Abstract
During the solution structure determination of the Escherichia coli quorum-sensing protein SdiA in the presence of N-octanoyl-L-homoserine lactone (HSL), NMR signals were detected in 13C-filter-13C-filter spectra for the bound HSL molecule. An additional set of coupled signals, independent of those of HSL were also detected, indicating the presence of another unlabeled molecule, also bound to the labeled SdiA. Analysis of the NMR spectrum of this ligand and of the mass spectrum of the dissociated components indicates that the ligand is most likely xylose. Further analysis of xylose-bound SdiA defines a site close to the C-terminus, remote from the HSL binding site. These observations provide an example of the sensitivity of high-resolution NMR experiments and their ability to detect, identify and map the adventitious binding of a small organic molecule to a protein.
The E. coli protein SdiA shows a marked sequence homology to well-known quorum-sensing proteins, such as LuxR of the luminescent bacterium Vibrio fischeri and TraR of the crown gall bacterium Agrobacterium tumefaciens.1 These proteins detect the production of small molecules, termed autoinducers, produced by other members of the same species. The detection mechanism for the TraI/TraR system includes a “folding switch”,2 where TraR is unfolded in the absence of the autoinducer, but folds in its presence, thereby enabling it to perform its function in the activation of genes.
The sequence homology of SdiA from E. coli and other intestinal bacteria to these highly specific quorum sensing proteins is intriguing, since the E. coli genome does not contain a gene for the production of an endogenous autoinducer.1 It is thought that the function of SdiA in these bacteria may be as a sensor for autoinducers produced by other species of bacteria.3 Recent NMR studies on SdiA confirmed that it too shows the “folding switch” behavior, but with a wide variety of autoinducer molecules.4 The structure of SdiA is very similar to that of TraR, but the bound autoinducer molecule is relatively disordered, consistent with the postulated role of SdiA in the detection of autoinducers from other organisms.4 Herein, we describe an additional site on this protein which binds the monosaccharide xylose and speculate as to the origin of this compound.
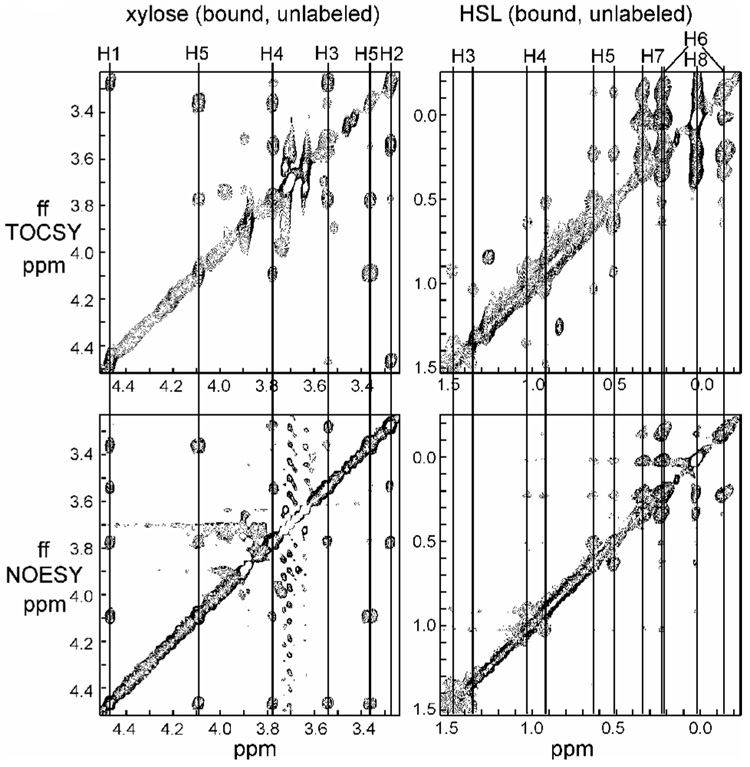
The solution structure of SdiA was calculated4 in the presence of the autoinducer molecule N-octanoyl-L-homoserine lactone (HSL). In order to define the bound conformation of HSL, 13C-filter-13C-filter TOCSY and NOESY spectra5 were acquired for unlabeled HSL bound to 15N,13C-labeled SdiA. Analysis of these spectra showed that the NMR signals of the acyl chain of the bound HSL increased in linewidth from the 8-CH3 group to the 3-CH2, with those of the 2-CH2 apparently broadened beyond detection. This observation is consistent with the presence of disorder in the acyl chain. However, a very clear set of coupled signals was also observed in the region 3–4.5 ppm (Fig. 1). These signals were initially assigned to the lactone ring of HSL, but the chemical shifts and coupling patterns were quite inconsistent with those of the unbound HSL. In particular, there was clear evidence for the presence of six coupled protons, where only five are expected for the lactone moiety. As well, these signals are relatively sharp, where following the trend of the acyl chain, one would expect that the lactone signals should be broad.
