wt. These results show that cyclin B2 is not essential for lymphocyte proliferation.
Generation of Cyclin B1-Null Mice.
We next used homologous recombination in ES cells to generate mice lacking functional cyclin B1. The targeting vector replaced the 3′ end of the coding region from the middle of exon 5 to the middle of exon 9 (thereby deleting the cyclin box) with a selection cassette consisting of an internal ribosome entry site-β-galactosidase-neo fusion protein. Expression of this protein relies on the promoter and the polyadenylation signal of the endogenous cyclin B1 gene. ES cells were electroporated with this construct as described for the cyclin B2 gene and in Materials and Methods. DNA prepared from G-418 resistant colonies was digested with NcoI and screened by Southern hybridization with the same intron 3 probe as was used for the cloning of the gene (Fig. 3A). The wt allele yielded a 9-kb band and the targeted allele a 5-kb band (Fig. 3B). As previously observed (32), the promoterless selection method used here dramatically enhanced the rate of homologous versus random integration, and ≈80% of the clones had the desired replacement.
Two of the targeted clones were separately injected into blastocysts as described above, and both gave rise to several germline transmitting, high contribution chimeras. The heterozygous offspring of these chimeras did not suffer from any obvious abnormalities. Five mating couples of heterozygous mice were set up and a total of 18 litters gave rise to 124 offspring, of which 81 were heterozygotes, 43 were wt, and none were homozygous B1-null pups. These results show that, unlike cyclin B2, cyclin B1 is an essential gene and that B1−/− mice do not survive to term. We have not yet established the embryonic stage at which these mice fail to develop further. So far, analysis of 10-day embryos from heterozygous crosses showed no homozygous B1-null in a sample of 20 (8 wt, 12 heterozygotes), so the failure in development probably occurred earlier.
Cyclin B1 and B2 Have Distinct Expression Patterns in the Testis.
In earlier studies of B-type cyclins in the mouse testis, Chapman and Wolgemuth investigated the distribution of the mRNA for cyclin B1 and B2 (4, 6), and also of B1 protein (41) at different stages of spermatogenesis. We stained paraffin sections of testis with anti-cyclin B1 or anti-cyclin B2 antibodies to further investigate this issue (Fig. 5). We found that cyclin B1 is present in primary spermatogonia at all stages of meiotic prophase. The strongest signal was obtained from cells in zygotene, when homologous chromosomes pair, although it was readily detectable in pachytene cells. Cyclin B1 was destroyed at meiotic anaphase I, resynthesized (to a lower level) and destroyed again at second meiotic anaphase. In contrast, cyclin B2 was barely detectable until early pachytene, but the staining dramatically increased to higher levels than cyclin B1 in mid-to late pachytene cells. Like cyclin B1, cyclin B2 disappeared at anaphase (Fig. 5C). Neither of the B-type cyclins could be detected in the round, postmeiotic spermatids (Fig. 5A). The patterns of expression of cyclin B1 we observed are consistent with those previously reported (41) and probably explain how cyclin B1 can compensate for the loss of cyclin B2 in spermatogenesis. No obvious morphological changes were detected in the sections of testis from B2-null males. The absence of cyclin protein after the end of meiosis II metaphase, despite the high levels of mRNA reported by Chapman and Wolgemuth (41) is consistent with our previous observation that B-type cyclins are highly unstable in G1 and G0 cells (40).
Cyclin B2 Is Associated with Subcellular Membranes and Cyclin B1 Also Is Found Free in the Cytoplasm.
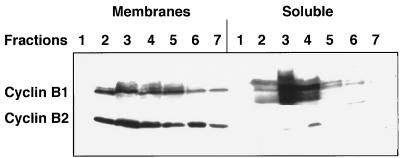
It has been reported that cyclin B1 and B2 differ in their subcellular localization in tissue culture cells (15). This difference might be relevant to the capability of cyclin B1 to compensate for the loss of cyclin B2 in the nullizygous mutant mice. We therefore examined the subcellular localization of the two B-type cyclins in cell-free extracts of testis. Mouse testes were homogenized in buffer and the postnuclear extract loaded on a sucrose gradient (0.25–2.0 M) and centrifuged for 16 hr at 105 × g. The fractions of this gradient were further separated into particulate matter (membranes) that were sedimented by high speed centrifugation, and soluble proteins, which were recovered by methanol-chloroform precipitation. Both cyclins were present in most of the gradient fractions, but while cyclin B1 was found equally in the soluble and membrane fractions, cyclin B2 was restricted to the membranes (Fig. 6). This result, which agrees with previous immunofluorescence studies (15), again suggests how cyclin B1 might compensate for the loss of cyclin B2.
