Abstract
Little is known about the division of eukaryotic cell organelles and up to now neither in animals nor in plants has a gene product been shown to mediate this process. A cDNA encoding a homolog of the bacterial cell division protein FtsZ, an ancestral tubulin, was isolated from the eukaryote Physcomitrella patens and used to disrupt efficiently the genomic locus in this terrestrial seedless plant. Seven out of 51 transgenics obtained were knockout plants generated by homologous recombination; they were specifically impeded in plastid division with no detectable effect on mitochondrial division or plant morphology. Implications on the theory of endosymbiosis and on the use of reverse genetics in plants are discussed.
Chloroplasts and mitochondria are remnants of free-living prokaryotes and, like these, multiply by constriction division (1). While several genes essential for bacterial cytokinesis have been identified (2, 3), no gene product has been found to mediate organelle division in a eukaryote. The best studied bacterial cell division protein is FtsZ, which is homologous to the eukaryotic cytoskeleton element tubulin, and forms the dividing ring during bacterial cytokinesis (4–6). This protein, first identified from Escherichia coli, is not only conserved in cyanobacteria, the progenitors of chloroplasts, but in all eubacteria and in archaebacteria (7–9). So far, a nuclear-encoded cDNA from only one eukaryote (Arabidopsis thaliana) has been reported to be homologous at the protein level to FtsZ, and its in vitro translation product was imported into isolated pea chloroplasts (10). However, the involvement of this gene in eukaryotic organelle division has not been demonstrated (11).
Because homologous recombination in plant nuclear DNA occurs at marginal frequencies, up to now plant genes have had to be silenced by antisense approaches, leading to varied and unstable phenotypes (12). However, recent targeting experiments with unidentified nuclear DNA have revealed that in Physcomitrella patens, a seedless terrestrial plant, homologous recombination is more frequent than illegitimate recombination (13), an important feature also found in yeast that led to its use as a model organism in molecular biology (14).
We report here on the isolation of a cDNA from the moss Physcomitrella, PpftsZ, with homology at the protein level to bacterial FtsZ proteins, on the efficient targeted disruption of the corresponding genomic locus as well as on the role of this nuclear gene in eukaryotic organelle division.
MATERIALS AND METHODS
Plant Material and Growth Conditions.
Physcomitrella patens (Hedw.) B.S.G. has been characterized previously (15). Plants were grown axenically under standard conditions (agitated liquid Knop medium, mg × L −1 250 mg KH2PO4, 250 mg MgSO4 × 7H2O, 250 mg KCl, 1000 mg Ca(NO3)2 × 4H2O, 12.5 mg FeSO4 × 7H2O, pH 5.8) in a growth chamber under controlled conditions (25 ± 1°C; light provided from above by two fluorescent tubes, Philips TL 65W/25; light flux of 55 μmol s−1 m−2 outside the flasks, light-dark regime of 16:8 h). Plants were subcultured in 10 day-intervals.
Protoplast Isolation, Transformation, and Regeneration.
Protoplasts were isolated and regenerated as described (16). Freshly isolated protoplasts were counted and resuspended at 1.2 × 106 ml−1 in 3M-medium (15 mM MgCl2/0.1% Mes/0.48 M mannitol, pH 5.6; see ref. 13). Protoplasts were transformed with linearized DNA. This DNA was isolated and purified using Qiagen (Chatsworth, CA) Tip-500 columns and resuspended in 0.1 M Ca(NO3)2 at a concentration of 0.5 μg μl−1. For transformation 100 μl of DNA-solution was transferred to a sterile glass tube; then 250 μl of the protoplast-suspension was added to the tube and mixed gently, followed by the addition of 350 μl PEG-solution (40% PEG4000 in 3M-medium, pH 6.0). The transformation mix was incubated at room temperature for 30 min with occasional gentle mixing. Subsequently, the solution was diluted every 5 min by adding 3M-medium: 1, 2, 3, and finally 4 ml. Protoplasts were centrifuged (5 min, 70 × g), resuspended in regeneration medium (16), transferred to 3-cm Petri dishes, and incubated for 24 h with a light flux of 4.9 μmol s−1/m−2. Thereafter, cultures were transferred to normal growth conditions (light flux of 46.8 μmol s−1/m−2; light-dark regime of 16:8 h; 25 ± 1°C) for 5–6 days, and regenerating protoplasts were transferred to solidified Knop medium overlaid with a cellophane disc. After 10 days the cellophane overlays were transferred to Knop medium supplemented with 50 mg L−1 G418 (GIBCO/BRL). Resistant plants were isolated 50 days after transformation.