Figure 1.
Portions of the 13C-filter-13C-filter NOESY and TOCSY spectra of SdiA showing the resonances of a bound, unlabeled molecule other than HSL.
We considered the possibility that the lactone had been chemically modified during the expression and purification process. However, this was ruled out by mass spectrometry of the bound HSL dissociated from the protein. A sample of HSL labeled with 13C in the lactone moiety was examined in the presence of unlabeled SdiA; this gave rise to very broad but identifiable cross peaks in the 13C-¹H HSQC spectrum,4 none of which corresponded to the mystery peaks. On the basis of this data, we concluded that these peaks must arise from a different molecule, also bound to SdiA.
Several characteristics of the unknown bound molecule can be immediately observed: firstly, it must be bound relatively tightly to the protein, since it was retained through a purification procedure that included two chromatography columns;6 secondly, since it gives NMR signals in the filter-filter spectrum, it is not 13C-labeled. It cannot therefore have been synthesized by the bacteria from labeled 13C-glucose as the SdiA protein was, but must have been derived from some other source. Indeed, the NMR spectra shown in Fig. 1 are consistent with a monosaccharide moiety, with the resonances and their coupling pattern corresponding almost exactly to the published spectrum of β-xylopyranose.7
As independent confirmation of this assignment, the additional bound ligand was also analyzed by GC/MS. Identification was performed by a computer search algorithm comparing the fragmentation pattern of eluting sample peaks with the fragmentation patterns of known compounds within the NIST mass spectral library. In addition, validation of the unknown compound as xylose also was given by comparison of retention time and mass spectral fragmentation pattern with an authentic xylose sample.
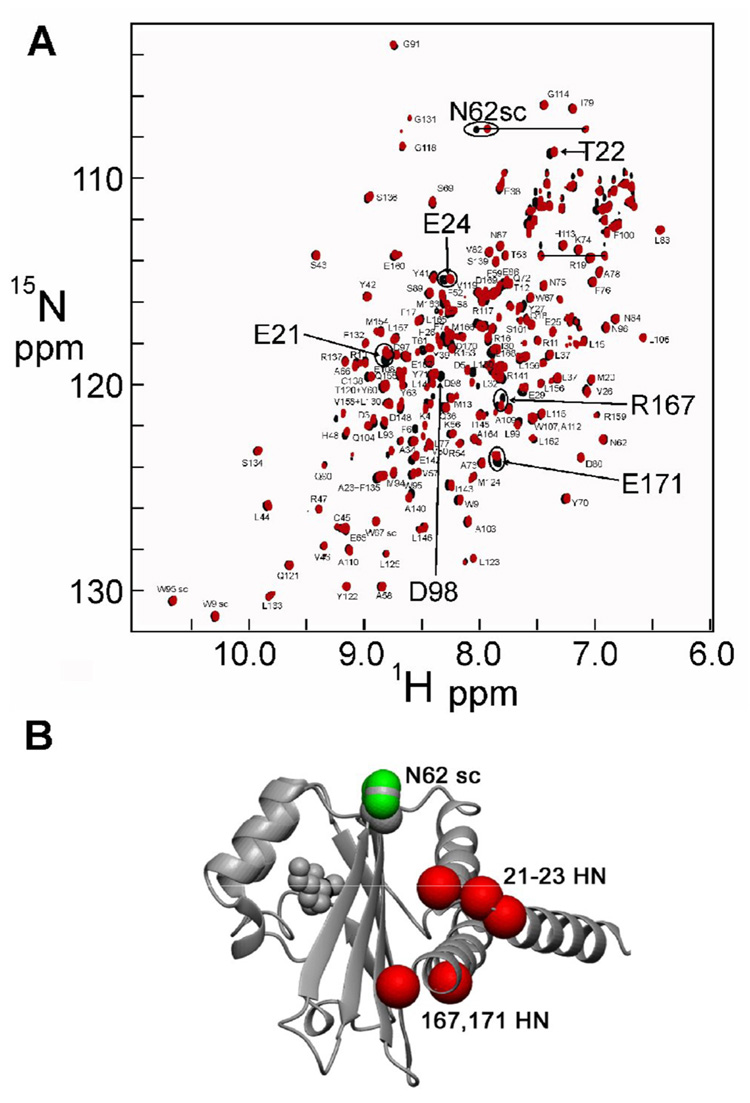
Despite the fact that xylose must have significant affinity for SdiA to remain bound throughout expression and purification, addition of D- or L-xylose to a solution of the refolded protein produced no change in the NMR spectrum, possibly indicating that it binds to the unfolded or partly folded protein during expression in the context of the bacterial cell. Since the in vitro refolding of the protein is done by dialysis from a high concentration of denaturant, in the presence of the autoinducer molecule, our inability to observe binding of xylose may simply be due to the solution conditions necessary for the folding reaction to occur in vitro. The 15N HSQC spectra of the HSL complex of SdiA derived from the soluble fraction of the expression medium and from the refolded material are virtually identical, but there are some small, specific differences (Figure 2A). If these differences are mapped to the solution structure of SdiA, they delineate a possible binding site near the C-terminus of the molecule (Figure 2B).
Figure 2.
(a) 15N-¹H HSQC spectra of SdiA, refolded in the presence of HSL (black) and purified from the soluble fraction of the bacterial expression system (red). (b) Positions of the shifted HSQC cross peaks of Figure 3 on the structure of SdiA.