Figure 6.
Cyclin B1 and B2 differ in their subcellular localization. Mouse testes were homogenized in a Hepes-sucrose buffer and nuclei were removed by low speed centrifugation. The postnuclear extract was loaded on a 4 ml sucrose step gradient and centrifuged overnight in a Beckman SW55 rotor at 30,000 rpm (105 g). The fractions collected from the gradient were diluted and spun for 45 min in a Beckman TL100 benchtop ultracentrifuge. The membranous pellet was dissolved in SDS/PAGE sample buffer. The protein in the supernatant was precipitated with methanol and chloroform, dried, and dissolved in sample buffer. The fractions were analyzed by immunoblotting with anti-cyclin B1 and B2 antibodies.
DISCUSSION
In this paper, we show that cyclin B1 is an essential gene in the mouse, whereas cyclin B2 appears to be dispensable. Several other cyclins have previously been “knocked out” in mice. In the case of cyclins D1 and D2, the mice developed relatively normally but showed defects in specific tissues: cyclin D1-null mice suffer from retinal atrophy and defective mammary gland development (33, 42), cyclin D2-null mice are sterile due to defects in granulosa cells in females and hypoplastic testes in males (43), attesting to compensation in all other tissues where these cyclins are usually expressed. In contrast, cyclin A2 (the somatic version of cyclin A) proved to be an essential gene whose homozygous loss led to early embryonic lethality (44). These observations are somewhat reminiscent of the situation in S. cerevisiae, where the different Clbs and Clns are at least partly functionally redundant (19–21, 23, 45), although Cdc13, one of three B-type cyclins in S. pombe, is an essential gene (26). We imagine that while the different B-type cyclins normally each target Cdc2 to certain favored substrates or subcellular organelles, they are capable of phosphorylating the entire spectrum of essential substrates. We were unable to detect any cell cycle abnormalities in lymphocytes derived from cyclin B2-null mice; there was no obvious lengthening of G2 phase, for example.
Although mutant mice lacking cyclin B2 proved to be viable, and both sexes were fertile, the sizes of the litters of homozygous mating pairs were consistently smaller than those of their heterozygous littermates (data not shown). These smaller litter sizes might either be due to lower fertility of the males or the females or to embryonic morbidity. The latter explanation would be consistent with the underrepresentation of B2-null mutants in the litters of matings between heterozygotes. A somewhat reduced body size of the B2-null mice also suggests that possession of cyclin B2 confers growth advantages that are evident even in small scale experiments under optimal laboratory conditions. In contrast to cyclin B2, we find that the cyclin B1 gene is essential in the mouse. This can be understood in terms of the consistently higher levels of cyclin B1 expressed in most proliferating cells and its localization to both membranes and cytoplasm, a wider range of subcellular sites than is found for cyclin B2. We are currently investigating the stage at which cyclin B1-null embryos die and whether this is also the stage at which some of the cyclin B2-null embryos fail to develop further. We are also engaged in mating pairs of B1+/−/B2+/− animals to see if it is possible to obtain B2-null/B1+/− offspring, with the thought that animals with half the normal level of cyclin B1 and no cyclin B2 might show a more severe phenotype than B2-null animals with normal levels of cyclin B1.
Acknowledgments
We would like to thank Peter Hagger and Tracy Crafton for excellent and dedicated animal care, Andor Sebesteny for the anatomical and Gill Williams for the histological examination, George Elia and his department for the histological work, D. Wolgemuth for the cyclin B2 probe, I. Chambers for pGT1.8IresBgeo, Gladys Lee and Michal Kubelka for help in preparing the anti-cyclin B2 antisera, and all the members of the Hunt laboratory. M.B. was supported by European Molecular Biology organization and Imperial Cancer Research Fund postdoctoral fellowships and also thanks B’nai Brith for their generous support.