Isolation of PpftsZ.
Using 5 μg of polyadenylated RNA from 9d old plants a cDNA library was constructed with the help of a ZAP-cDNA Gigapack II Gold Cloning kit (Stratagene) as described previously (15). From this library a 219-bp DNA fragment could be amplified by using degenerate primers and PCR characteristics as described in ref. 17 for the isolation of ftsZ from the soil bacterium Rhizobium meliloti. This PCR-fragment was used as a probe to isolate PpftsZ from the Physcomitrella cDNA library according to standard procedures (18). The cDNAs were subjected to automatic sequencing using the Taq-Dye-Primer Cycle Sequencing kit on an automated sequencer (type 373A, Applied Biosystems). Clones were sequenced from both directions with appropriate overlaps by using primers specific for the T3- and T7-promoters, and for PpftsZ-sequences. Sequence data were analyzed with dnasis (Pharmacia) and homology searches were performed with fasta (http://www2.ebi.ac.uk/fasta3/). Alignments of amino acid sequences were made using clustal w (http://alfredo.wustl.edu/msa/clustal_old.cgi), and the search for protein motifs was performed with prosite (http://www.tokyo-center.genome.ad.jp/SIT/MOTIF.html).
Analysis of Transgenic Plants.
DNA and RNA were isolated from Physcomitrella according to ref. 19. A 1,173 bp PpftsZ subclone and the nptII cassette, indicated in Fig. 2, were used as probes in Northern experiments. The presence of the nptII cassette as well as integration of the transforming linear DNA into the PpftsZ locus were analyzed by PCR. Pairs of PCR primers were specific for nptII sequences (PT 1: GAGGCTATTCGGCTATGACTG and PT2: ATCGGGAGCGGCGATACCGTA), 5′-integration site (ppf4: GGAGCTGACATGGTTTTCGT and RT1: TGTCGTGCTCCACCATGTTG) and 3′-integration site (RT4: GTTGA GCAT- ATAAGAAACCC and ppf5: AACCCATACTTAACTAGGCA), respectively (refer to Fig. 2). Plant tissues were analyzed with the help of a Dialux 20-microscope (Leitz). Electron microscopy was performed according to ref. 20.
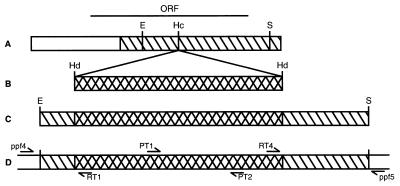
Figure 2.
Schematic representation of the cloning strategy to generate, and of PCR primer specifications to analyze ΔPpftsZ knockout plants. (A) The 1775 bp PpftsZ cDNA (open bar) comprises an ORF for 378 amino acids (ORF). A 1,173 bp subclone (hatched bar) was used for subsequent experiments. (B) A HindIII (Hd) fragment (cross-hatched bar) containing the 35S promoter-driven nptII gene as a selectable marker (23) was cloned into the HincII (Hc) digested subclone (C) The hybrid EcoRI/SacI fragment (E/S), containing the nptII cassette flanked by 247 and 658 bp PpftsZ cDNA sequence, was subsequently used for transformation of Physcomitrella protoplasts. (D) Pairs of PCR primers were specific for nptII sequences (PT 1/PT2), 5′-integration site (ppf4/RT1) and 3′-integration site (RT4/ppf5), respectively. Dimensions are drawn to scale.
RESULTS AND DISCUSSION
Isolation of PpftsZ, a Eukaryotic ftsZ Homolog.