The source of a significant amount of xylose in the culture medium is a mystery. The yield of pure SdiA from the soluble fraction is approximately 5 mg/L of medium, translating to a final concentration in the bacterial medium of at least 0.23 µM, not counting losses during purification. Thus, the minimum amount of xylose required to bind to the amount of soluble SdiA in 1 L of medium would be 0.23 µM (~42 mg), assuming 1:1 stoichiometry of binding. For reference, the carbon-containing components of the medium, together with their concentrations at the beginning of bacterial growth, are shown in Table 1. It is unlikely that such a significant amount of xylose would be present as a contaminant in any of these reagents, which are of the highest grade and purity. Furthermore, the solvent water is triple distilled and deionized. A contaminant in the purification columns can be ruled out, since the bound xylose appears only in the soluble fraction of the expressed protein and is absent from the refolded protein, which is purified on identical columns. Thus, we must consider that the xylose arises through chemical or biochemical transformation of a component of the culture medium. Only carbon-containing compounds need be considered, and of these, the glucose can be dismissed, since it is 13C-labeled, and the xylose bound to SdiA is unlabeled. The remaining compounds include two antibiotics, kanamycin and carbenicillin, the components of the added vitamin mixture, and the additional components, HSL, added to promote the formation of soluble SdiA, and isopropyl-thiogalactopyranoside (IPTG), added to activate the lac promoter for the expression of SdiA from the plasmid-borne gene.
Table 1.
Carbon-containing compounds in the bacterial expression medium.
| Component | Concentration (µM) |
|---|---|
| Carbenicillin | 250 |
| Kanamycin | 60 |
| Glucose (13C6) | 22,000 |
| D-biotin | 4.1 |
| Choline chloride | 7.2 |
| Folic acid | 0.2 |
| Myo-inositol | 11 |
| Niacinamide | 8.2 |
| D-pantothenic acid | 4.2 |
| Pyridoxine HCl | 4.9 |
| Riboflavin | 0.27 |
| Thiamine HCl | 3.0 |
| HSL | 400 |
| IPTG | 500 |
As stated, the absolute minimum concentration of xylose in the expression medium is 0.23 µM. All of the carbon-containing compounds in Table 1 have comparable or greater initial concentrations than this. However, the vitamins are present in concentrations that are several orders of magnitude lower than those of the four most likely components (e.g., carbenicillin, kanamycin, HSL, and IPTG). In addition, the vitamins are present from the beginning of the bacterial growth in the expression medium, and would presumably be considerably depleted, if not completely absent, by the time that the expression of SdiA was initiated. The two antibiotics, HSL and IPTG, however, are present in much higher concentrations in the stationary phase bacterial expression medium as the protein is expressed. The antibiotics will be metabolized by the bacteria, since they provide the selection method by which the plasmid containing the gene of interest is forced to remain in the bacteria. The HSL is also potentially capable of being transformed by the bacteria, but this cannot proceed to 100%, as HSL is present intact in the soluble SdiA. IPTG, the inducer of expression, is present in the highest concentration, and is added immediately before expression of the SdiA.
