Abstract
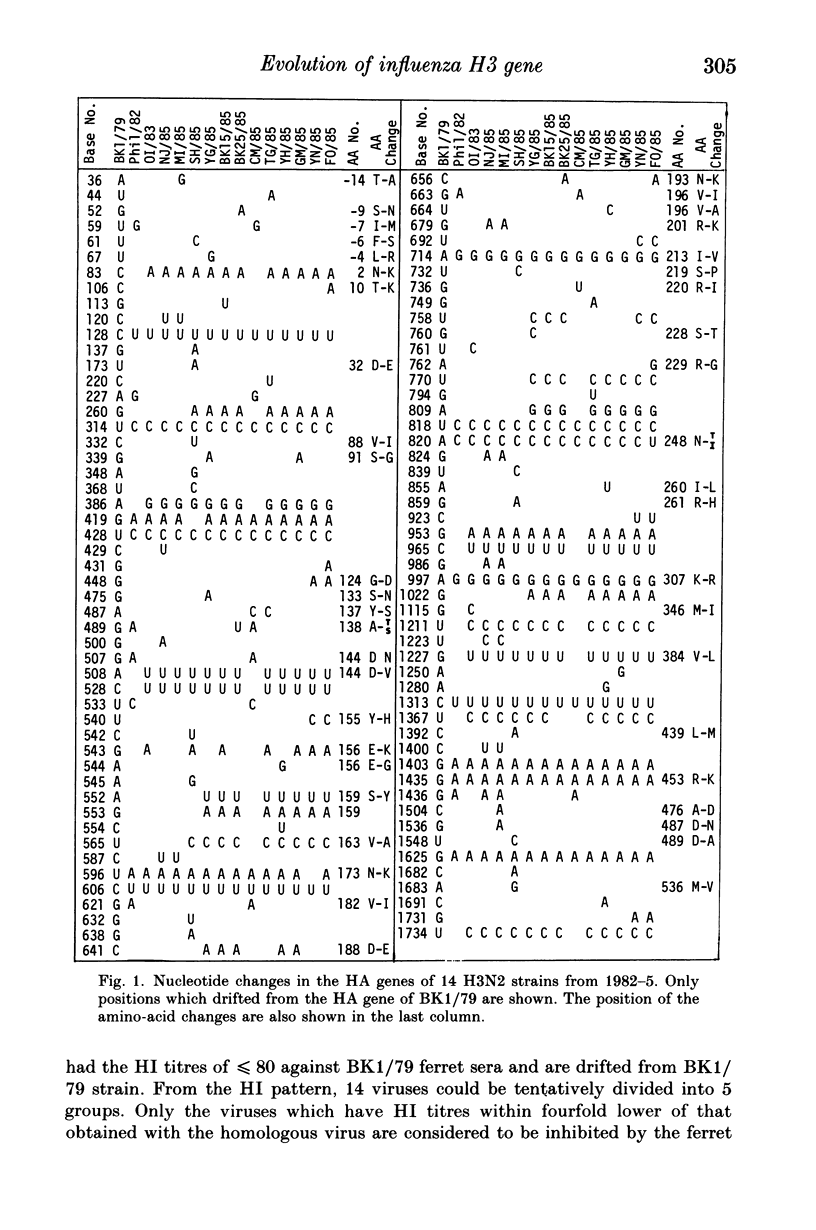
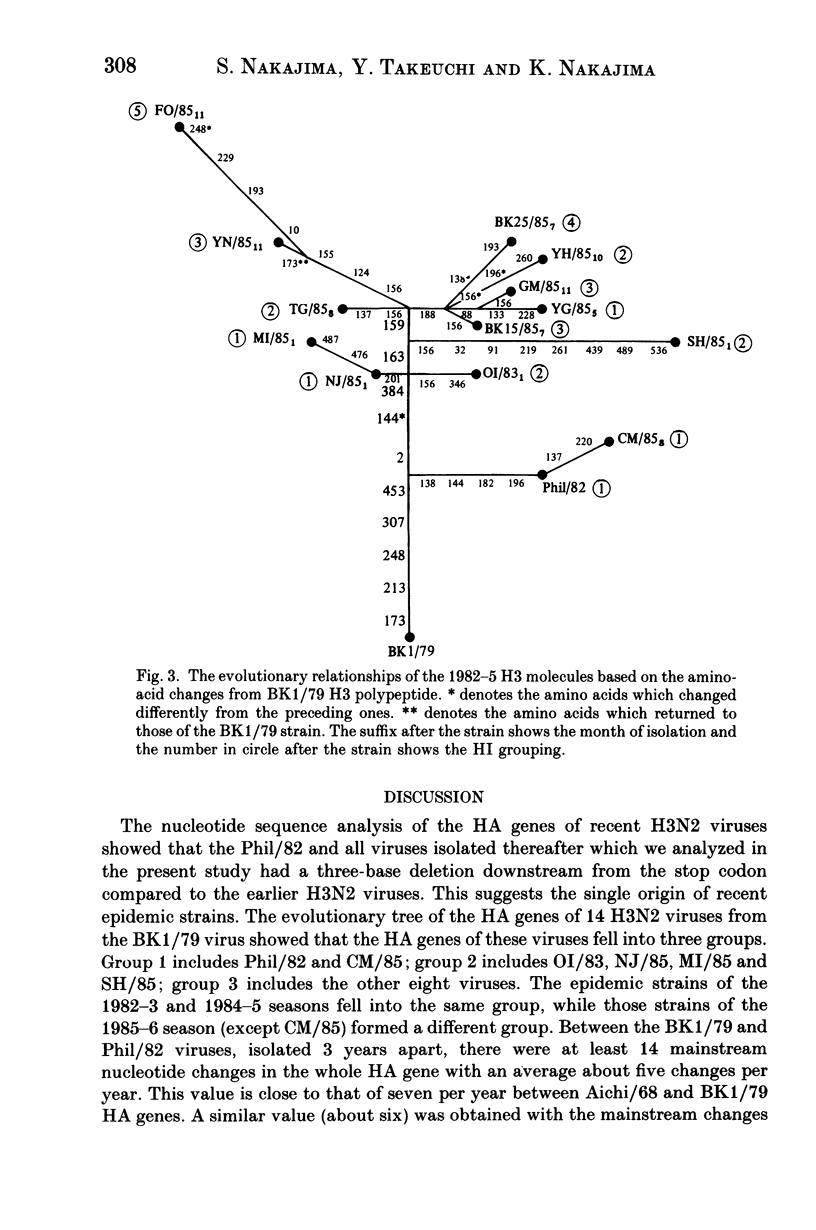
The nucleotide sequences of the haemagglutinin (HA) genes of influenza A (H3N2) isolates from the 1985-6 season in Japan along with those of several viruses isolated between 1982-5 from other countries were analyzed to determine the origin of the 1985-6 Japanese strains. The HA genes of these viruses consisted of 1762 nucleotides and had a three-nucleotide deletion downstream from the stop codon when compared to the sequences of earlier Hong Kong H3N2 viruses. An evolutionary tree of the HA genes of these viruses was drawn using the A/Bangkok/1/79 sequence as the starting point. Eight strains isolated from Asian and Pacific regions including Japan in the 1985-6 season (one in May) had the HA genes located closely on the evolutionary tree but away from those of the isolates in North America and Europe during the 1984-5 season, and a common ancestry for these viruses was suggested.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Both G. W., Sleigh M. J. Complete nucleotide sequence of the haemagglutinin gene from a human influenza virus of the Hong Kong subtype. Nucleic Acids Res. 1980 Jun 25;8(12):2561–2575. doi: 10.1093/nar/8.12.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Both G. W., Sleigh M. J. Conservation and variation in the hemagglutinins of Hong Kong subtype influenza viruses during antigenic drift. J Virol. 1981 Sep;39(3):663–672. doi: 10.1128/jvi.39.3.663-672.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Both G. W., Sleigh M. J., Cox N. J., Kendal A. P. Antigenic drift in influenza virus H3 hemagglutinin from 1968 to 1980: multiple evolutionary pathways and sequential amino acid changes at key antigenic sites. J Virol. 1983 Oct;48(1):52–60. doi: 10.1128/jvi.48.1.52-60.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima S., Brown D. J., Ueda M., Nakajima K., Sugiura A., Pattnaik A. K., Nayak D. P. Identification of the defects in the hemagglutinin gene of two temperature-sensitive mutants of A/WSN/33 influenza virus. Virology. 1986 Oct 30;154(2):279–285. doi: 10.1016/0042-6822(86)90454-x. [DOI] [PubMed] [Google Scholar]