From polyadenylated RNA of 9-day-old Physcomitrella patens (Hedw.) B.S.G., a seedless terrestrial plant, a cDNA library was established in λ ZAPII. This library was used to isolate a DNA fragment with homology to bacterial ftsZ genes in a PCR-approach utilizing degenerate primers. Primer design and PCR conditions were identical to those described for the isolation of ftsZ from the soil bacterium Rhizobium meliloti (17). This PCR fragment was used as a probe to isolate a 1,775-bp cDNA, PpftsZ, coding for 378 amino acids, representing the second known eukaryotic ftsZ homolog. Based on homology searches in the EST database, a ftsZ homolog had been identified previously from the dicotyledonous plant Arabidopsis thaliana (10). The deduced amino acid sequence of this clone had an N-terminal extension, which may serve as a leader peptide in organellar import (21), and the in vitro translation product was shown to be imported into isolated pea chloroplasts. Thus, this gene was named cpftsZ and it was concluded that cpFtsZ may be involved in plastid division (10). However, a biological function for cpFtsZ in Arabidopsis has not been established yet (11). Unlike cpFtsZ, the protein coded for by the eukaryotic ftsZ homolog from Physcomitrella lacks an obvious N-terminal extension that might serve as a leader peptide in organellar import (Fig. 1). When compared with FtsZ from E. coli, an extension of 10 amino acids becomes obvious, the function of which remains to be elucidated. However, the prokaryotic as well as the two different eukaryotic FtsZ proteins share extensive sequence homologies (Fig. 1), namely two conserved boxes, motif FTSZ_1 (PROSITE PS01134), a motif of as yet unknown function, and motif FTSZ_2 (PROSITE PS01135), including the TUBULIN motif (PROSITE PS00227). This “tubulin signature motif” is conserved in the products of all prokaryotic ftsZ genes (4) and is one of the many reasons to identify ftsZ as the gene coding for an ancestral tubulin and possibly a prokaryotic cytoskeleton element (4–6, 11). Interestingly, all known prokaryotic FtsZ proteins match imperfectly to the corresponding motif of eukaryotic tubulin (GGGTGSG) in that the serine is substituted by a threonine (4), while for both known eukaryotic ftsZ genes the amino acid sequence matches perfectly to this tubulin signature motif (Fig. 1).
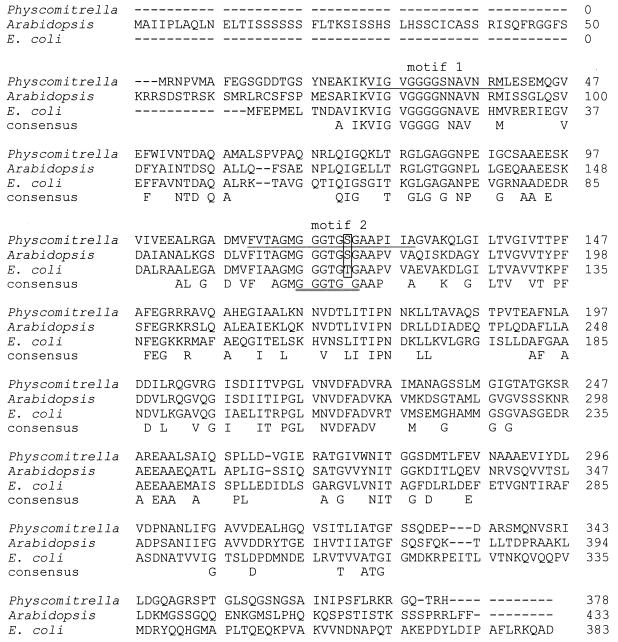
Figure 1.