Bacteria, particularly enteric bacteria such as E. coli, can subsist on many varieties of carbon-containing compounds, including sugars, sugar phosphates, sugar alcohols and acids. However, each bacterial species has preferred substrates, and in a mixture of possible nutrients, will utilize the preferred substrates first, and will subsequently use the less-preferred substrates when the preferred ones are used up. This may result in the observation of two distinct growth curves, with a lag period in between, depending upon whether additional genes for transport and metabolism of the new substrate must be activated. This was first described by Monod in 1942, 8 and later reviewed by Roseman and Meadow.9
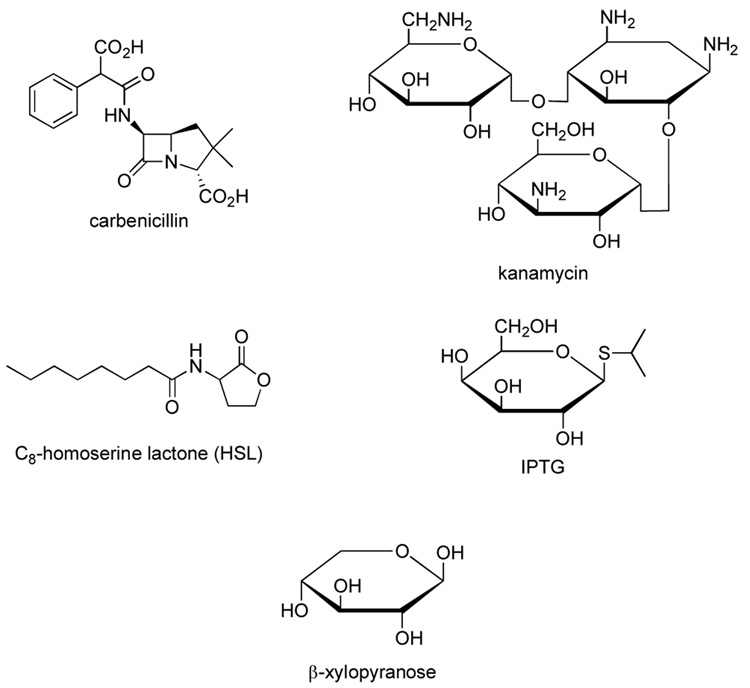
These ideas can be applied to our observations on the SdiA expression system. By the time that expression of SdiA is initiated by the addition of IPTG and HSL to the medium, the bacterial cells have been grown to late-log or stationary phase (OD600 ~ 0.8) at 37 °C. In order to maximize the production of soluble protein, these cells are left at 15 °C overnight,6 during which the 13C-glucose in the medium is used by the bacteria to make SdiA. At some stage, this substrate will be depleted, and the bacteria is forced to utilize alternative substrate to continue their metabolism. Four possible alternate substrates were identified: kanamycin, carbenicillin, HSL and IPTG (Figure 3). Carbenicillin contains a classic β-lactam structure. Presumably this compound is deactivated by hydrolysis of the lactam by the lactamase enzyme whose gene is present on the expression plasmid. However, it is very unlikely that carbenicillin is the precursor of the observed xylose, since, apart from the phenyl group, there is no contiguous carbon chain longer than 3 atoms. Kanamycin (3-D-glucosamine-2-deoxystreptamine-6-D-glucosamine) is a better prospect, as it contains two 6-carbon amino-sugar moieties, which could be decarboxylated and deaminated after an initial hydrolysis step to give 5-carbon sugars. The plasmid-borne kanamycin-resistance gene codes for an enzyme that inactivates the antibiotic by phosphorylation, acetylation or adenylation. The most common enzyme, APH(3’)II inactivates kanamycin by phosphorylation of the 3’-OH of the glucosamine.10 Conversion of the 6-carbon amino sugar moieties of the kanamycin to a specific 5-carbon sugar, xylose, has not been reported. However, enzymes that could catalyze individual steps of a synthesis, including hydrolysis and decarboxylation, are well known in bacteria.