ABBREVIATIONS
- wt
wild type
- ES
embryonic stem
References
- 1.Labbé J C, Capony J P, Caput D, Cavadore J C, Derancourt J, Kaghad M, Lelias J M, Picard A, Dorée M. EMBO J. 1989;8:3053–3058. doi: 10.1002/j.1460-2075.1989.tb08456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Draetta G, Luca F, Westendorf J, Brizuela L, Ruderman J, Beach D. Cell. 1989;56:829–838. doi: 10.1016/0092-8674(89)90687-9. [DOI] [PubMed] [Google Scholar]
- 3.Gautier J, Matsukawa T, Nurse P, Maller J. Nature (London) 1989;339:626–629. doi: 10.1038/339626a0. [DOI] [PubMed] [Google Scholar]
- 4.Chapman D L, Wolgemuth D J. Mol Reprod Dev. 1992;33:259–269. doi: 10.1002/mrd.1080330305. [DOI] [PubMed] [Google Scholar]
- 5.Pines J, Hunter T. Cell. 1989;58:833–846. doi: 10.1016/0092-8674(89)90936-7. [DOI] [PubMed] [Google Scholar]
- 6.Chapman D L, Wolgemuth D J. Development (Cambridge, UK) 1993;118:229–240. doi: 10.1242/dev.118.1.229. [DOI] [PubMed] [Google Scholar]
- 7.Minshull J, Blow J J, Hunt T. Cell. 1989;56:947–956. doi: 10.1016/0092-8674(89)90628-4. [DOI] [PubMed] [Google Scholar]
- 8.Kreutzer M A, Richards J P, De Silva Udawatta M N, Temenak J J, Knoblich J A, Lehner C F, Bennett K L. J Cell Sci. 1995;108:2415–2424. doi: 10.1242/jcs.108.6.2415. [DOI] [PubMed] [Google Scholar]
- 9.Gallant P, Nigg E A. EMBO J. 1994;13:595–605. doi: 10.1002/j.1460-2075.1994.tb06297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tombes R M, Peloquin J G, Borisy G G. Cell Regul. 1991;2:861–874. doi: 10.1091/mbc.2.11.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ookata K, Hisanaga S, Okumura E, Kishimoto T. J Cell Sci. 1993;105:873–81. doi: 10.1242/jcs.105.4.873. [DOI] [PubMed] [Google Scholar]
- 12.Ookata K, Hisanaga S, Bulinski J C, Murofushi H, Aizawa H, Itoh T J, Hotani H, Okumura E, Tachibana K, Kishimoto T. J Cell Biol. 1995;128:849–862. doi: 10.1083/jcb.128.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maldonado-Codina G, Glover D M. J Cell Biol. 1992;116:967–976. doi: 10.1083/jcb.116.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pines J, Hunter T. J Cell Biol. 1991;115:1–17. doi: 10.1083/jcb.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackman M, Firth M, Pines J. EMBO J. 1995;14:1646–1654. doi: 10.1002/j.1460-2075.1995.tb07153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi H, Minshull J, Ford C, Golsteyn R, Poon R, Hunt T. J Cell Biol. 1991;114:755–765. doi: 10.1083/jcb.114.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gautier J, Minshull J, Lohka M, Glotzer M, Hunt T, Maller J L. Cell. 1990;60:487–494. doi: 10.1016/0092-8674(90)90599-a. [DOI] [PubMed] [Google Scholar]
- 18.Surana U, Robitsch H, Price C, Schuster T, Fitch I, Futcher A B, Nasmyth K. Cell. 1991;65:145–161. doi: 10.1016/0092-8674(91)90416-v. [DOI] [PubMed] [Google Scholar]
- 19.Richardson H, Lew D J, Henze M, Sugimoto K, Reed S I. Genes Dev. 1992;6:2021–2034. doi: 10.1101/gad.6.11.2021. [DOI] [PubMed] [Google Scholar]
- 20.Kuhne C, Linder P. EMBO J. 1993;12:3437–3447. doi: 10.1002/j.1460-2075.1993.tb06018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fitch I, Dahmann C, Surana U, Amon A, Nasmyth K, Goetsch L, Byers B, Futcher B. Mol Biol Cell. 1992;3:805–818. doi: 10.1091/mbc.3.7.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grandin N, Reed S I. Mol Cell Biol. 1993;13:2113–2125. doi: 10.1128/mcb.13.4.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwob E, Nasmyth K. Genes Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- 24.Fisher D, Nurse P. Semin Cell Biol. 1995;6:73–78. doi: 10.1016/1043-4682(95)90003-9. [DOI] [PubMed] [Google Scholar]