Alignment of amino acid sequences of two eukaryotic and one prokaryotic FtsZ proteins in the one-letter-code. Aligned are PpFtsZ from the moss Physcomitrella patens (GenBank AJ001586), cpFtsZ from the dicotyledon Arabidopsis thaliana (SwissProt Q42545) and FtsZ from the enterobacterium Escherichia coli (SwissProt P06138). Where applicable a consensus is given below the alignment. Two PROSITE motifs, FTSZ 1 and FTSZ 2, are marked by underlining the Physcomitrella sequence once. A third PROSITE motif, the tubulin signature motif, is marked by underlining the consensus sequence twice. An amino acid position is boxed within this motif, where serines in the eukaryotic proteins match perfectly to the tubulin signature motif while a threonine in the prokaryotic protein deviates from this consensus motif. Note the different lengths of the N-terminal extensions of the two eukaryotic FtsZ proteins as compared with the bacterial sequence.
A Strategy to Elucidate the Biological Function of PpFtsZ.
A straightforward way to test the biological role of a novel gene is to knock out its function and analyze the resulting phenotype. Utilizing efficient homologous recombination, this approach of reverse genetics has long been used in yeast (14) and more recently in mouse embryonic stem cells (22). Recent targeting experiments with three different but unidentified loci have revealed that in Physcomitrella nuclear DNA homologous recombination is more frequent than illegitimate recombination; for the first time making the approach of reverse genetics feasible in terrestrial plants (13).
To generate ΔPpftsZ knockout plants, a 1,173-bp subclone of PpftsZ (Fig. 2A) was used in subsequent experiments. We inserted a selectable marker gene, a 35S promoter-driven nptII gene (23), into the coding region (Fig. 2B) and used linearized mutated PpftsZ, comprising 905 bp of PpftsZ sequence interrupted by the nptII cassette (Fig. 2C), to transform 1.2 × 106 Physcomitrella protoplasts. Subsequent regeneration of protoplasts into plants and growth of these plants on selective media yielded 51 stably transformed, G418-resistant Physcomitrella plants that grew like wild type.
Efficient Targeted Knockout Establishes the Function of PpFtsZ.
Upon scrutiny of these transgenics with a light microscope, 44 were indistinguishable from wild type, possessing about 50 lens-shaped chloroplasts per cell in all inspected tissues (Fig. 3 A–C). In contrast, the cells of seven transgenics (14%) appeared to be filled with one huge chloroplast each, an observation made with every inspected tissue (Fig. 3 D–F). Analysis of the different cells with electron microscopy provided evidence that the huge chloroplasts differed from lens-shaped wild-type plastids significantly in length but not in diameter (Fig. 3G), thereby resembling the filamentous E. coli mutants from which all fts-genes had been isolated (7). From this result we concluded that the giant chloroplasts have arisen from plastids inhibited in constriction division. In contrast, form and number of mitochondria were not affected in transgenics with macrochloroplasts (Fig. 3G), indicating that the constriction division of this organelle was not affected by PpFtsZ.
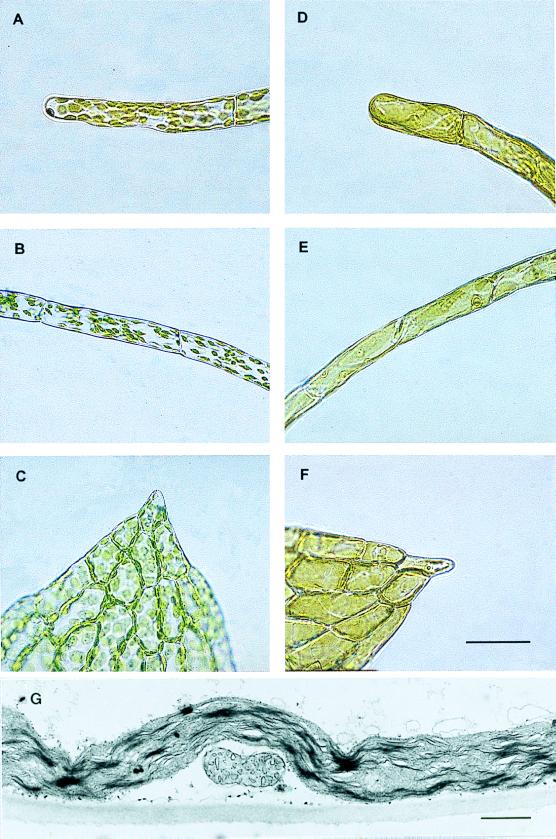
Figure 3.