Figure 3.
Molecular structures of carbenicillin, kanamycin, HSL and IPTG. The structure of xylose is given for comparison.
Another possible unlabeled substrate that is present in relatively large quantities in the culture medium is IPTG. Like kanamycin, there is no reported breakdown of this galactose derivative to xylose, but enzymes are known that could, for example, perform an epimerization of galactose to glucose, following a hydrolysis step to remove the isopropylthiol group. Oxidation of the UDP derivative of glucose to glucuronate by UDP glucose dehydrogenase, followed by decarboxylation to xylose by UDP glucuronate decarboxylase is a possibility for the derivation of xylose from the added IPTG.
In conclusion, we have shown by filtered NMR spectra that there is an unlabeled molecule bound to the labeled SdiA protein expressed in E. coli and isolated from the soluble fraction in the bacteria. This bound molecule has been identified on the basis of its NMR spectrum and GC-MS analysis to be xylose, and it appears to be bound relatively tightly in a specific area of the SdiA molecule. In the absence of an obvious source for a significant amount of unlabeled xylose in the expression medium, we conclude that this observation may be related to utilization of alternative substrates by the E. coli expression system as the 13C-labeled glucose is depleted in the medium. However, the current experimental data cannot mechanistically discern between de novo synthesis and macromolecule degradation as the route of xylose production.11 Although the physiological significance of this observation is unclear, this work provides an illustration of the sensitivity of NMR experiments and their role in detecting molecular interactions that would otherwise be totally overlooked.
Acknowledgments
We thank Linda Tennant for technical assistance and Gerard Kroon for help with NMR experiments. Maria Yamout provided invaluable help with protein expression and purification and helpful suggestions. We are grateful to Dr. Michael Burkhart for helpful discussion. This work was supported by grant GM57374 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Miller MB, Bassler BL. Annu. Rev. Microbiol. 2001;55:165. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 2.Zhu J, Winans SC. Proc. Natl. Acad. Sci. USA. 2001;98:1507. doi: 10.1073/pnas.98.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Michael B, Smith JN, Swift S, Heffron F, Ahmer BM. J. Bacteriol. 2001;183:5733. doi: 10.1128/JB.183.19.5733-5742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ahmer BMM. Molec. Microbiol. 2004. 52;933 doi: 10.1111/j.1365-2958.2004.04054.x. [DOI] [PubMed] [Google Scholar]
- 4.Yao Y, Martinez-Yamout MA, Dickerson TJ, Brogan AP, Wright PE, Dyson HJ. J. Mol. Biol. 2006;355:262. doi: 10.1016/j.jmb.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 5.Zwahlen C, Legault P, Vincent SJF, Greenblatt J, Konrat R, Kay LE. J. Am. Chem. Soc. 1997;119:6711. [Google Scholar]
- 6.Yao Y, Martinez-Yamout MA, Dyson HJ. J. Biomol. NMR. 2005;31:373. doi: 10.1007/s10858-005-2470-0. [DOI] [PubMed] [Google Scholar]
- 7.Benesi AJ, Falzone CJ, Banerjee S, Farber GK. Carbohydrate Res. 1994;258:27. [Google Scholar]
- 8.Monod J. Recherches sur la Croissance des Cultures Bacteriennes. Ph.D. Thesis. University of Paris; 1942. [Google Scholar]
- 9.Roseman S, Meadow ND. J. Biol. Chem. 1990;265:2993. [PubMed] [Google Scholar]
- 10.Wright GD, Berghuis AM, Mobashery S. Adv. Exp. Med. Biol. 1998;456:27. [PubMed] [Google Scholar]
- 11.Yuan J, Rabinowitz JD. J. Am. Chem. Soc. 2007 ASAP Article. [Google Scholar]