- 25.Fisher D L, Nurse P. EMBO J. 1996;15:850–860. [PMC free article] [PubMed] [Google Scholar]
- 26.Booher R, Beach D. EMBO J. 1988;7:2321–2327. doi: 10.1002/j.1460-2075.1988.tb03075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mondesert O, McGowan C H, Russell P. Mol Cell Biol. 1996;16:1527–1533. doi: 10.1128/mcb.16.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Connolly T, Beach D. Mol Cell Biol. 1994;14:768–776. doi: 10.1128/mcb.14.1.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warren A J, Colledge W H, Carlton M B, Evans M J, Smith A J, Rabbitts T H. Cell. 1994;78:45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- 30.Hanley-Hyde J, Mushinski J, Sadofsky M, Huppi K, Krall M, Kozak C, Mock B. Genomics. 1992;13:1018–1030. doi: 10.1016/0888-7543(92)90015-k. [DOI] [PubMed] [Google Scholar]
- 31.Lock L, Pines J, Hunter T, Gilbert D, Gopalan G, Jenkins N, Copeland N, Donovan P. Genomics. 1992;13:415–424. doi: 10.1016/0888-7543(92)90262-q. [DOI] [PubMed] [Google Scholar]
- 32.Mountford P, Zevnik B, Duwel A, Nichols J, Li M, Dani C, Robertson M, Chambers I, Smith A. Proc Natl Acad Sci USA. 1994;91:4303–4307. doi: 10.1073/pnas.91.10.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fantl V, Stamp G, Andrews A, Rosewell I, Dickson C. Genes Dev. 1995;9:2364–2372. doi: 10.1101/gad.9.19.2364. [DOI] [PubMed] [Google Scholar]
- 34.Kontgen F, Stewart C L. Methods Enzymol. 1993;225:878–890. doi: 10.1016/0076-6879(93)25055-7. [DOI] [PubMed] [Google Scholar]
- 35.Gown A M, de Wever N, Battifora H. Appl Immunohistochem. 1993;1:256–266. [Google Scholar]
- 36.Crompton T, Gilmour K C, Owen M J. Cell. 1996;86:243–251. doi: 10.1016/s0092-8674(00)80096-3. [DOI] [PubMed] [Google Scholar]
- 37.Crompton T. Growth Factors. 1991;4:109–116. doi: 10.3109/08977199109000262. [DOI] [PubMed] [Google Scholar]
- 38.Nugent J H, Alfa C E, Young T, Hyams J S. J Cell Sci. 1991;99:669–674. doi: 10.1242/jcs.99.3.669. [DOI] [PubMed] [Google Scholar]
- 39.Jeffrey P D, Russo A A, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich N P. Nature (London) 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 40.Brandeis M, Hunt T. EMBO J. 1996;15:5280–5289. [PMC free article] [PubMed] [Google Scholar]
- 41.Chapman D L, Wolgemuth D J. Dev Biol. 1994;165:500–506. doi: 10.1006/dbio.1994.1270. [DOI] [PubMed] [Google Scholar]
- 42.Sicinski P, Donaher J L, Parker S B, Li T, Fazeli A, Gardner H, Haslam S Z, Bronson R T, Elledge S J, Weinberg R A. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 43.Sicinski P, Donaher J L, Geng Y, Parker S B, Gardner H, Park M Y, Robker R L, Richards J S, McGinnis L K, Biggers J D, et al. Nature (London) 1996;384:470–474. doi: 10.1038/384470a0. [DOI] [PubMed] [Google Scholar]
- 44.Murphy M, Stinnakre M-G, Senamaud-Beaufort C, Sweeney C, Kubelka M, Carrington M, Bréchot C, Sobczak-Thépot J. Nat Genet. 1997;15:83–86. doi: 10.1038/ng0197-83. [DOI] [PubMed] [Google Scholar]
- 45.Nasmyth K. Curr Opin Cell Biol. 1993;5:166–179. doi: 10.1016/0955-0674(93)90099-c. [DOI] [PubMed] [Google Scholar]
- 46.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1977;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Page R D. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]