Phenotypes of transgenic Physcomitrella patens generated by the construct outlined in Fig. 2C. Light microscopy from different tissues from transgenic plants with wild type-like plastids (A–C), and macrochloroplasts (D–F), respectively. The tissues depicted are chloronema (A and D), caulonema (B and E), and leaves (C and F). See ref. 30 for a review on moss development and tissue definition. Bar = 100 μm. As judged by transmission electron microscopy, division of chloroplasts was impeded in ΔPpftsZ knockouts, whereas form and number of mitochondria remained unchanged (G). (Bar = 1 μm.)
Parts of the nptII gene could be amplified by the PCR from DNA of every transgenic but not from DNA of the wild type (Fig. 4A). To analyze these integrations of the nptII gene, two additional sets of primers were generated. In each set one primer was specific for a PpftsZ sequence outside of the transformation construct whereas the second primer was specific for an nptII sequence (Fig. 2D). With these primers no DNA could be amplified from wild type or from transgenics with wild type-like plastids. In contrast, with each set of primers one specific fragment was amplified from every transgenic with macrochloroplasts, indicating integration of nptII into the PpftsZ locus in these plants (Fig. 4 B and C). The primer pair ppf4/RT1 generated a 373-bp PCR fragment, and the primer pair RT4/ppf5 generated a 826 bp PCR fragment, with the two spanning the 3′- and the 5′-integration site, respectively. These fragments were sequenced directly from three different transgenics and gave identical results. Comparison to the sequence of the entire PpftsZ demonstrated base pair-specific integration of the transforming DNA into the natural PpftsZ locus. Thus, ≈905 bp of homologous sequence was sufficient to mediate homologous recombination into the genomic locus in seven out of 51 transgenic plants (14%) tested.
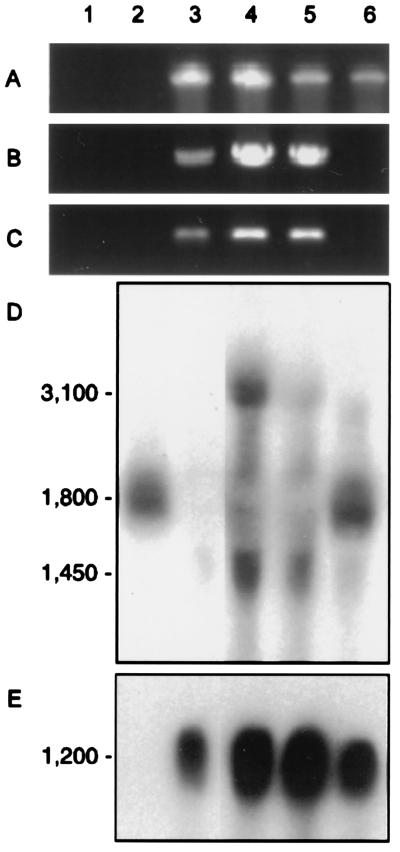
Figure 4.
Molecular analyses of transgenic Physcomitrella generated by the construct outlined in Fig. 2C. Analyzed were water (lane 1), untransformed wild type (lane 2), three different transgenics with macrochloroplasts (lanes 3–5; compare Fig. 3 D–G), and a transgenic with wild type-like plastids (lane 6; compare Fig. 3 A–C), respectively. (A–C) DNA gel analysis of representative PCR reactions. Each pair of primers was specific for sequences from nptII (A), 5′-integration site (B) and 3′-integration site (C), respectively. See Fig. 2D for primer specifications. Sizes of PCR fragments were 700 bp (A), 373 bp (B), and 826 bp (C). (D) Representative RNA blot analysis. About 10 μg of total RNA (19) was isolated from plants and probed with the 1,173 bp PpftsZ subclone indicated in Fig. 2A. (E) RNA blot analysis. The same filter was reprobed with the nptII cassette indicated in Fig. 2B. Transcript sizes in nucleotides.