- Nakajima S., Nakajima K., Takeuchi Y., Sugiura A. Influenza surveillance based on oligonculeotide mapping of RNA of H1N1 viruses prevalent in Japan, 1978-1979. J Infect Dis. 1980 Oct;142(4):492–502. doi: 10.1093/infdis/142.4.492. [DOI] [PubMed] [Google Scholar]
- Palese P., Schulman J. L. Differences in RNA patterns of influenza A viruses. J Virol. 1976 Mar;17(3):876–884. doi: 10.1128/jvi.17.3.876-884.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond F. L., Caton A. J., Cox N. J., Kendal A. P., Brownlee G. G. Antigenicity and evolution amongst recent influenza viruses of H1N1 subtype. Nucleic Acids Res. 1983 Oct 25;11(20):7191–7203. doi: 10.1093/nar/11.20.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond F. L., Caton A. J., Cox N. J., Kendal A. P., Brownlee G. G. The antigenicity and evolution of influenza H1 haemagglutinin, from 1950-1957 and 1977-1983: two pathways from one gene. Virology. 1986 Jan 30;148(2):275–287. doi: 10.1016/0042-6822(86)90325-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood P. A. An antigenic map of the haemagglutinin of the influenza Hong Kong subtype (H3N2), constructed using mouse monoclonal antibodies. Mol Immunol. 1984 Jul;21(7):663–671. doi: 10.1016/0161-5890(84)90052-x. [DOI] [PubMed] [Google Scholar]
- Underwood P. A. Mapping of antigenic changes in the haemagglutinin of Hong Kong influenza (H3N2) strains using a large panel of monoclonal antibodies. J Gen Virol. 1982 Sep;62(Pt 1):153–169. doi: 10.1099/0022-1317-62-1-153. [DOI] [PubMed] [Google Scholar]
- Verhoeyen M., Fang R., Jou W. M., Devos R., Huylebroeck D., Saman E., Fiers W. Antigenic drift between the haemagglutinin of the Hong Kong influenza strains A/Aichi/2/68 and A/Victoria/3/75. Nature. 1980 Aug 21;286(5775):771–776. doi: 10.1038/286771a0. [DOI] [PubMed] [Google Scholar]
- Wiley D. C., Wilson I. A., Skehel J. J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981 Jan 29;289(5796):373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- Wilson I. A., Skehel J. J., Wiley D. C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981 Jan 29;289(5796):366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]