In Northern blot experiments, one PpftsZ transcript of ≈1,800 nucleotides was detected at low abundance in wild type and in transgenics with wild type-like plastids. In contrast, in transgenics with macrochloroplasts the same probe detected either no transcript or aberrant transcripts of ≈3,100 and 1,450 nucleotides at low abundance, indicating disruption of the PpftsZ locus (Fig. 4D). The major nptII transcript of 1,200 nucleotides was present at high abundance in every transgenic but not in wild type (Fig. 4E). Obviously, knockout plants with aberrant PpftsZ-transcripts were not uniform in these Northern blot analyses. Most likely these differences in transcript amounts from the disrupted PpftsZ locus resulted from the insertion of multiple copies of the transgene into one locus, a feature that is well-documented for PEG-mediated transformation of Physcomitrella protoplasts (13, 24, 25).
However, integration-specific PCR fragments, and alterations in PpftsZ transcription were correlated in all cases tested with loss of chloroplast division. Thus, we have generated ΔPpftsZ knockouts by homologous recombination at a relative frequency among transgenics of ≈14%. Gene disruption led to altered transcription of PpftsZ and subsequently resulted in plants with macrochloroplasts, identifying PpFtsZ as the first known organellar division protein in any eukaryote. As it is a homolog of bacterial cell division proteins, we here present the first molecular evidence that prokaryotic cytokinesis and eukaryotic organelle division are conserved and take this as a further strong support for the theory of endosymbiosis.
It is noteworthy that division of mitochondria was obviously not affected by this ΔPpftsZ knockout, demonstrating that division of this organelle is not regulated by ancestral tubulin, although FtsZ appears to be responsible for the constriction division of all eubacteria and archaebacteria tested so far (11). Thus, two or more FtsZ homologs may exist in a eukaryotic cell. However, the only other known eukaryotic FtsZ homolog, cpFtsZ from Arabidopsis, differs from PpFtsZ from Physcomitrella in having an obvious leader peptide to mediate import into chloroplasts (Fig. 1) and, thus, is most probably likewise not involved in mitochondrial division. Moreover, according to the sequence of the entire yeast genome and the sequence of ≈90% of its mitochondrial DNA, yeast does not possess any FtsZ homologs (11). Therefore, it seems unlikely that mitochondria divide by the help of this ancestral tubulin. The acquisition of plastids by the “domestication” of free-living cyanobacteria is a relatively recent event compared with the acquisition of mitochondria by the “domestication” of free-living purple bacteria (1). Our data now suggest that the establishment of endosymbiosis leading to mitochondria is an evolutionarily old process during which the bacterial division mechanism has been modified or substituted. In contrast, based on recent biochemical data from this laboratory, probably other proteins in addition to FtsZ are conserved between bacterial cell division and plastid division in eukaryotes (26).
The Exceptional Plant Physcomitrella.
Recently it was found with unidentified nuclear DNA that in the seedless plant Physcomitrella homologous recombination occurs more frequently than illegitimate recombination (13). Using genomic DNA to disrupt the locus of a desaturase in Physcomitrella we could confirm this observation (T. Girke, H. Schmidt, R.R., and E. Heinz, unpublished data). In the present study with 905 bp of sequence we obtained homologous recombination at a relative frequency after selection of ≈14%. As this sequence was derived from a cDNA and the presence of introns in the genomic PpftsZ cannot be excluded, the real target for homologous recombination might have been even smaller than the 247 and 658 bp of 5′- and 3′-flanking regions. However, with a clearly visible cellular phenotype, a frequency of 14% among selected transgenics is a very convenient rate to screen for. Such a high rate for gene targeting has not been found in any other plant before.
Nevertheless, one knockout event in 750 transgenic Arabidopsis plants was reported recently (27), although the authors did not present a phenotype for this plant. Moreover, this report, based on a single event, cannot provide any accurate frequency value for homologous recombination in seed plants until this result has been adequately repeated. According to a conflicting recent report from another laboratory there was not a single event of homologous recombination in 18,974 transformants although a positive-negative selection and homologous genomic DNA of up to 23 kb had been used (28).
Why does homologous recombination occur orders of magnitude more often in Physcomitrella than in other plants? As its genome is about three times larger than that of Arabidopsis but smaller than that of Phaseolus or Nicotiana (G. Gorr and R.R., unpublished data), there is obviously no correlation to genome size. Moreover, recent expressed sequence tags data from this laboratory have also given no indication for major differences in gene conservation or codon usage between Physcomitrella and seed plants (18). Physcomitrella is a moss, and therefore spends most of its lifecycle in the haploid phase while seed plants are at least diploid. As yeast with its efficient homologous recombination is also haploid, it has been suggested that there is a connection between haploidy and efficient homologous recombination in eukaryotes (13). This appears to be a simplistic argumentation as Volvox, for instance, is a haploid eukaryote as well but has no efficient homologous recombination system when compared with yeast or to Physcomitrella (29). This hypothesis can, however, be tested very easily as the regeneration of haploid seed plants is a standard procedure, at least in Nicotiana.
In our opinion there are two characteristics different between Physcomitrella and seed plants that more likely account for their differences in homologous recombination frequencies. First, the tissue for transformation in Physcomitrella is not only haploid, but also gametophytic, clearly an important difference. Haploidy of a tissue does not constitute its gametophytic character (in haploid regenerants of seed plants) nor does diploidy of the tissue constitute its sporophytic character (in somatic hybrids of mosses). From this it becomes obvious that differences in gene regulation do exist between the gametophytic and the sporophytic phase of a plant (see refs. 18 and 30 for a detailed discussion). Thus, it may be worthwhile to apply targeting experiments to the gametophytic tissue of seed plants. Secondly, the moss tissue used preferentially for the transformation procedure—protonema—is, at least under our culture conditions, very synchronously dividing and for most of the day arrested at the G2/M-boundary of the cell-cycle (G. Gorr and R.R., unpublished data). In contrast, most of the tissues used for the transformation of seed plants are not synchronized and are not regularly arrested at the G2/M-boundary. It might well be that the occurrence of factors mediating efficient homologous recombination is restricted to the transition point from the G2-phase of the cell cycle to mitosis when homologous chromosomes may be associated.
However, at present Physcomitrella is the only terrestrial plant with an efficient gene targeting system, and the use of reverse genetics in this organism will help to address a broad range of fundamental questions in a heretofore unavailable way.
Acknowledgments
We thank Eberhard Schäfer, Gunther Neuhaus, Rainer Hertel, Susan Reynolds, and Randall Cassada for helpful comments on drafts of the manuscript. We appreciate stimulating discussions with Thomas Girke and Holger Puchta, as well as with colleagues in the European Community-funded project EUROMOSS, namely Didier Schaefer, Michel Laloue, Enzo Russo, and David Cove, who shared unpublished results. R.R. is indebted to the Deutsche Forschungsgemeinschaft for a Heisenberg Fellowship (Re 837/3–1) and for funding this project (Re 837/4–1). This article is based in part on a doctoral study by R.S. in the Faculty of Biology, University of Hamburg.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ001586).
References
- 1.Gray M W. Int Rev Cytol. 1992;141:233–357. doi: 10.1016/s0074-7696(08)62068-9. [DOI] [PubMed] [Google Scholar]
- 2.Donachie W D. Annu Rev Microbiol. 1993;47:199–230. doi: 10.1146/annurev.mi.47.100193.001215. [DOI] [PubMed] [Google Scholar]
- 3.Rothfield L I, Zhao C-R. Cell. 1996;84:183–186. doi: 10.1016/s0092-8674(00)80971-x. [DOI] [PubMed] [Google Scholar]
- 4.de Boer P, Crossley R, Rothfield L. Nature (London) 1992;359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- 5.Erickson H P. Cell. 1995;80:367–370. doi: 10.1016/0092-8674(95)90486-7. [DOI] [PubMed] [Google Scholar]
- 6.Rothfield L I, Justice S S. Cell. 1997;88:581–584. doi: 10.1016/s0092-8674(00)81899-1. [DOI] [PubMed] [Google Scholar]
- 7.Lutkenhaus J. Trends Genet. 1990;6:22–25. doi: 10.1016/0168-9525(90)90045-8. [DOI] [PubMed] [Google Scholar]
- 8.Doherty H M, Adams D G. Gene. 1995;163:93–96. doi: 10.1016/0378-1119(95)00416-4. [DOI] [PubMed] [Google Scholar]
- 9.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 10.Osteryoung K W, Vierling E. Nature (London) 1995;376:473–474. doi: 10.1038/376473b0. [DOI] [PubMed] [Google Scholar]
- 11.Erickson H P. Trends Cell Biol. 1997;7:362–67. doi: 10.1016/S0962-8924(97)01108-2. [DOI] [PubMed] [Google Scholar]
- 12.Puchta H, Hohn B. Trends Plant Sci. 1996;1:340–348. doi: 10.1016/S1360-1385(03)00004-9. [DOI] [PubMed] [Google Scholar]
- 13.Schaefer D G, Zryd J-P. Plant J. 1997;11:1195–1206. doi: 10.1046/j.1365-313x.1997.11061195.x. [DOI] [PubMed] [Google Scholar]
- 14.Struhl K. Nature (London) 1983;305:391–397. doi: 10.1038/305391a0. [DOI] [PubMed] [Google Scholar]
- 15.Reski R, Faust M, Wang X-H, Wehe M, Abel W O. Mol Gen Genet. 1994;244:352–359. doi: 10.1007/BF00286686. [DOI] [PubMed] [Google Scholar]
- 16.Rother S, Hadeler B, Orsini J M, Abel W O, Reski R. J Plant Physiol. 1994;143:72–77. [Google Scholar]
- 17.Margolin W, Corbo J C, Long S R. J Bacteriol. 1991;173:5822–5830. doi: 10.1128/jb.173.18.5822-5830.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reski R, Reynolds S, Wehe M, Kleber-Janke T, Kruse S. Bot Acta. 1998;111:141–151. [Google Scholar]
- 19.Reski R, Wehe M, Hadeler B, Marienfeld J R, Abel W O. J Plant Physiol. 1991;138:236–243. [Google Scholar]
- 20.Kasten B, Buck F, Nuske J, Reski R. Planta. 1997;201:261–272. doi: 10.1007/s004250050065. [DOI] [PubMed] [Google Scholar]
- 21.Keegstra K, Olsen L J, Theg S M A. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:471–501. [Google Scholar]
- 22.Capecchi M R. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 23.Töpfer R, Schell J, Steinbiss H-H. Nucleic Acids Res. 1988;16:8725. doi: 10.1093/nar/16.17.8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaefer D, Zryd J-P, Knight C D, Cove D J. Mol Gen Genet. 1991;226:418–424. doi: 10.1007/BF00260654. [DOI] [PubMed] [Google Scholar]
- 25.Reutter K, Reski R. Plant Tissue Culture and Biotechnology. 1996;2:142–147. [Google Scholar]
- 26.Kasten B, Reski R. J Plant Physiol. 1997;150:137–140. [Google Scholar]
- 27.Kempin S A, Liljegren S J, Block L M, Rounsley SD, Yanofsky M F, Lam E. Nature (London) 1997;389:802–803. doi: 10.1038/39770. [DOI] [PubMed] [Google Scholar]
- 28.Thykjaer T, Finnemann J, Schauser L, Christensen L, Poulsen C, Stougaard J. Plant Mol Biol. 1997;35:523–530. doi: 10.1023/a:1005865600319. [DOI] [PubMed] [Google Scholar]
- 29.Hallmann A, Rappel A, Sumper M. Proc Natl Acad Sci USA. 1997;94:7469–7474. doi: 10.1073/pnas.94.14.7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reski R. Bot Acta. 1998;111:1–15. [Google Scholar]